Introduction
Glioma is the most common form of brain cancer
(1), and the prognosis of glioma is
relatively poor (2). Glioma cells
infiltrate the healthy tissues surrounding the main tumor mass,
limiting the survival of patients with glioma (3). Despite advances in technology and
medicine, the incidence and mortality rates of glioma still
continue to increase (4).
Furthermore, the incidence of glioma is ~22/100,000 people, and the
5-year survival rate is ~10% (5). In
recent years, dysregulation of oncogenes and tumor suppressor genes
during glioma progression has been observed (6). Glioma treatment outcomes may be
improved by the identification of potential therapeutic targets
based on an improved understanding of the molecular signaling
pathways involved in glioma progression.
MicroRNAs (miRNAs) are a class of small and
non-coding RNA molecules that are 22 nucleotides long. miRNAs serve
as post-transcriptional regulators for gene expression by targeting
the 3′-untranslated region (3′UTR) of their target mRNAs, resulting
in mRNA degradation or inhibition of protein translation (7). miRNAs have been widely reported to be
involved in the progression of various types of cancer, as they can
function as oncogenes or tumor suppressors (8,9).
Recently, certain aberrantly expressed miRNAs have been identified
in glioma that serve crucial roles in tumor progression. For
instance, downregulation of miR-449 was detected in glioma tissues
and demonstrated to be involved in the tumor progression by
targeting flotillin 2, which was associated with poor prognosis in
patients with glioma (10).
Upregulation of miR-6807-3p was observed in glioma specimens;
miR-6807-3p promoted tumor progression and suppressed apoptosis by
targeting dachshund family transcription factor 1 (11). In addition, a microRNA microarray
assay indicated that miR-769-3p was differentially expressed in
glioma tissues compared with matched normal tissues (12), and the dysregulation of miR-769-3p
has been reported in several types of human cancer, such as
colorectal cancer and hepatocellular carcinoma (13,14). A
recent study on the role of the p53 R273H mutation in the tumor
microenvironment has revealed that miR-769-5p is closely associated
with p53, suggesting its potential role in pulmonary metastasis
(15). However, the exact clinical
and functional roles of miR-769-3p in glioma have not been
previously investigated.
The present study aimed to examine the expression
levels and clinical roles of miR-769-3p in patients with glioma and
glioma cell lines and to further explore the potential underlying
molecular mechanisms.
Materials and methods
Patients and sample collection
A total of 113 patients, who were pathologically
diagnosed with glioma and underwent resection of the primary tumor
between August 2010 and January 2012 in Weihai Central Hospital
(Weihai, China), were enrolled in the study. Paired samples of
glioma and adjacent healthy tissues (>3 cm from cancer tissues)
were collected. All tissue samples were immediately snap-frozen in
liquid nitrogen and stored at −80°C for future use. In addition,
preoperative serum samples were collected from the patients, and 95
additional serum samples were collected from healthy volunteers
undergoing routine physical examination at the same hospital to
serve as the control group. The healthy control group had a similar
age and sex ratio to the patient group and had no history of
cancer. The blood samples were centrifuged at 1,000 × g for 20 min
at 4°C within 1 h after collection, and the plasma was stored at
−80°C until further processing. None of the patients received
radiotherapy or chemotherapy prior to sample collection. The
present study was approved by the Ethics Committee of Weihai
Central Hospital, and written informed consent was obtained from
each participant.
Cell culture and transfection
Human glioma cell lines LN-229, A-172, T98G and
SHG-44, as well as normal human astrocytes (NHAs) were obtained
from the American Type Culture Collection (Manassas). All cell
lines were cultured in Dulbecco's modified Eagle's medium (DMEM;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.) and maintained in a
humidified incubator at 37°C with 5% CO2.
The miR-769-3p mimic, miR-769-3p inhibitor and the
corresponding negative controls (mimic NC and inhibitor NC) were
synthesized and purified by Shanghai GenePharma Co., Ltd. The
sequence were as follows: miR-769-3p mimic,
5′-UUGGUUCUGGGGCCUCUAGGGUC-3′; mimic NC,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-769-3p inhibitor,
5′-GACCCUAGAGGCCCCAGAACCAA-3′; inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′. Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect
the miRNAs (100 nM) into glioma cells (A-172 and SHG-44) according
to the manufacturer's protocol. Cells were harvested for subsequent
experiments following 48 h incubation at 37°C.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the tissue or serum
samples and cells using TRIzol® Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. RNA quality and concentration were assessed using a
NanoDrop ND1000 Spectrophotometer (Thermo Fisher Scientific, Inc.).
RNA was reverse transcribed to cDNA using a miRcute miRNA cDNA
first-strand synthesis kit (Tiangen Biotech Co., Ltd.) according to
the manufacturer's instructions. SYBR-Green I Master Mix kit
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform
qPCR. U6 was used as an internal control, and the relative
expression levels were estimated using the 2−ΔΔCq method
(16). The primer sequences used
were: miR-769-3p forward, 5′-TCGGCAGGCTGGGATCTCCGGGG-3′ and
reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
MTT assay
The proliferation of glioma cells was determined
using MTT assay. Once A-172 and SHG-44 cells were transfected with
miR-769-3p mimic or inhibitor, or their negative controls for 48 h,
cells were harvest for subsequent experiments. The transfected
cells were seeded in 96-well plates (1×104 cells/well)
and incubated in a humidified incubator with 5% CO2 at
37°C for 16–48 h. Subsequently, 20 µl 5% MTT solution was added
into each well, followed by an additional 4-h incubation with 5%
CO2 at 37°C in the dark. The medium was discarded, and
100 µl dimethyl sulfoxide was added to stop the reaction. Cell
proliferation was estimated by measuring the optical density at 490
nm using a microplate reader.
Transwell assay
Cell migration and invasion were analyzed using a
Transwell chambers with a pore size of 8 µm (Corning, Inc.). For
the invasion analysis, the membranes were pre-treated with Matrigel
(Corning, Inc.), whereas uncoated membranes were used for the
migration assay. Once A-172 and SHG-44 cells were transfected with
miR-769-3p mimic or inhibitor, or their negative controls for 48 h,
the cells (1×105) were collected, resuspended in 200 µl
serum-free DMEM and seeded into the upper chamber, whereas 300 µl
DMEM containing 10% FBS was added into the lower chamber to act as
a chemoattractant. Following incubation for 48 h at 37°C, cells
remaining on the upper surface of the membranes were removed with a
cotton swab. The cells that had migrated into the lower chambers
were fixed in 4% paraformaldehyde at room temperature for 30 min
and stained with 0.5% crystal violet at room temperature for 30
min. The number of migrated cells was counted using a light
microscope (magnification, ×200; Olympus Corporation).
