Introduction
Malignant pleural mesothelioma (MPM) is a rare and
aggressive malignancy arising from the mesothelial cells lining the
pleural cavity. There is a clear association between occupational
or environmental asbestos exposure, and the development of MPM,
with a latency period of about 40 years before disease
presentation. Global incidence of MPM has risen steadily over the
past decade, and it is predicted to reach the highest peak in 2020
(1,2). MPM is a heterogeneous tumor, including
three main histological subtypes: Epithelioid (60–80%), sarcomatoid
(<10%) and mixed (10–15%) (3,4).
The definitive MPM diagnosis is mainly based on
histopathological examinations of pleural tissues, which could not
be sufficiently clear to discriminate MPM neither from secondary
tumors involving the pleura nor from benign pleural proliferations
(3). Particularly, the differential
diagnosis of MPM and benign pleural lesions is a hard task to
accomplish, and currently the only criterion to certainly determine
the malignancy is the presence of stromal or lung invasion
(5). However, it is not always
possible to estimate whether stromal invasion is present or not,
according to quantitative and qualitative parameters of pleural
biopsies and their representativeness of the whole lesion (4). Moreover, for many patients pleural
biopsies are not available and diagnosis has to be made on
cytological specimens from pleural effusions, whose diagnostic
sensitivity is variable ranging from 20 to 70% (6).
A variety of ancillary tests, mostly based on the
evaluation of immunohistochemical markers, have been claimed to be
useful for separating benign from malignant mesothelial
proliferations either on pleural tissues or effusions (7). However, the majority of these markers
did not achieve sufficient diagnostic accuracy. Recently, the
deletion of the cyclin dependent kinase inhibitor 2A
(CDKN2A) gene, better known as p16, and loss BRCA1
associated protein 1 (BAP1) protein have shown an excellent
specificity in separating MPM from pleural mesothelial hyperplasia
(MH) (8–12).
p16 is a tumor suppressor gene which is
located in chromosome 9p21.3, it regulates cell cycle, and its
inactivation results in the enhancement of cell proliferation.
Inactivation of p16 can occur through a homozygous deletion,
point mutations or methylation changes. Homozygous deletion of
p16, detectable by Fluorescent in Situ Hybridization (FISH),
is very common in malignant mesotheliomas, but it has never been
described in benign mesothelial proliferations, indicating a
specificity of 100% (8–13). Unfortunately, not all mesotheliomas
harbor p16 alterations, and consequently, the sensitivity
for epithelioid/biphasic (mixed) and sarcomatoid MPM ranges from
approximately 45 to 85% and 50 to 100%, respectively (11).
BAP1 is a nuclear ubiquitin hydrolase that functions
as tumor suppressor; it controls DNA repair, apoptosis promotion,
and expression of genes related to cell cycle and cell
proliferation. The expression of BAP1 is frequently lost in MPM due
to point mutations or chromosomal losses (3p21.1). The lack of
immunohistochemical staining is highly specific for MPM, but it is
observed only in 60–70 and 15% of epithelioid/mixed and sarcomatoid
mesotheliomas respectively (8,13).
Although the combination of BAP1 and p16 can
increase their diagnostic sensitivity, the absence of p16
deletion or BAP1 loss does not allow to rule out MPM.
In this context, in a previous study (14) our group developed and tested a new
tool for MPM differential diagnosis, based on the expression
profile of 117 genes that had been reported as deregulated in MPM,
including BAP1 and p16. In detail, gene expression
levels were determined using the NanoString System (NanoString
Technologies) and samples were classified as malignant or benign by
the Uncorrelated Shrunken Centroid (USC) classification algorithm.
In our precedent study, the USC identified two classification
models (22 genes and 40 genes), both able to properly classify all
the analyzed pleural samples (14).
The aim of this study was to directly compare the
performance of the tool previously identified by our group with
BAP1 immunohistochemistry (IHC) and p16 FISH, in order to
evaluate whether it could really improve the differential diagnosis
between benign and malignant mesothelial proliferations. In detail,
we performed p16 FISH and BAP1 IHC on the same series of
epithelioid MPM and benign pleural lesions, previously analyzed by
our system, and assessed the diagnostic performance of each
method.
