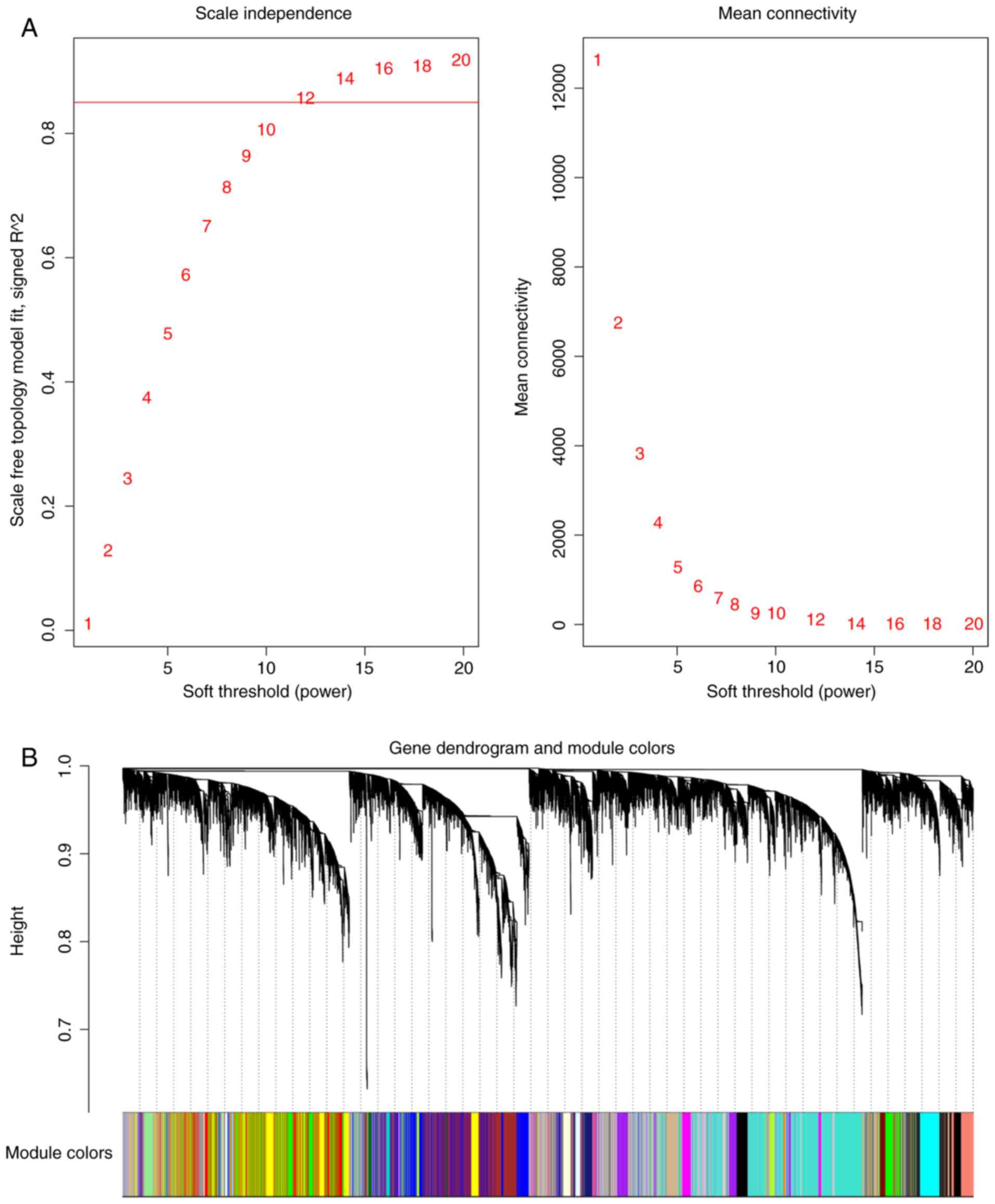
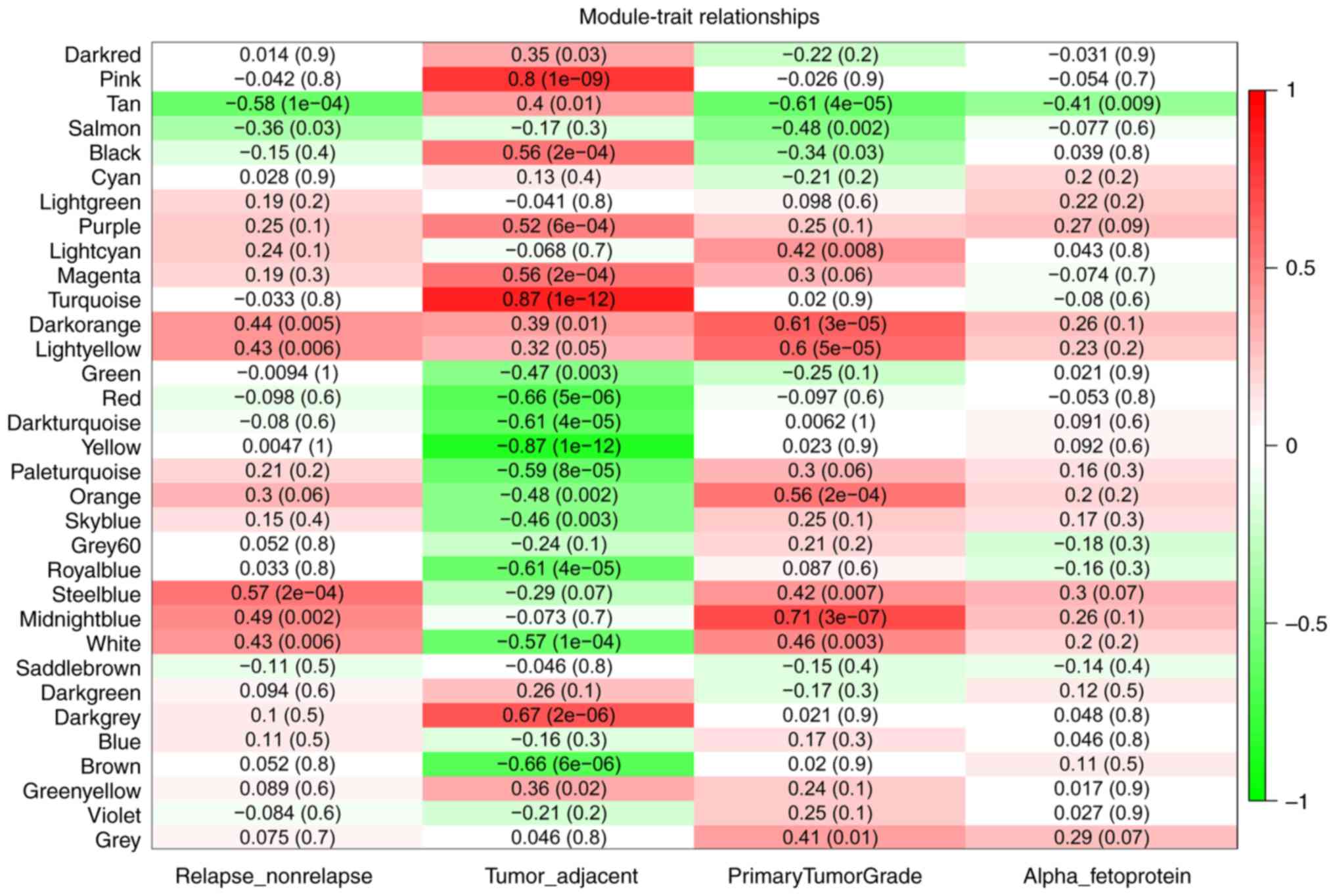
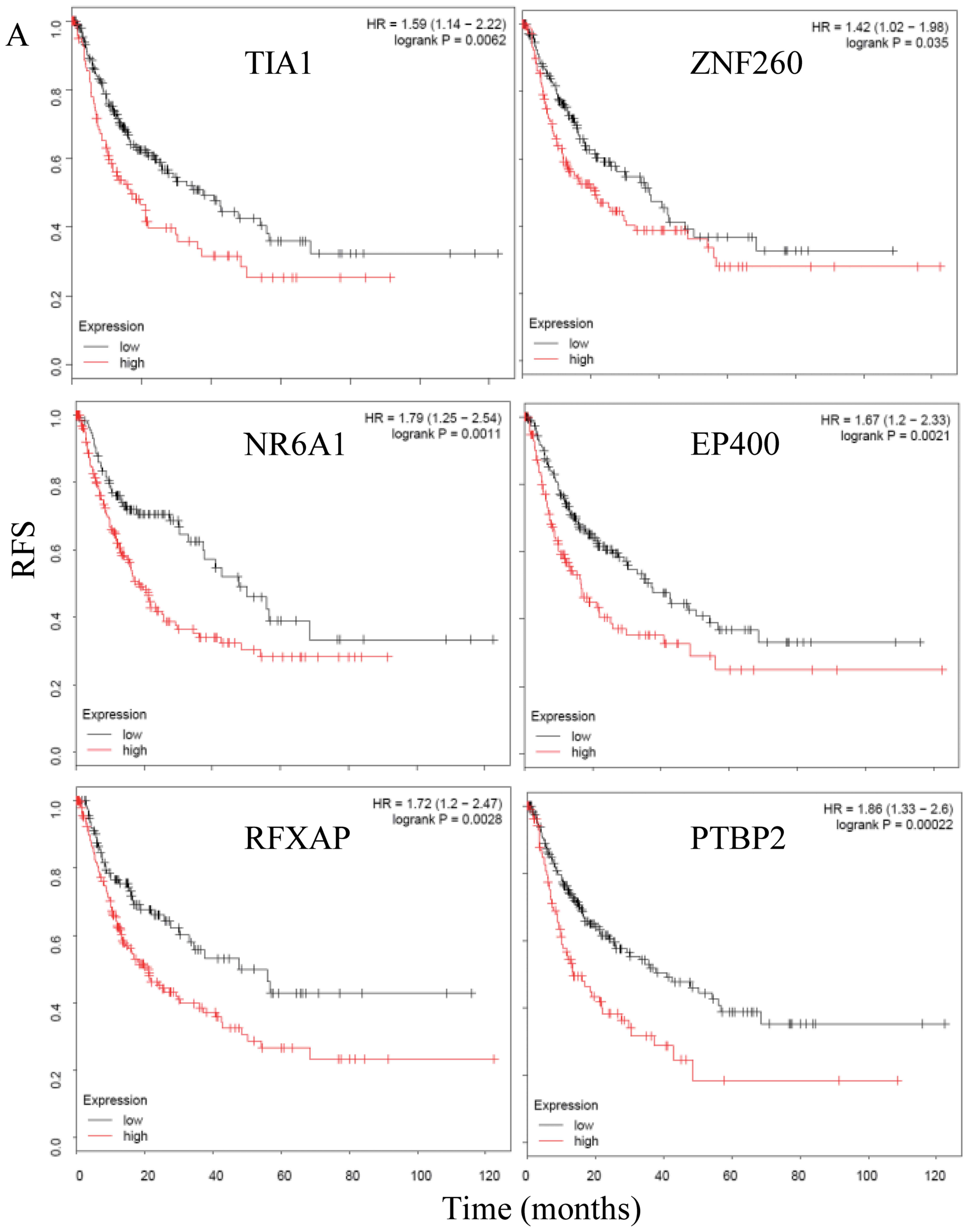
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jung KH, Zhang J, Zhou C, Shen H, Gagea M,
Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK and Beretta L:
Differentiation therapy for hepatocellular carcinoma: Multifaceted
effects of miR-148a on tumor growth and phenotype and liver
fibrosis. Hepatology. 63:864–879. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Llovet JM, Fuster J and Bruix J;
Barcelona-Clínic Liver Cancer Group, : The Barcelona approach:
Diagnosis, staging, and treatment of hepatocellular carcinoma.