Luciferase reporter assay
A putative binding site in the 3′UTR of zinc finger
E-box binding homeobox 2 (ZEB2) was identified for miR-769-3p by
TargetScan (http://www.targetscan.org) analysis.
The luciferase reporter gene assay was performed to determine
whether ZEB2 was indeed a target gene of miR-769-3p. Briefly, the
miR-769-3p-binding site in the ZEB2 3′UTR (wild type or mutant) was
cloned downstream of the firefly luciferase gene in a pGL3-promoter
vector. The luciferase activity was measured using a
Dual-Luciferase Reporter Assay System (Promega Corporation).
Renilla luciferase activity was used for normalization.
Western blot assay
Proteins were extracted using RIPA lysis buffer
(Beyotime Institute of Biotechnology), separated by SDS-PAGE and
transferred onto PVDF membranes (EMD Millipore). The membranes were
blocked with 5% non-fat dried milk and incubated with primary
antibodies. The antibodies targeting β-catenin and cyclin D1 were
purchased from Cell Signaling Technology, Inc. Quantitative
densitometric analysis of the immunoblotting images was performed
using ImageJ software (version 1.8.0; National Institutes of
Health).
Statistical analysis
Data were analyzed using SPSS 18.0 (SPSS, Inc.) and
GraphPad Prism 5.0 (GraphPad Software, Inc.). Data are expressed as
the mean ± SD. Student's t-test was used to analyze the differences
between two groups, whereas one-way ANOVA followed by Tukey's
multiple comparison test was applied to analyze multiple groups for
statistical significance. The associations between miR-769-3p
expression and clinicopathological characteristics of patients with
glioma were determined by χ2 test. Receiver operating
characteristic (ROC) curve was used to assess the diagnostic
specificity and sensitivity of miR-769-3p levels. The 5-year
survival rate of patients was calculated by Kaplan-Meier analysis
with log-rank test. Cox regression analysis was used to further
determine the prognostic value of miR-769-3p levels in patients
with glioma. P<0.05 was considered to indicate a statistically
significant difference.
Results
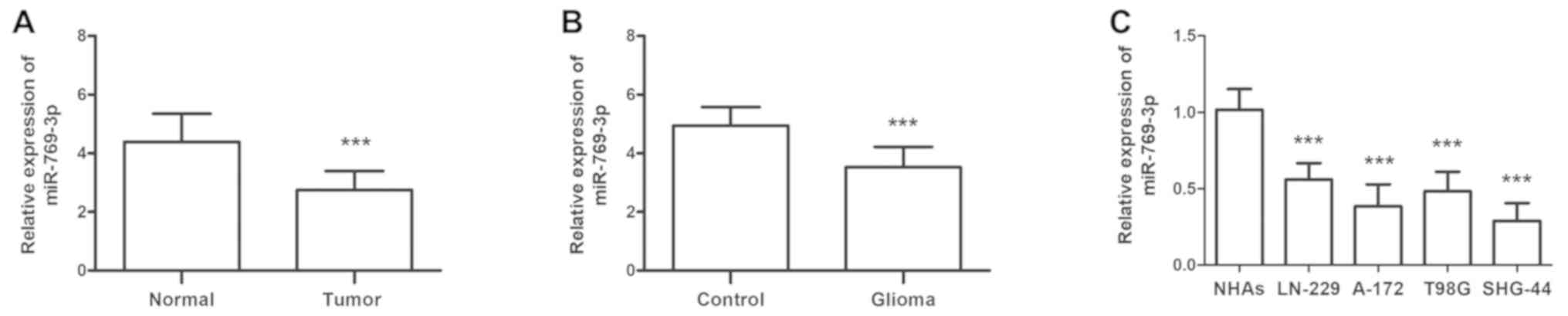
Expression levels of miR-769-3p in
glioma miR-769-3p expression levels were initially detected in 113
paired glioma and adjacent healthy tissues
The results of the RT-qPCR analysis demonstrated
that miR-769-3p levels were significantly decreased in glioma
compared with adjacent healthy tissues (P<0.001; Fig. 1A). Similar results were observed for
patient serum miR-769-3p levels compared with serum samples from
healthy individuals (P<0.001; Fig.
1B). The expression levels of miR-769-3p were also investigated
in four glioma cell lines (LN-229, A-172, T98G and SHG-44); the
results revealed that compared with the levels in normal NHAs,
miR-769-3p expression levels were significantly lower in glioma
cell lines (all P<0.001; Fig.
1C).
Association between miR-769-3p
expression and clinicopathological characteristics of patients with
glioma
To investigate the association of miR-769-3p levels
with the clinicopathological characteristics of patients with
glioma, the patients were classified into low and high expression
groups according to the mean value of miR-769-3p levels in glioma
tissues or serum (Table I). The
χ2 test was used to identify the differences in
clinicopathological characteristics between the two groups. The
results demonstrated that miR-769-3p levels in the serum and glioma
tissues were significantly associated with the World Health
Organization (WHO) grade (P<0.01) (17) and Karnofsky performance score (KPS;
P<0.05) (18). No similar results
were observed for other clinicopathological parameters, including
age, sex and tumor size (all P>0.05; Table I).
 | Table I.Association of tissue and serum
miR-769-3p with the clinicopathological characteristics of patients
with glioma. |
Table I.