Materials and methods
Patients
Pleural tissues from 54 patients, comprising 34
epithelioid MPM and 20 pleural MH were analyzed in this study. All
patients underwent surgical resection at the Unit of Thoracic
Surgery of the University Hospital of Pisa from January 2012 to
December 2015. This study was conducted retrospectively conforming
to the principles of the Helsinki Declaration of 1975. Clinical
information, including patient sex and age, is reported in Table I.
 | Table I.Clinicopathological characteristics
of patients. |
Table I.
Clinicopathological characteristics
of patients.
| A, Epithelioid
malignant pleural mesothelioma |
|---|
|
|---|
| Clinicopathological
characteristics | n. cases (%) |
|---|
| Age (N=34) |
|
|
Range | 40–85 years |
|
Median | 68.5 years |
| Sex (N=34) |
|
|
Male | 24 (70.6) |
|
Female | 10 (29.4) |
| Type of specimen
(N=34) |
|
|
Pleurectomy/decortication | 28 (82.4) |
| Pleural
biopsy | 6 (17.6) |
|
| B, Mesothelial
hyperplasia |
|
|
Clinicopathological
characteristics | n. cases
(%) |
|
| Age (N=20) |
|
|
Range | 18–85 years |
|
Median | 51.5 years |
| Sex (N=20) |
|
|
Male | 15 (75.0) |
|
Female | 5 (25.0) |
| Type of specimen
(N=20) |
|
| Lung
atypical resection | 9 (45.0) |
| Pleural
biopsy | 11 (55.0) |
Among the 34 patients with epithelioid MPM, 28
(82.4%) had a pleurectomy/decortication, whereas the remaining 6
patients (17.6%) had video-toracoscopic pleural biopsy. Regarding
the 20 MH patients, the histological diagnosis of MH was an
incidental finding associated with bollous emphysema and pleural
inflammatory effusion.
All tumor samples were formalin-fixed and paraffin
embedded (FFPE) for microscopic examination. Histological diagnosis
and pathological features were reviewed by two pathologists (GA and
GF) according to the WHO 2015 histological and immunohistochemical
criteria (15). The most
representative paraffin block of each tumor was selected for BAP1
immunohistochemistry and p16 FISH analyses.
Gene expression analysis
Gene expression analysis was performed in our
previous study (14) using an
nCounter custom codeset including 117 MPM target genes and 6
housekeeping genes, synthesized by NanoString Technologies
(NanoString Technologies).
Briefly in the previous work, for each case RNA was
purified from four FFPE tissue sections using Qiagen RNeasy FFPE
kit (Qiagen) according to manufacturers' instructions. A total of
150 ng RNA was used for NanoString analysis, which was performed in
accordance to manufacturers' protocol (NanoString Technologies).
Then, for each sample the background noise was calculated on the
basis of 8 spike-in negative controls included in the panel.
Moreover, the raw NanoString counts of all genes underwent a
technical and biological normalization using the nSolver software
version 2.5 (NanoString Technologies). The technical normalization,
based on 6 spike-in positive controls included in the panel, allows
to check on technical variability. On the other hand, the
biological normalization, based on the housekeeping genes, allow to
correct for differences in RNA input. Only samples which passed
both the normalization steps were considered for further statistics
and bioinformatics analyses (14).
Immunohistochemistry
IHC was performed on 4 µm thick tissue sections that
were deparaffinized in xylene and rehydrated using a graded series
of ethanol solutions. Sections were then subjected to
immunohistochemical staining with a mouse monoclonal primary
anti-BAP1 antibody (clone C-4, Santa Cruz Biotechnology; 1: 100
dilution) using the UltraView DAB IHC Detection kit (Ventana
Medical System, Inc.). Immunostaining was performed as a fully
automated assay using BenchMark ULTRA automated slide stainer
(Ventana Medical System, Inc.). Counterstaining was performed with
hematoxylin. In all cases, the immunohistochemical evaluation was
performed independently by two pathologists (GA and GF) who were
blinded to the clinicopathological characteristics of the
patients.