Liver Transpl. 10:S115–S120. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Welker MW, Bechstein WO, Zeuzem S and
Trojan J: Recurrent hepatocellular carcinoma after liver
transplantation - an emerging clinical challenge. Transpl Int.
26:109–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Darnell JE Jr: Transcription factors as
targets for cancer therapy. Nat Rev Cancer. 2:740–749. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malz M, Pinna F, Schirmacher P and
Breuhahn K: Transcriptional regulators in hepatocarcinogenesis-key
integrators of malignant transformation. J Hepatol. 57:186–195.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huh HD, Kim DH, Jeong HS and Park HW:
Regulation of TEAD transcription factors in cancer biology. Cells.
8:6002019. View Article : Google Scholar
|
|
9
|
Luo D, Wang Z and Wu J, Jiang C and Wu J:
The role of hypoxia inducible factor-1 in hepatocellular carcinoma.
Biomed Res Int. 2014:4092722014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin D and Wu J: Hypoxia inducible factor
in hepatocellular carcinoma: A therapeutic target. World J
Gastroenterol. 21:12171–12178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Méndez-Blanco C, Fondevila F,
García-Palomo A, González-Gallego J and Mauriz JL: Sorafenib
resistance in hepatocarcinoma: Role of hypoxia-inducible factors.
Exp Mol Med. 50:1342018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Matteis S, Scarpi E, Granato AM,
Vespasiani-Gentilucci U, La Barba G, Foschi FG, Bandini E, Ghetti
M, Marisi G, Cravero P, et al: Role of SIRT-3, p-mTOR and HIF-1α in
hepatocellular carcinoma patients affected by metabolic
dysfunctions and in chronic treatment with metformin. Int J Mol
Sci. 20:2019. View Article : Google Scholar
|
|
13
|
Wen Y, Zhou X, Lu M, He M, Tian Y, Liu L,
Wang M, Tan W, Deng Y, Yang X, et al: Bclaf1 promotes angiogenesis
by regulating HIF-1α transcription in hepatocellular carcinoma.
Oncogene. 38:1845–1859. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang G and Zhang G: Upregulation of FoxP4
in HCC promotes migration and invasion through regulation of EMT.
Oncol Lett. 17:3944–3951. 2019.PubMed/NCBI
|
|
15
|
Wang Q, Tan YX, Ren YB, Dong LW, Xie ZF,
Tang L, Cao D, Zhang WP, Hu HP and Wang HY: Zinc finger protein
ZBTB20 expression is increased in hepatocellular carcinoma and
associated with poor prognosis. BMC Cancer. 11:2712011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kan H, Huang Y, Li X, Liu D, Chen J and
Shu M: Zinc finger protein ZBTB20 is an independent prognostic
marker and promotes tumor growth of human hepatocellular carcinoma
by repressing FoxO1. Oncotarget. 7:14336–14349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Z, Sun B, Li Y, Zhao X, Zhao X, Gu Q,
An J, Dong X, Liu F and Wang Y: ZEB2 promotes vasculogenic mimicry
by TGF-β1 induced epithelial-to-mesenchymal transition in
hepatocellular carcinoma. Exp Mol Pathol. 98:352–359. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu D, Liu G, Liu Y, Saiyin H, Wang C, Wei
Z, Zen W, Liu D, Chen Q, Zhao Z, et al: Zinc finger protein 191
inhibits hepatocellular carcinoma metastasis through discs large
1-mediated yes-associated protein inactivation. Hepatology.
64:1148–1162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yi PS, Wu B, Deng DW, Zhang GN and Li JS:
Positive expression of ZNF689 indicates poor prognosis of
hepatocellular carcinoma. Oncol Lett. 16:5122–5130. 2018.PubMed/NCBI
|
|
20
|
Xiang Q, Zhou D, He X, Fan J, Tang J, Qiu
Z, Zhang Y, Qiu J, Xu Y and Lai G: The zinc finger protein GATA4
induces MET and cellular senescence through the NF-kappaB pathway
in hepatocellular carcinoma. J Gastroenterol Hepatol. 2019.