Association of tissue and serum
miR-769-3p with the clinicopathological characteristics of patients
with glioma.
|
|
| Tissue miR-769-3p
expression |
| Serum miR-769-3p
expression |
|
|---|
|
|
|
|
|
|
|
|---|
| Characteristic | Total (n=113) | Low (n=69) | High (n=44) | P-value | Low (n=59) | High (n=54) | P-value |
|---|
| Age, years |
|
≤60 | 60 | 37 | 23 |
| 30 | 30 |
|
|
>60 | 53 | 32 | 21 | 0.888 | 29 | 24 | 0.616 |
| Sex |
|
Male | 59 | 36 | 23 |
| 31 | 28 |
|
|
Female | 54 | 33 | 21 | 0.992 | 28 | 26 | 0.941 |
| Tumor size, cm |
|
<5.0 | 57 | 30 | 27 |
| 27 | 30 |
|
|
≥5.0 | 56 | 39 | 17 | 0.064 | 32 | 24 | 0.298 |
| WHO grade |
|
I–II | 58 | 28 | 30 |
| 23 | 35 |
|
|
III–IV | 55 | 41 | 14 | 0.004b | 36 | 19 | 0.006b |
| KPS |
|
<80 | 58 | 41 | 17 |
| 37 | 21 |
|
|
≥80 | 55 | 28 | 27 | 0.031a | 22 | 33 | 0.011a |
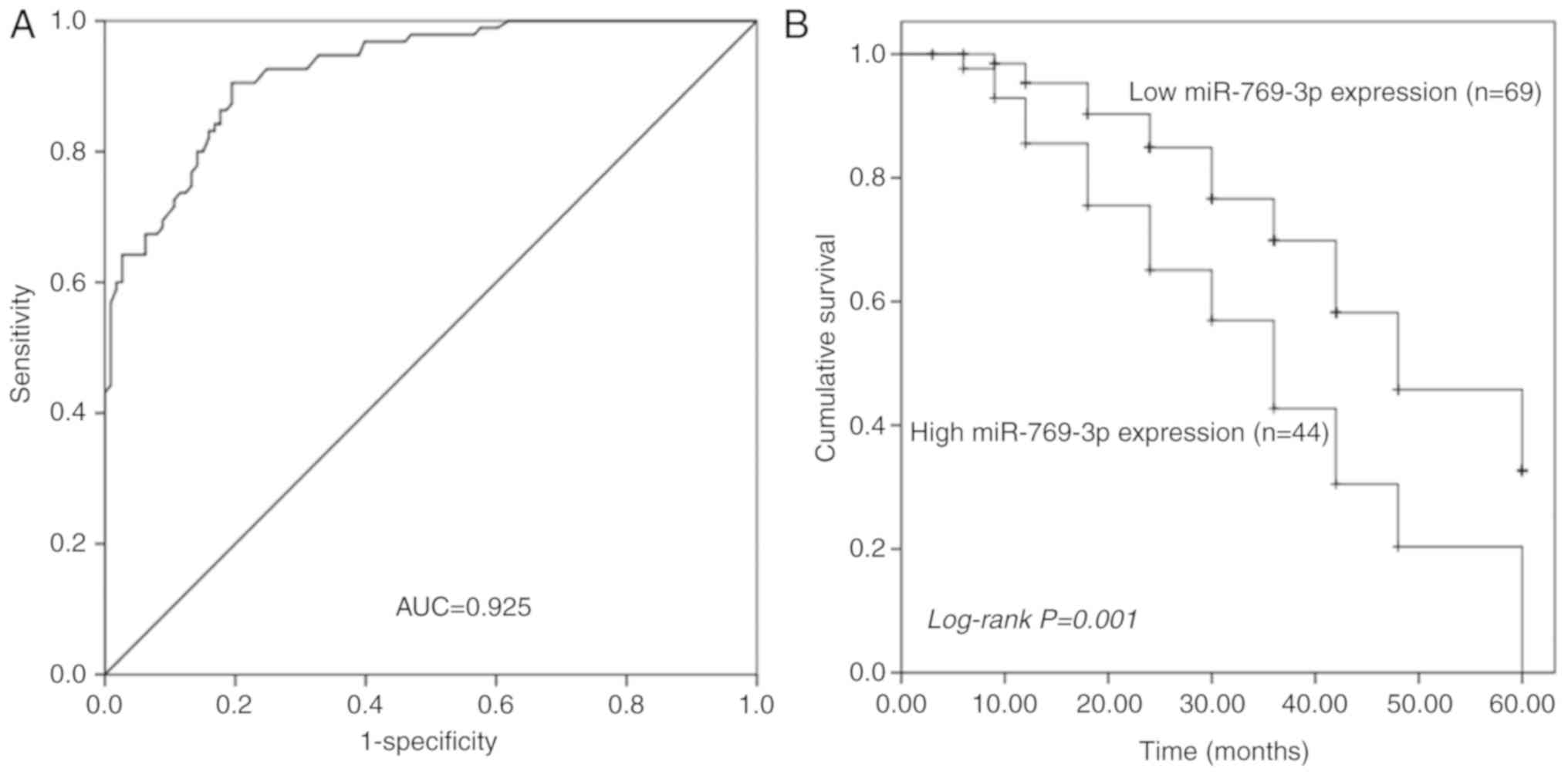
ROC analysis of the diagnostic value
of serum miR-769-3p level for glioma
The ROC curve is a graphical representation that
reflects the association between the sensitivity and specificity of
a laboratory test. The ROC curve analysis revealed that serum
miR-769-3p levels could reliably distinguish patients with glioma
from healthy individuals with an area under the curve value of
0.925. At the optimal cut-off value of 4.085, the sensitivity and
specificity were 90.5 and 80.5%, respectively (Fig. 2A).
Prognostic value of tissue miR-769-3p
level in patients with glioma
Kaplan-Meier analysis was used to assess the
prognostic value of tissue miR-769-3p level in patients with
glioma. The results demonstrated that patients with a high
miR-769-3p expression in the glioma tissue exhibited relatively
poor overall survival compared with those in the low expression
group (log-rank P=0.001; Fig. 2B).
In addition, the clinical parameters and tissue miR-769-3p level
were included in the multivariate Cox analysis to determine their
influence on the overall survival of patients with glioma. The
results demonstrated that the tissue miR-769-3p expression level
[hazard ratio (HR), 2.556; 95% CI, 1.449–4.508; P=0.001] and the
WHO grade (HR, 0.544; 95% CI, 0.314–0.943; P=0.030) were
independent prognostic factors for glioma (Table II).
 | Table II.Multivariate Cox regression analysis
of miR-769-3p expression and clinicopathological characteristics in
patients with glioma. |
Table II.
Multivariate Cox regression analysis
of miR-769-3p expression and clinicopathological characteristics in
patients with glioma.
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Variable | HR | 95% CI | P-value |
|---|
| miR-769-3p
expression (low vs. high) | 2.556 | 1.449–4.508 | 0.001b |
| Age, years (≤60 vs.
>60) | 1.111 | 0.647–1.907 | 0.703 |
| Sex (male vs.
female) | 1.472 | 0.850–2.548 | 0.167 |
| Tumor size, cm
(<5.0 vs. ≥5.0) | 0.747 | 0.436–1.281 | 0.289 |
| WHO grade (I–II vs.