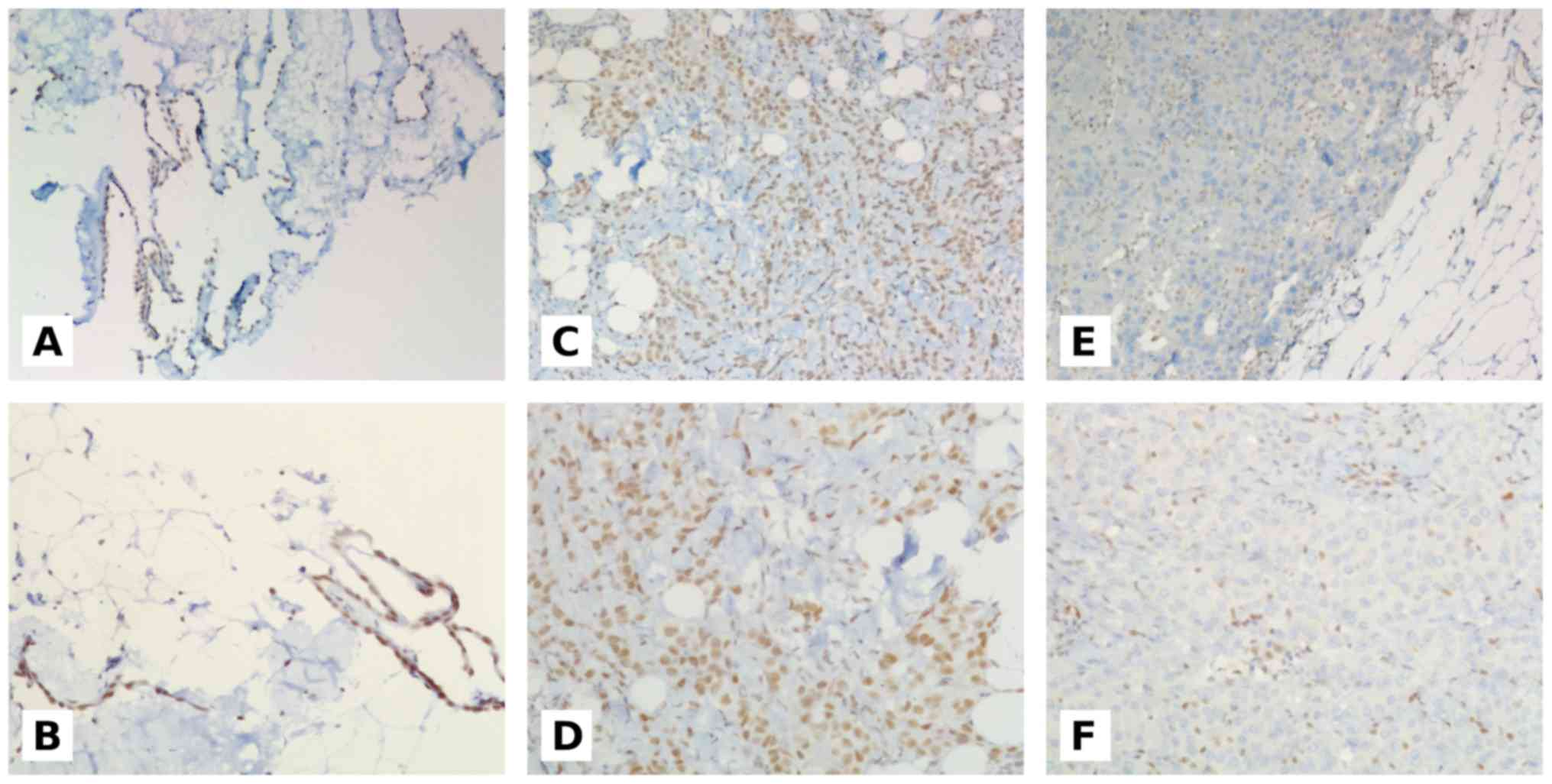
Only nuclear expression of BAP1 was considered for
evaluation and was scored as positive if there was unambiguous
presence of BAP1 expression in mesothelial nuclei without
percentage or intensity cutoff values (16–18). The
negative controls were carried out by omitting the primary
antibody. All the analyzed samples showed internal positive
controls represented by non-mesothelial BAP1-reactive cells such as
fibroblasts, lymphocytes, histiocytes, endothelial cells and
pneumocytes. Examples of BAP1 immunostaining are reported in
Fig. 1.