View Article : Google Scholar
|
|
21
|
Wang N, Wang S, Yang SL, Liu LP, Li MY,
Lai PBS and Chen GG: Targeting ZBP-89 for the treatment of
hepatocellular carcinoma. Expert Opin Ther Targets. 22:817–822.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang T, Huang Y, Liu W, Meng W, Zhao H,
Yang Q, Gu SJ, Xiao CC, Jia CC, Zhang B, et al: Overexpression of
zinc finger protein 687 enhances tumorigenic capability and
promotes recurrence of hepatocellular carcinoma. Oncogenesis.
6:e3632017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu X, Zhou Y, Miao R, Chen W, Qu K, Pang Q
and Liu C: Transcriptional modules related to hepatocellular
carcinoma survival: Coexpression network analysis. Front Med.
10:183–190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yin L, Cai Z, Zhu B and Xu C:
Identification of key pathways and genes in the dynamic progression
of HCC based on WGCNA. Genes (Basel). 9:922018. View Article : Google Scholar
|
|
25
|
Xu W, Rao Q, An Y, Li M and Zhang Z:
Identification of biomarkers for Barcelona Clinic Liver Cancer
staging and overall survival of patients with hepatocellular
carcinoma. PLoS One. 13:e02027632018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li B, Pu K and Wu X: Identifying novel
biomarkers in hepatocellular carcinoma by weighted gene
co-expression network analysis. J Cell Biochem. Feb 11–2019.(Epub
ahead of print). doi: 10.1002/jcb.28420.
|
|
27
|
Zhang C, Peng L, Zhang Y, Liu Z, Li W,
Chen S and Li G: The identification of key genes and pathways in
hepatocellular carcinoma by bioinformatics analysis of
high-throughput data. Med Oncol. 34:1012017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Skinkyte-Juskiene R, Kogelman LJA and
Kadarmideen HN: Transcription factor co-expression networks of
adipose RNA-Seq data reveal regulatory mechanisms of obesity. Curr
Genomics. 19:289–299. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xue F, Higgs BW, Huang J, Morehouse C, Zhu
W, Yao X, Brohawn P, Xiao Z, Sebastian Y, Liu Z, et al: HERC5 is a
prognostic biomarker for post-liver transplant recurrent human
hepatocellular carcinoma. J Transl Med. 13:3792015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Collado-Torres L, Nellore A, Kammers K,
Ellis SE, Taub MA, Hansen KD, Jaffe AE, Langmead B and Leek JT:
Reproducible RNA-seq analysis using recount2. Nat Biotechnol.
35:319–321. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng G, Tu K, Yang Q, Xiong Y, Wei C, Xie
L, Zhu Y and Li Y: ITFP: An integrated platform of mammalian
transcription factors. Bioinformatics. 24:2416–2417. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marbach D, Lamparter D, Quon G, Kellis M,
Kutalik Z and Bergmann S: Tissue-specific regulatory circuits
reveal variable modular perturbations across complex diseases. Nat
Methods. 13:366–370. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Neph S, Stergachis AB, Reynolds A,
Sandstrom R, Borenstein E and Stamatoyannopoulos JA: Circuitry and
dynamics of human transcription factor regulatory networks. Cell.
150:1274–1286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stergachis AB, Neph S, Sandstrom R, Haugen
E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee
K, et al: Conservation of trans-acting circuitry during mammalian
regulatory evolution. Nature. 515:365–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang C, Xuan Z, Zhao F and Zhang MQ:
TRED: A transcriptional regulatory element database, new entries
and other development. Nucleic Acids Res. 35:D137–D140. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Han H, Shim H, Shin D, Shim JE, Ko Y, Shin
J, Kim H, Cho A, Kim E, Lee T, et al: TRRUST: A reference database
of human transcriptional regulatory interactions. Sci Rep.