III–IV) | 0.544 | 0.314–0.943 | 0.030a |
| KPS (<80 vs.
>80) | 0.696 | 0.398–1.215 | 0.202 |
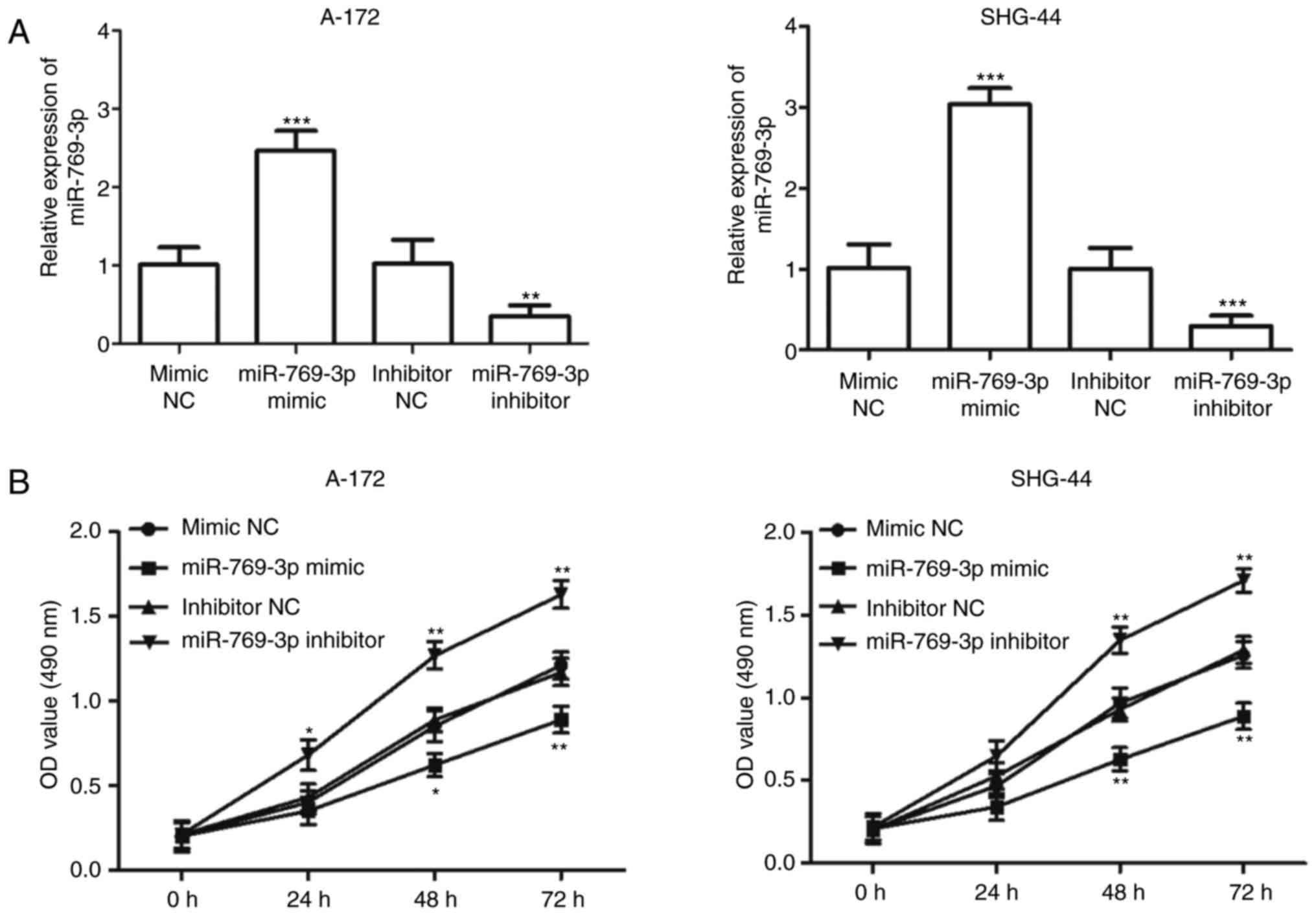
Effects of miR-769-3p on cell
proliferation, migration and invasion in glioma cells
Since miR-769-3p expression was downregulated to a
greater extent in A-172 and SHG-44 cells compared with the other
glioma cell lines (P<0.001; Fig.
1B), the two cell lines were used in subsequent experiments.
miR-769-3p was overexpressed or downregulated in A-172 and SHG-44
cells by transfection with miR-769-3p mimic or miR-769-3p
inhibitor, respectively. The transfection efficiency was determined
by RT-qPCR, and the results indicated that transfection with the
miR-769-3p mimic resulted in an increase in miR-769-3p expression
levels, whereas transfection with the miR-769-3p inhibitor
significantly decreased its expression (P<0.01; Fig. 3A). The MTT assay results revealed
that the overexpression of miR-769-3p significantly inhibited cell
proliferation in A-172 and SHG-44 cells, whereas silencing of
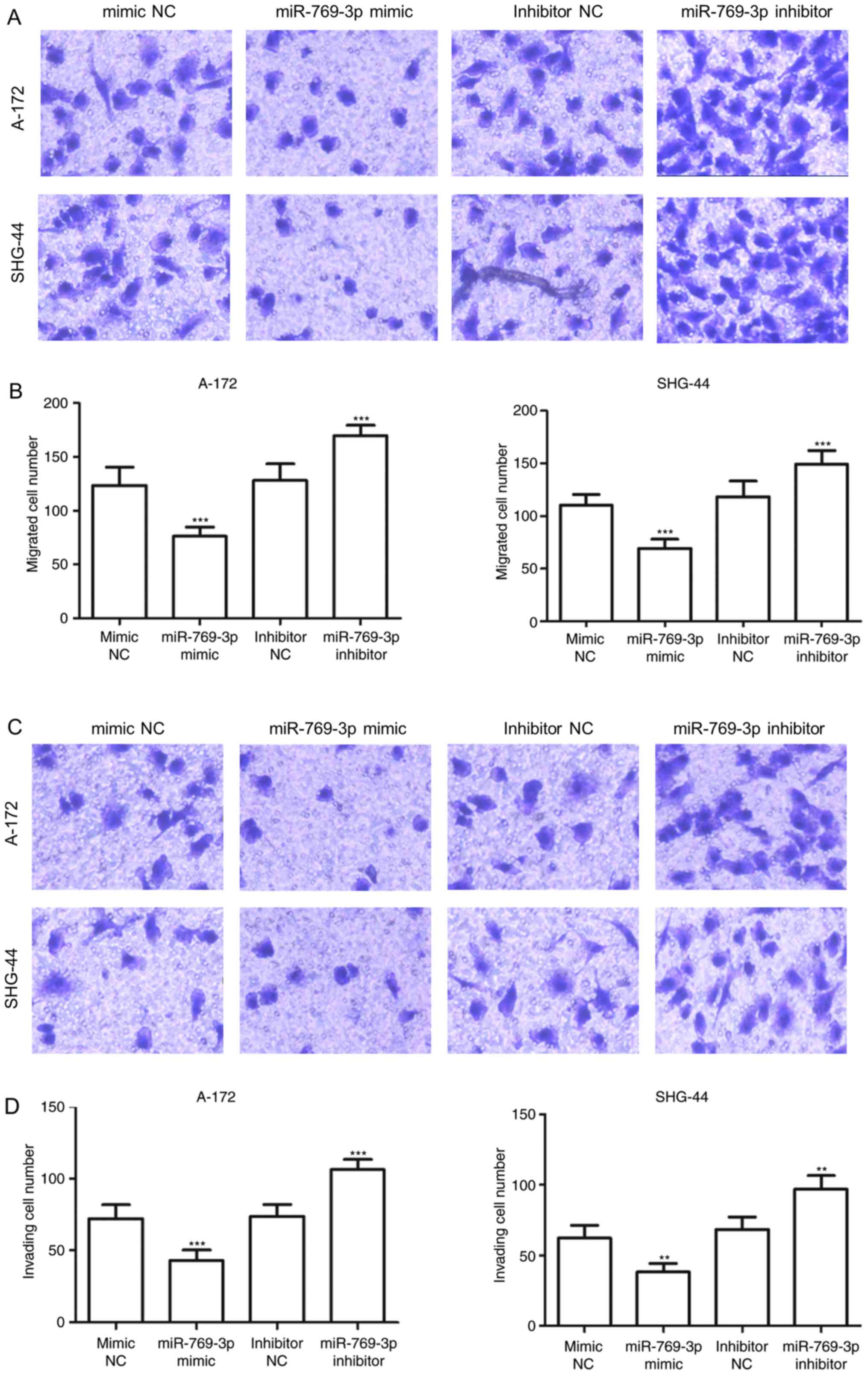
miR-769-3p enhanced cell proliferation (P<0.05; Fig. 3B). The results of the Transwell assay
demonstrated that in A-172 and SHG-44 cells, the number of migrated