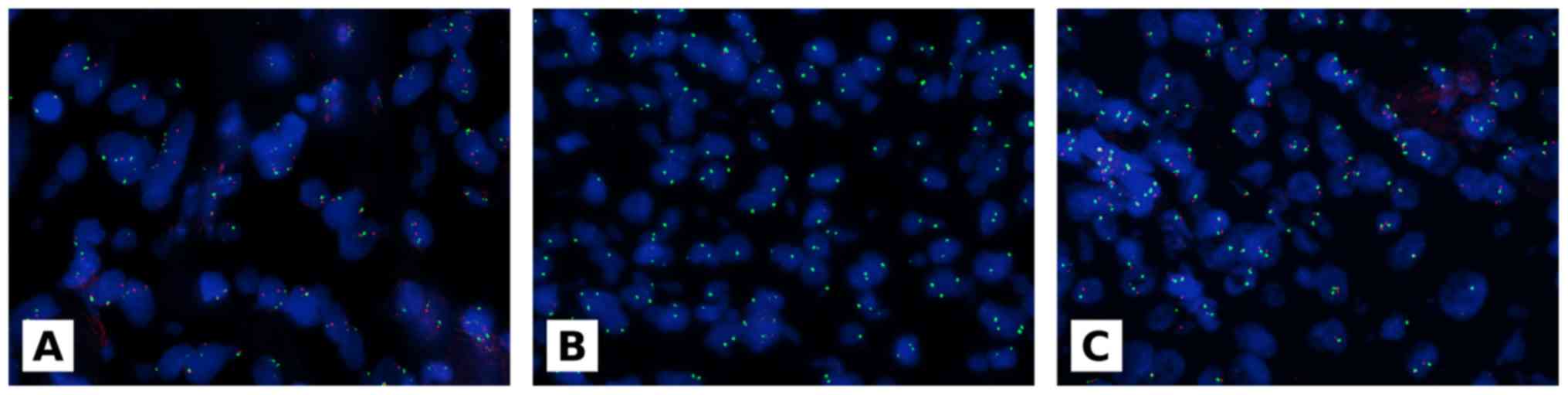
p16 fluorescence in situ
hybridization
p16 deletion was evaluated by FISH using the
Vysis LSI CDKN2A(p16) spectrum orange/CEP 9 spectrum green kit
(Abbott Molecular) according to the manufacturer's recommendations
and as previously described (19,20).
FISH was performed on 4 to 6 µm thick paraffin
sections of MPM and MH tissues. Before hybridization, paraffin
sections were deparaffinized in xylene (3 times, 10 min each),
dehydrated by two washing steps of 5 min each in 100% ethanol and
two washing steps of 5 min each in 96% ethanol, and air-dried at
room temperature. Tissue sections were then transferred to a
pretreatment solution at 80°C for 15 min, followed by a 3 min wash
in purified water, and incubated in a protease solution for 10 min
at 37°C to digest proteins. After a brief washing in purified
water, the slides were sequentially dehydrated in 70, 85, and 100%
alcohol and air-dried at room temperature. Tissue sections were
placed in a Hybrite (Abbott Molecular) for 3 min at 73°C to
denature DNA, and probe hybridization was carried out overnight at
37°C. Tissue sections were washed in 0.1% NP40/2× SSC at 76°C for 4
min and then washed in 0.1% NP40/2× SSC at room temperature for 1
min. Slides were mounted with 1.5 µg/ml
4′,6-diamidino-2-phenylindole. Tumor samples were scored by 2
independent investigators (GA and AP) who were blinded to the
clinicopathological characteristics of the patients and to the
immunohistochemical results. Normal cells present in the samples
negative for the deletion, such as lymphocytes, fibroblasts,
histiocytes, endothelial cells, and pneumocytes, were used as
internal positive controls. At least 60 non-overlapping and
well-delineated cells were scored for each case. Each specimen was
evaluated by the average and the maximum numbers of copies of the
p16 gene per cell and the average ratio of the gene to CEP 9
copy numbers. Homozygous deletion was defined by loss of both
p16 gene orange signals when more than 11% of tumor nuclei
showed at least 1 signal CEP 9 green signal (18). Examples of p16 FISH test are
reported in Fig. 2.