5:114322015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mason MJ, Fan G, Plath K, Zhou Q and
Horvath S: Signed weighted gene co-expression network analysis of
transcriptional regulation in murine embryonic stem cells. BMC
Genomics. 10:3272009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Reimand J, Arak T, Adler P, Kolberg L,
Reisberg S, Peterson H and Vilo J: g:Profiler-a web server for
functional interpretation of gene lists (2016 update). Nucleic
Acids Res. 44:W83–W89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Reimand J, Arak T and Vilo J:
g:Profiler--a web server for functional interpretation of gene
lists (2011 update). Nucleic Acids Res. 39:W307–W315. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Raudvere U, Kolberg L, Kuzmin I, Arak T,
Adler P, Peterson H and Vilo J: g:Profiler: A web server for
functional enrichment analysis and conversions of gene lists (2019
update). Nucleic Acids Res. 47:W191–W198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W and Sun SH: Oncofetal long
noncoding RNA PVT1 promotes proliferation and stem cell-like
property of hepatocellular carcinoma cells by stabilizing NOP2.
Hepatology. 60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
von Mering C, Jensen LJ, Snel B, Hooper
SD, Krupp M, Foglierini M, Jouffre N, Huynen MA and Bork P: STRING:
Known and predicted protein-protein associations, integrated and
transferred across organisms. Nucleic Acids Res. 33:D433–D437.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yuan L, Qian G, Chen L, Wu CL, Dan HC,
Xiao Y and Wang X: Co-expression network analysis of biomarkers for
adrenocortical carcinoma. Front Genet. 9:3282018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen PF, Wang F, Nie JY, Feng JR, Liu J,
Zhou R, Wang HL and Zhao Q: Co-expression network analysis
identified CDH11 in association with progression and prognosis in
gastric cancer. Onco Targets Ther. 11:6425–6436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Janky R, Verfaillie A, Imrichová H, Van de
Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval
Sanchez M, Potier D, et al: iRegulon: From a gene list to a gene
regulatory network using large motif and track collections. PLoS
Comput Biol. 10:e10037312014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yin L, Guo X, Zhang C, Cai Z and Xu C: In
silico analysis of expression data during the early priming stage
of liver regeneration after partial hepatectomy in rat. Oncotarget.
9:11794–11804. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Menyhárt O, Nagy Á and Győrffy B:
Determining consistent prognostic biomarkers of overall survival
and vascular invasion in hepatocellular carcinoma. R Soc Open Sci.
5:1810062018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang X, Hua L, Yan D, Zhao F, Liu J, Zhou
H, Liu J, Wu M, Zhang C, Chen Y, et al: Overexpression of PCBP2
contributes to poor prognosis and enhanced cell growth in human
hepatocellular carcinoma. Oncol Rep. 36:3456–3464. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Golob-Schwarzl N, Krassnig S, Toeglhofer
AM, Park YN, Gogg-Kamerer M, Vierlinger K, Schröder F, Rhee H,
Schicho R, Fickert P and Haybaeck J: New liver cancer biomarkers:
PI3K/AKT/mTOR pathway members and eukaryotic translation initiation
factors. Eur J Cancer. 83:56–70. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Poon RT: Prevention of recurrence after
resection of hepatocellular carcinoma: A daunting challenge.
Hepatology. 54:757–759. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Vilarinho S and Calvisi DF: New advances
in precision medicine for hepatocellular carcinoma recurrence
prediction and treatment. Hepatology. 60:1812–1814. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shimagaki T, Yoshizumi T, Harimoto N,
Yoshio S, Naito Y, Yamamoto Y, Ochiya T, Yoshida Y, Kanto T and
Maehara Y: MicroRNA-125b expression and intrahepatic metastasis are
predictors for early recurrence after hepatocellular carcinoma
resection. Hepatol Res. 48:313–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chuma M, Toyoda H, Matsuzaki J, Saito Y,
Kumada T, Tada T, Kaneoka Y, Maeda A, Yokoo H, Ogawa K, et al:
Circulating microRNA-1246 as a possible biomarker for early tumor
recurrence of hepatocellular carcinoma. Hepatol Res. 49:810–822.
2019.PubMed/NCBI
|
|
61
|
Song S, Yang Y, Liu M, Liu B, Yang X, Yu
M, Qi H, Ren M, Wang Z, Zou J, et al: MiR-125b attenuates human
hepatocellular carcinoma malignancy through targeting SIRT6. Am J
Cancer Res. 8:993–1007. 2018.PubMed/NCBI
|
|
62
|
Zhang Q, Cao LY, Cheng SJ, Zhang AM, Jin
XS and Li Y: p53-induced microRNA-1246 inhibits the cell growth of
human hepatocellular carcinoma cells by targeting NFIB. Oncol Rep.