cells was significantly lower in the miR-769-3p mimics group
compared with that in the mimic NC group, but higher in the
miR-302b inhibitor group compared with that in the inhibitor NC
group (P<0.001; Fig. 4A and B).
Additionally, overexpression of miR-769-3p significantly reduced
the number of invasive cells, whereas silencing of miR-769-3p
increased the number of invasive cells (P<0.01; Fig. 4C and D).
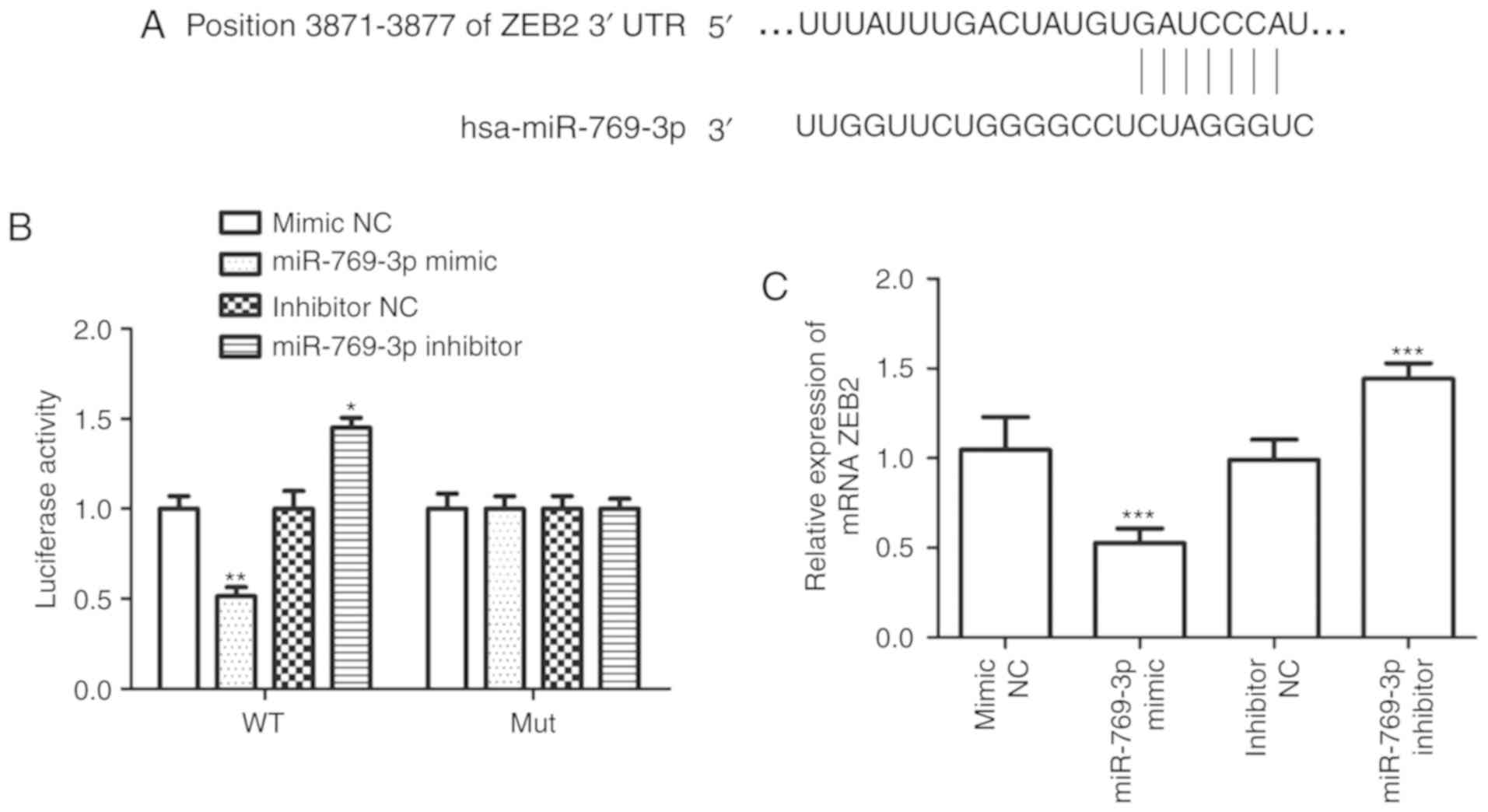
ZEB2 is a target gene of miR-769-3p in
glioma
TargetScan was used to identify the target genes of
miR-769-3p and revealed that ZEB2 mRNA contained seven matched
nucleotides with miR-769-3p at position 3871–3877 in the 3′UTR
(Fig. 5A). The results of the
luciferase reporter assay demonstrated that miR-769-3p mimic
transfection attenuated the luciferase activity of ZEB2 3′UTR,
whereas silencing of miR-769-3p promoted ZEB2 3′UTR luciferase
activity; this was not observed when the mutant ZEB2 3′UTR was
expressed (Fig. 5B). In addition,
RT-qPCR analysis of ZEB2 mRNA expression levels revealed that
miR-769-3p overexpression decreased ZEB2 expression, whereas
opposite results were observed when miR-769-3p expression was
silenced (Fig. 5C).