Statistical analyses
In our previous work, we have identified two
classification models (22-gene and 40-gene reported in Table SI), both able to properly classify
all the analyzed cases. In this study, the normalized expression
levels of the genes included in the two classifiers were selected
(14). A partial least square model
was used to classify samples with both the 22-gene and 40-gene
classifiers by the procedure of the caret R package version 6.0–78.
A bootstrap resampling (n=2000) was used to assess the area under
the curve (AUC). Also for BAP1, p16 and their combination,
AUC was calculated after bootstrap resampling (n=2000) by the
procedure of the pROC R package version 1.10.0. Positive predictive
value (PPV) and negative predictive value (NPV) were assessed for
BAP1, p16, the combination of BAP1 and p16 and both
gene-classifiers, using the prevalence of our series (0.6296). The
association between loss of BAP1 expression and p16 deletion
was tested by Fisher's exact test.
Results
BAP1 IHC and p16 FISH results
BAP1 nuclear expression was observed in all 20 MH
cases. Among the 34 MPM cases, 12 showed positive neoplastic cells
nuclei, whereas 22 lost BAP1 expression.
As regards p16, all 20 MH cases were negative
for p16 deletion. p16 homozygous deletion was
observed in 18 out of 34 MPM cases, whereas 16 out of 34 were
negative for the deletion. Furthermore, there was no association
between BAP1 loss and p16 deletion. Details are reported in
Tables II and III.
 | Table II.BAP1 IHC and p16 FISH
results. |
Table II.
BAP1 IHC and p16 FISH
results.
| Test | Mesothelial
hyperplasia N=20 (%) | Malignant pleural
mesothelioma N=34 (%) |
|---|
| BAP1 |
|
|
|
Positive | 20 (100) | 12 (35.3) |
|
Negative | 0 | 22 (64.7) |
| p16 |
|
|
|
Negative for deletion | 20 (100) | 16 (47.1) |
|
Positive for deletion | 0 | 18 (52.9) |
 | Table III.Association between BAP1 IHC and
p16 FISH. |
Table III.
Association between BAP1 IHC and
p16 FISH.
| Mesothelial
lesion | Only BAP1 loss
(%) | Only p16
deletion (%) | Both BAP1 loss and
p16 deletion (%) | Neither BAP1 loss
nor p16 deletion (%) |
|---|
| Mesothelial
hyperplasia (N=20) | 0 | 0 | 0 | 20 (100) |
| Malignant pleural
mesothelioma (N=34) | 8 (23.52) | 14 (41.17) | 4 (11.74) | 8 (23.52) |
Comparison among BAP1 IHC, p16 FISH,
and gene expression panel
The AUC for BAP1 and p16 was 0.8235 and
0.7647 respectively. Although the combination of BAP1 and
p16 produced a higher AUC than those obtained with a single
biomarker (0.8824), both gene-classifiers reached better AUC,
0.9996 and 0.9990 for the 22-gene and 40-gene classifier
respectively. Sensitivity, specificity, PPV and NPV of BAP1,
p16 and gene-classifiers in discriminating MH and MPM are
summarized in Table IV.
 | Table IV.Performance of BAP1, p16
(alone and in combination) and gene classifiers. |
Table IV.
Performance of BAP1, p16
(alone and in combination) and gene classifiers.
| Test | Sensitivity (95%
CI) | Specificity (95%
CI) | AUC (95%
CI) | PPV | NPV |
|---|
| BAP1 | 0.6471
(0.4706–0.7941) | 1 | 0.8235
(0.7353–0.8971) | 1 | 0.6250 |
| p16 | 0.5294
(0.3529–0.7059) | 1 | 0.7647
(0.6765–0.8529) | 1 | 0.5556 |
| BAP1 and
p16 | 0.7647
(0.6176–0.8824) | 1 | 0.8824
(0.8088–0.9412) | 1 | 0.7144 |
| 22 genes | 0.9784
(0.9018–1) | 0.9987
(0.9682–1) | 0.9996
(0.9945–1) | 0.9992 | 0.9645 |
| 40 genes | 0.9701
(0.8817–1) | 0.9957
(0.9338–1) | 0.9990
(0.9894–1) | 0.9974 | 0.9515 |
Discussion
Differential diagnosis between epithelioid MPM and
reactive MH is one of the most challenging diagnostic issues. To
date, the best criterion to ascertain the malignancy of pleural
lesions is the presence of stromal or lung invasion, which is not
always easy to evaluate (3,15,21). So
the analyses of p16 gene deletion and BAP1 loss of
expression are recommended (8–13).