33:1335–1341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai
C, Lu S, Han Q and Zhao RC: MicroRNA-1246 enhances migration and
invasion through CADM1 in hepatocellular carcinoma. BMC Cancer.
14:6162014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chan HM, Narita M, Lowe SW and Livingston
DM: The p400 E1A-associated protein is a novel component of the p53
-->p21 senescence pathway. Genes Dev. 19:196–201. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shi M, Chen MS, Sekar K, Tan CK, Ooi LL
and Hui KM: A blood-based three-gene signature for the non-invasive
detection of early human hepatocellular carcinoma. Eur J Cancer.
50:928–936. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Montalvá EM, Cantos M, Boscà A, Rubín A,
Vinaixa C, Granero P, Maupoey J and López-Andújar R: Prognostic
value of pre-transplantation serum alpha-fetoprotein levels in
hepatocellular carcinoma recurrence. Transplant Proc. 48:2966–2968.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tsukamoto M, Nitta H, Imai K, Higashi T,
Nakagawa S, Okabe H, Arima K, Kaida T, Taki K, Hashimoto D, et al:
Clinical significance of half-lives of tumor markers α-fetoprotein
and des-γ-carboxy prothrombin after hepatectomy for hepatocellular
carcinoma. Hepatol Res. 48:E183–E193. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Carrascoso I, Alcalde J, Tabas-Madrid D,
Oliveros JC and Izquierdo JM: Transcriptome-wide analysis links the
short-term expression of the b isoforms of TIA proteins to
protective proteostasis-mediated cell quiescence response. PLoS
One. 13:e02085262018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tak H, Kang H, Ji E, Hong Y, Kim W and Lee
EK: Potential use of TIA-1, MFF, microRNA-200a-3p, and microRNA-27
as a novel marker for hepatocellular carcinoma. Biochem Biophys Res
Commun. 497:1117–1122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Subramaniam K, Ooi LL and Hui KM:
Transcriptional down-regulation of IGFBP-3 in human hepatocellular
carcinoma cells is mediated by the binding of TIA-1 to its AT-rich
element in the 3′-untranslated region. Cancer Lett. 297:259–268.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yao Y, Mao W, Dong M, Yang D, Li W and
Chen Y: Serum insulin-like growth factor-1 (IGF-1): A novel
prognostic factor for early recurrence of hepatocellular carcinoma
(HCC). Clin Lab. 63:261–270. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Li Z, Shao C, Kong Y, Carlock C, Ahmad N
and Liu X: DNA damage response-independent role for MDC1 in
maintaining genomic stability. Mol Cell Biol. 37:2017. View Article : Google Scholar
|
|
73
|
Lee JH, Park SJ, Hariharasudhan G, Kim MJ,
Jung SM, Jeong SY, Chang IY, Kim C, Kim E, Yu J, et al: ID3
regulates the MDC1-mediated DNA damage response in order to
maintain genome stability. Nat Commun. 8:9032017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang S, Zou Z, Luo X, Mi Y, Chang H and
Xing D: LRH1 enhances cell resistance to chemotherapy by
transcriptionally activating MDC1 expression and attenuating DNA
damage in human breast cancer. Oncogene. 37:3243–3259. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhou H, Qin Y, Ji S, Ling J, Fu J, Zhuang
Z, Fan X, Song L, Yu X and Chiao PJ: SOX9 activity is induced by
oncogenic Kras to affect MDC1 and MCMs expression in pancreatic
cancer. Oncogene. 37:912–923. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bailey SD, Zhang X, Desai K, Aid M,
Corradin O, Cowper-Sal Lari R, Akhtar-Zaidi B, Scacheri PC,
Haibe-Kains B and Lupien M: ZNF143 provides sequence specificity to
secure chromatin interactions at gene promoters. Nat Commun.