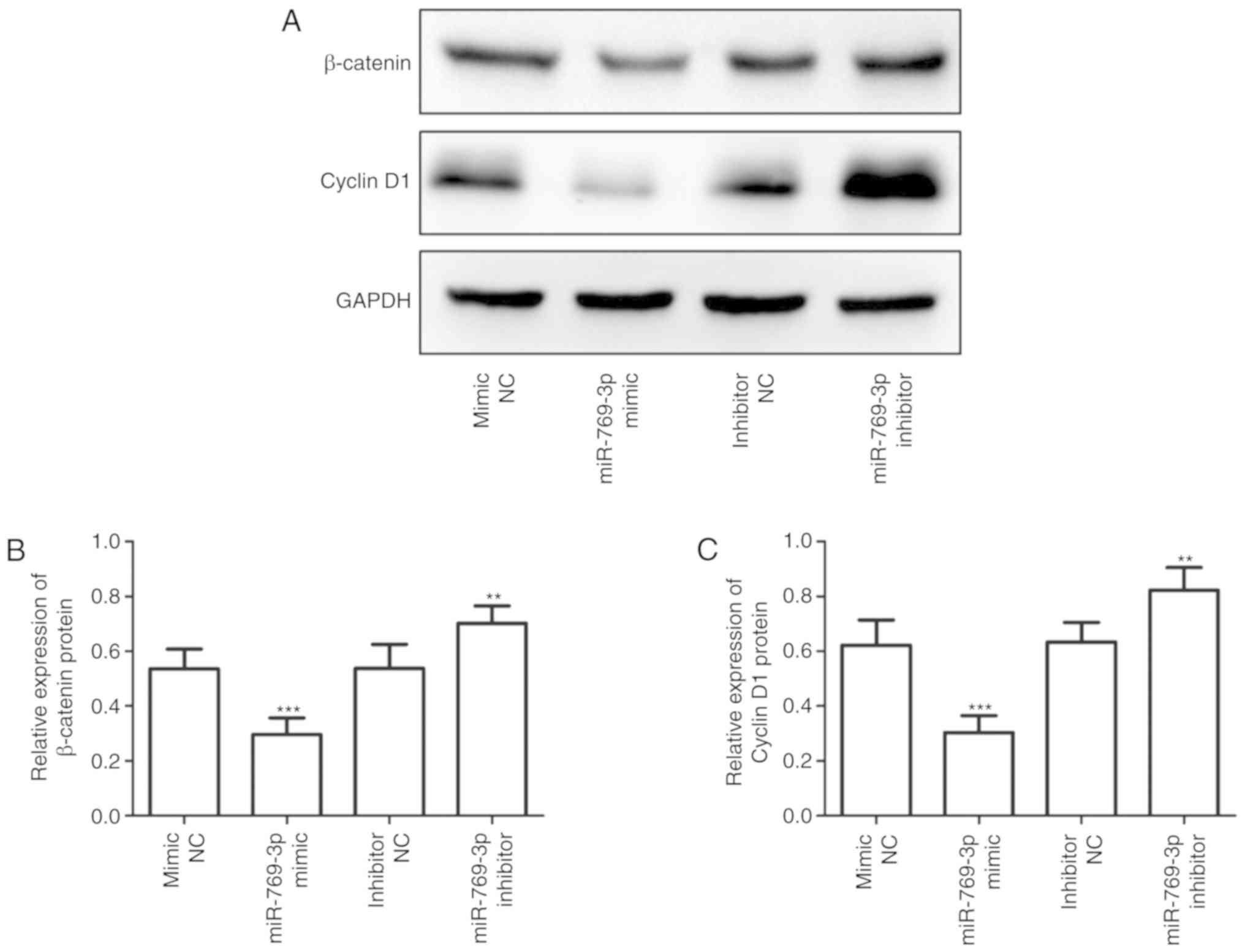
miR-769-3p is involved in the
regulation of the Wnt/β-catenin signaling pathway
Western blotting was performed to determine whether
miR-769-3p may influence the levels of Wnt signaling-related
proteins. As presented in Fig. 6,
overexpression of miR-769-3p significantly suppressed the
expression of Wnt signaling-related proteins β-catenin and cyclin
D1, whereas silencing of miR-769-3p exhibited the opposite
result.
Discussion
Glioma is the most common primary malignant tumor of
the brain (19). The majority of
patients with glioma experience recurrence with poor prognosis,
leading to challenges in the treatment of malignant glioma
(20). The common treatment methods
for glioma include radiotherapy, chemotherapy and surgery (21). However, due to the specific location
of these tumors, the application of radiotherapy and chemotherapy
is limited (22). Thus, it is of
great significance to identify glioma-related therapeutic targets
to improve patient outcomes. miRNAs have been demonstrated to be
involved in different types of human cancer, including glioma
(23). Certain miRNAs have been
identified to be aberrantly expressed in glioma and to be involved
in tumor progression (24).
The dysregulation of miR-769-3p has been reported in
several types of cancer, such as colorectal cancer and melanoma
(13,25). In the present study, the expression
level of miR-769-3p was determined to be downregulated in glioma,
which was consistent with a previous study (12). Additionally, the present result
suggested that miR-769-3p levels were significantly associated with
WHO grade and KPS in patients with glioma. As the alteration of
miR-769-3p expression in glioma was identified, its clinical value
was further examined in glioma diagnosis and prognosis in the
present study. The ROC curve analysis revealed that serum
miR-769-3p levels could reliably distinguish patients with glioma
from healthy individuals. Considering these results, it may be
concluded that miR-769-3p may be a tumor suppressor gene and serve
as a potential diagnostic factor for glioma. Additionally,
Kaplan-Meier survival analysis indicated that low expression levels
of miR-769-3p were significantly associated with poor overall
survival of patients with glioma. Several clinicopathological
variables have been previously used to predict prognosis for
patients with glioma, such as the WHO grade and KPS score (26). In the present study, the WHO grade
and KPS score of the enrolled patients with glioma were analyzed,
and the results demonstrated that miR-769-3p exhibited a
significant association with the WHO grade and KPS score. In
addition, Cox regression analysis results confirmed that miR-769-3p
levels and the WHO grade were independent prognostic factors for
glioma. These results suggested a potential prognostic value for
miR-769-3p in glioma; therefore, miR-769-3p may be a novel
diagnostic and prognostic factor for glioma. To further explore the
functional role of miR-769-3p in glioma, MTT and Transwell in
vitro assays were performed. The results demonstrated that
overexpression of miR-769-3p inhibited cell proliferation,
migration and invasion in glioma cells, suggesting that miR-769-3p
may act as a tumor suppressor and inhibit glioma growth and
metastasis.
ZEB2, also termed SIP1, is a member of zinc-finger
E-box binding proteins (27). ZEB2
is located in the nucleus and is reported to serve as a DNA-binding
transcriptional repressor by interacting with activated Smad
proteins (28). A previous study has
suggested a promoting role of ZEB2 in epithelial-mesenchymal
transition, which is involved in tumor metastasis (29). In the present study, an important
molecular link was observed between miR-769-3p and ZEB2, as
miR-769-3p negatively regulated the level of ZEB2 in glioma cells.
Previous studies have demonstrated that aberrant expression of ZEB2
is associated with the development and progression of various types
of cancer, such as gastric, breast and liver cancer (30–32). In
addition, upregulation of ZEB2 has been detected in patients with
glioma, and ZEB2 has been demonstrated to effectively promote tumor
cell progression (33,34). Collectively, these data supported the
hypothesis of the present study that ZEB2 was a direct target gene
of miR-769-3p in glioma.
The Wnt/β-catenin pathway is considered to be one of
the most important molecular pathways involved in the development
of various types of human cancer, including glioma (35,36).
Downregulation of ZEB2 reduces cell proliferation and suppresses
the expression of β-catenin, c-Myc and cyclin D1 in glioma,
suggesting the involvement of ZEB2 in the regulation of the
Wnt/β-catenin pathway in patients with glioma (34). Therefore, the present study focused
on the association between miR-769-3p and the Wnt/β-catenin
pathway, and the results suggested that overexpression of
miR-769-3p suppressed the expression levels of Wnt
signaling-related proteins. Thus, the results of the present study
indicated that miR-769-3p inhibited glioma cell progression by
targeting ZEB2 and further affecting the Wnt/β-catenin signaling
pathway.
In conclusion, the results of the present study
suggested a crucial role for miR-769-3p in the occurrence and
development of glioma. These results indicate a potential clinical
value of miR-769-3p as an effective biomarker for the diagnosis and
prognosis of glioma. In addition, the results of the present study
demonstrated that miR-769-3p exerted its tumor suppressor role by
targeting ZEB2 and inhibiting the Wnt/β-catenin signaling pathway.
Therefore, miR-769-3p may be a novel therapeutic target for the
treatment of glioma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KW and QS initiated and designed this work and were
responsible for the data acquisition, data analysis and writing the
manuscript. SY and YG collected clinical tissues, performed RNA
extraction and RT-qPCR and data analysis. CZ performed the in
vitro experiments.
Ethics approval and consent to
participate
This study was completed with the approval of the
Ethics Committee of Weihai Central Hospital, and written informed
consent was collected from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liang J, Zhang XL, Li S, Xie S, Wang WF
and Yu RT: Ubiquitin-specific protease 22 promotes the
proliferation, migration and invasion of glioma cells. Cancer
Biomark. 23:381–389. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao W, Yin CY, Jiang J, Kong W, Xu H and
Zhang H: MicroRNA-153 suppresses cell invasion by targeting SNAI1
and predicts patient prognosis in glioma. Oncol Lett. 17:1189–1195.
2019.PubMed/NCBI
|
|
3
|
Yang D, Yuan Y, Zhang S, Zhao K, Li F, Ren
H, Zhang Z and Yu Y: Association between IL-13 Gene rs20541
polymorphism and glioma susceptibility: A meta-analysis. Oncol Res
Treat. 41:14–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hu S, Xu L, Li L, Luo D, Zhao H, Li D and
Peng B: Overexpression of lncRNA PTENP1 suppresses glioma cell
proliferation and metastasis in vitro. Onco Targets Ther.
12:147–156. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Z, Huang X, Li J, Fan H, Yang F,
Zhang R, Yang Y, Feng S, He D, Sun W and Xin T: Interleukin 10
promotes growth and invasion of glioma cells by up-regulating KPNA
2 in vitro. J Cancer Res Ther. 15:927–932. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hao B, Chen X and Cao Y: Yes-associated
protein 1 promotes the metastasis of U251 glioma cells by
upregulating Jagged-1 expression and activating the Notch signal
pathway. Exp Ther Med. 16:1411–1416. 2018.PubMed/NCBI
|
|
7
|
Jiang YR, Du JY, Wang DD and Yang X:
miRNA-130a improves cardiac function by down-regulating TNF-α
expression in a rat model of heart failure. Eur Rev Med Pharmacol
Sci. 22:8454–8461. 2018.PubMed/NCBI
|
|
8
|
Wu YF, Ou CC, Chien PJ, Chang HY, Ko JL
and Wang BY: Chidamide-induced ROS accumulation and
miR-129-3p-dependent cell cycle arrest in non-small lung cancer
cells. Phytomedicine. 56:94–102. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen X and Wang A: Clinical significance
of miR-195 in hepatocellular carcinoma and its biological function
in tumor progression. Onco Targets Ther. 12:527–534. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang S, Zheng S, Cheng H, Lin Y, Wen Y
and Lin W: Flot2 targeted by miR-449 acts as a prognostic biomarker
in glioma. Artif Cells Nanomed Biotechnol. 47:250–255. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu GF, Geng F, Xiao Z, Chen YS, Han Y, You
CY, Gong NL, Xie ZM and Pan M: MicroRNA-6807-3p promotes the
tumorigenesis of glioma by targeting downstream DACH1. Brain Res.