2:61862015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ishiguchi H, Izumi H, Torigoe T, Yoshida
Y, Kubota H, Tsuji S and Kohno K: ZNF143 activates gene expression
in response to DNA damage and binds to cisplatin-modified DNA. Int
J Cancer. 111:900–909. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Paek AR, Lee CH and You HJ: A role of
zinc-finger protein 143 for cancer cell migration and invasion
through ZEB1 and E-cadherin in colon cancer cells. Mol Carcinog. 53
(Suppl 1):E161–E168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kawatsu Y, Kitada S, Uramoto H, Zhi L,
Takeda T, Kimura T, Horie S, Tanaka F, Sasaguri Y, Izumi H, et al:
The combination of strong expression of ZNF143 and high MIB-1
labelling index independently predicts shorter disease-specific
survival in lung adenocarcinoma. Br J Cancer. 110:2583–2592. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Haibara H, Yamazaki R, Nishiyama Y, Ono M,
Kobayashi T, Hokkyo-Itagaki A, Nishisaka F, Nishiyama H, Kurita A,
Matsuzaki T, et al: YPC-21661 and YPC-22026, novel small molecules,
inhibit ZNF143 activity in vitro and in vivo. Cancer Sci.
108:1042–1048. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ruosi C, Colella G, Fazioli F, Miceli R,
Gallo M, Di Salvatore MG, Cimmino A and de Nigris F: Yin Yang I as
an epimodulator of miRNAs in the metastatic cascade. Crit Rev
Oncog. 22:99–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Cho AA and Bonavida B: Targeting the
overexpressed YY1 in cancer inhibits EMT and metastasis. Crit Rev
Oncog. 22:49–61. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Han J, Meng J, Chen S, Wang X, Yin S,
Zhang Q, Liu H, Qin R, Li Z, Zhong W, et al: YY1 complex promotes
quaking expression via super-enhancer binding during EMT of
hepatocellular carcinoma. Cancer Res. 79:1451–1464. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Dong S, Ma X, Wang Z, Han B, Zou H, Wu Z,
Zang Y and Zhuang L: YY1 promotes HDAC1 expression and decreases
sensitivity of hepatocellular carcinoma cells to HDAC inhibitor.
Oncotarget. 8:40583–40593. 2017.PubMed/NCBI
|
|
85
|
Kim JS, Son SH, Kim MY, Choi D, Jang IS,
Paik SS, Chae JH, Uversky VN and Kim CG: Diagnostic and prognostic
relevance of CP2c and YY1 expression in hepatocellular carcinoma.
Oncotarget. 8:24389–24400. 2017.PubMed/NCBI
|
|
86
|
Tsang DP, Wu WK, Kang W, Lee YY, Wu F, Yu
Z, Xiong L, Chan AW, Tong JH, Yang W, et al: Yin Yang 1-mediated
epigenetic silencing of tumour-suppressive microRNAs activates
nuclear factor-κB in hepatocellular carcinoma. J Pathol.
238:651–664. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang S, Jiang T, Feng L, Sun J, Lu H,
Wang Q, Pan M, Huang D, Wang X, Wang L and Jin H: Yin Yang-1
suppresses differentiation of hepatocellular carcinoma cells
through the downregulation of CCAAT/enhancer-binding protein alpha.
J Mol Med (Berl). 90:1069–1077. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Teng CF, Wu HC, Tsai HW, Shiah HS, Huang W
and Su IJ: Novel feedback inhibition of surface antigen synthesis
by mammalian target of rapamycin (mTOR) signal and its implication
for hepatitis B virus tumorigenesis and therapy. Hepatology.
54:1199–1207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Moldovan GL and D'Andrea AD: How the
fanconi anemia pathway guards the genome. Annu Rev Genet.
43:223–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ferroudj S, Yildiz G, Bouras M, Iscan E,
Ekin U and Ozturk M: Role of Fanconi anemia/BRCA pathway genes in
hepatocellular carcinoma chemoresistance. Hepatol Res.
46:1264–1274. 2016. View Article : Google Scholar : PubMed/NCBI
|