1708:47–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu F, Xiong Y, Zhao Y, Tao L, Zhang Z,
Zhang H, Liu Y, Feng G, Li B, He L, et al: Identification of
aberrant microRNA expression pattern in pediatric gliomas by
microarray. Diagn Pathol. 8:1582013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han C, Song Y and Lian C: MiR-769 inhibits
colorectal cancer cell proliferation and invasion by targeting
HEY1. Med Sci Monit. 24:9232–9239. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang CJ and Du HJ: Screening key miRNAs
for human hepatocellular carcinoma based on miRNA-mRNA functional
synergistic network. Neoplasma. 64:816–823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ju Q, Zhao L, Gao J, Zhou L, Xu Y, Sun Y
and Zhao X: Mutant p53 increases exosome-mediated transfer of
miR-21-3p and miR-769-3p to promote pulmonary metastasis. Chin J
Cancer Res. 31:533–546. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Komori T: The 2016 WHO classification of
tumours of the central nervous system: The major points of
revision. Neurol Med Chir (Tokyo). 57:301–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang F, Wu A, Wang Y and Liu J:
miR-490-3p functions as a tumor suppressor in glioma by inhibiting
high-mobility group AT-hook 2 expression. Exp Ther Med. 18:664–670.
2019.PubMed/NCBI
|
|
19
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chung DS, Shin HJ and Hong YK: A new hope
in immunotherapy for malignant gliomas: Adoptive T cell transfer
therapy. J Immunol Res. 2014:3265452014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Waghmare I, Roebke A, Minata M,
Kango-Singh M and Nakano I: Intercellular cooperation and
competition in brain cancers: Lessons from Drosophila and
human studies. Stem Cells Transl Med. 3:1262–1268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lima FR, Kahn SA, Soletti RC, Biasoli D,
Alves T, da Fonseca AC, Garcia C, Romao L, Brito J, Holanda-Afonso
R, et al: Glioblastoma: Therapeutic challenges, what lies ahead.
Biochim Biophys Acta. 1826:338–349. 2012.PubMed/NCBI
|
|
23
|
Wang R, Zuo X, Wang K, Han Q, Zuo J, Ni H,
Liu W, Bao H, Tu Y and Xie P: MicroRNA-485-5p attenuates cell
proliferation in glioma by directly targeting paired box 3. Am J
Cancer Res. 8:2507–2517. 2018.PubMed/NCBI
|
|
24
|
Wang LQ, Sun W, Wang Y, Li D and Hu AM:
Downregulation of plasma miR-124 expression is a predictive
biomarker for prognosis of glioma. Eur Rev Med Pharmacol Sci.
23:271–276. 2019.PubMed/NCBI
|
|
25
|
Qiu HJ, Lu XH, Yang SS, Weng CY, Zhang EK
and Chen FC: MiR-769 promoted cell proliferation in human melanoma
by suppressing GSK3B expression. Biomed Pharmacother. 82:117–123.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang W, Zhao W, Ge C, Li X, Yang X, Xiang
Y and Sun Z: Decreased let-7b is associated with poor prognosis in
glioma. Medicine (Baltimore). 98:e157842019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Epifanova E, Babaev A, Newman AG and
Tarabykin V: Role of Zeb2/Sip1 in neuronal development. Brain Res.
1705:24–31. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Zhang J, Zhang W, Liu Y, Wang K,
Zhang Y, Yang C, Li X, Shi J, Su L and Hu D: MicroRNA-192 regulates
hypertrophic scar fibrosis by targeting SIP1. J Mol Histol.
48:357–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji H, Sang M, Liu F, Ai N and Geng C:
miR-124 regulates EMT based on ZEB2 target to inhibit invasion and
metastasis in triple-negative breast cancer. Pathol Res Pract.
215:697–704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li H, Xu L, Zhao L, Ma Y, Zhu Z, Liu Y and
Qu X: Insulin-like growth factor-I induces epithelial to
mesenchymal transition via GSK-3βand ZEB2 in the BGC-823 gastric
cancer cell line. Oncol Lett. 9:143–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zou Y, Ouyang Q, Wei W, Yang S, Zhang Y
and Yang W: FAT10 promotes the invasion and migration of breast
cancer cell through stabilization of ZEB2. Biochem Biophys Res
Commun. 506:563–570. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang X, Xu X, Ge G, Zang X, Shao M, Zou
S, Zhang Y, Mao Z, Zhang J, Mao F, et al: miR498 inhibits the
growth and metastasis of liver cancer by targeting ZEB2. Oncol Rep.
41:1638–1648. 2019.PubMed/NCBI
|
|
33
|
Suzuki K, Kawataki T, Endo K, Miyazawa K,
Kinouchi H and Saitoh M: Expression of ZEBs in gliomas is
associated with invasive properties and histopathological grade.
Oncol Lett. 16:1758–1764. 2018.PubMed/NCBI
|
|
34
|
Qi S, Song Y, Peng Y, Wang H, Long H, Yu
X, Li Z, Fang L, Wu A, Luo W, et al: ZEB2 mediates multiple
pathways regulating cell proliferation, migration, invasion, and
apoptosis in glioma. PLoS One. 7:e388422012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gong X, Liao X and Huang M: LncRNA CASC7
inhibits the progression of glioma via regulating Wnt/β-catenin
signaling pathway. Pathol Res Pract. 215:564–570. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Yang Q, Cheng Y, Gao M, Kuang L
and Wang C: Myosin heavy chain 10 (MYH10) gene silencing reduces
cell migration and invasion in the glioma cell lines U251, T98G,
and SHG44 by inhibiting the Wnt/β-catenin pathway. Med Sci Monit.
24:9110–9119. 2018. View Article : Google Scholar : PubMed/NCBI
|