Overall, BAP1 and p16 examinations do not allow the
detection of all MPM cases, even combining the two assays, since
they are altered only in a proportion of mesotheliomas (8). In a previous study we identified two
classification models based on the expression profile of 22 and 40
genes specifically deregulated in MPM, which perfectly worked in
discriminating epithelioid MPM from benign lesions (14).
In the present study, we compared the performance of
our gene classifiers with BAP1 and p16 testing. We observed
that both BAP1 loss and p16 deletion were highly specific
for MPM, since they were never detected in benign lesions. However,
their AUC values were not completely satisfying (BAP1: 0.8235;
p16: 0.7647) particularly due to their low sensitivities, in
fact 8 MPM cases (23.5%) were negative for p16 deletion as
well as positive for BAP1 expression. As expected, combining BAP1
and p16 tests increased the diagnostic sensitivity, thus
improving the AUC (0.8824). All these results were in agreement
with other previously published studies (12,13,16,22,23).
Furthermore, we confirmed that there was no association between
BAP1 loss and p16 deletion (13,18,24).
In our series, both the 22- and 40-gene expression
classifiers outperformed BAP1 and p16 tests (AUC 22-gene
model: 0.9996; AUC 40-gene model: 0.9990).
BAP1 and p16 are undoubtedly valuable MPM
biomarkers, but, as confirmed in this study, a multi-marker
approach seemed to better overcome the great heterogeneity of this
tumor (7).
Our MPM tool requires a low input of starting
material comparable or even less than the one necessary for BAP1
and p16 evaluation. Moreover, our system allows to obtain a
faster analysis and an easier interpretation of results. In fact,
IHC and FISH tests require a tissue section for each marker and can
be influenced by several pre-analytical factors. Moreover, the
interpretation of FISH test can be quite challenging because it
requires highly skilled staff.
On the other hand, the MPM tool is highly
reproducible, and almost completely automatized (14,25). It
could also be even more informative, due to the inclusion of genes
with a crucial role in cancer development, and progression
(14); some of which also correlate
with MPM prognosis and are potential therapeutic targets (26,27).
Our gene expression classifiers proved a great
potential as a diagnostic tool. The encouraging results on
histological specimens suggest that a prospective validation is
warranted to concretely evaluate the use of the 117 gene panel in
the clinical context.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
GA, GF and RB contributed to study design, data
collection and interpretation. AMP contributed to statistical and
data analysis. GA, GF, AP and SR contributed to histological
revision of cases, immunohistochemistry and FISH analysis. AC, FM,
MCA, ML defined the study population, designed the study, drafted
and critical revised the manuscript. All authors discussed the
results and contributed to the final manuscript.
Ethics approval and consent to
participate
This study was retrospectively conducted in
accordance to the principles of the Helsinki Declaration of 1975
and was approved by Comitato Etico di Area Vasta Nord-Ovest per la
Sperimentazione Clinica. Only archival and anonymous samples were
included, no protected health information was used and informed
consents were obtained from patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Scherpereel A, Astoul P, Baas P, Berghmans
T, Clayson H, de Vuyst P, Dienemann H, Galateau-Salle F, Hennequin
C, Hillerdal G, et al: Guidelines of the European respiratory
society and the European society of thoracic surgeons for the
management of malignant pleural mesothelioma. Eur Respir J.
35:479–495. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sekido Y: Molecular pathogenesis of
malignant mesothelioma. Carcinogenesis. 34:1413–1419. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Husain AN, Colby TV, Ordóñez NG, Allen TC,
Attanoos RL, Beasley MB, Butnor KJ, Chirieac LR, Churg AM, Dacic S,
et al: Guidelines for pathologic diagnosis of malignant
mesothelioma 2017 update of the consensus statement from the
international mesothelioma interest group. Arch Pathol Lab Med.
142:89–108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alì G, Bruno R and Fontanini G: The
pathological and molecular diagnosis of malignant pleural
mesothelioma: A literature review. J Thorac Dis. 10 (Suppl
2):S276–S284. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galateau-Salle F, Churg A, Roggli V and
Travis WD; World Health Organization Committee for Tumors of the
Pleura, : The 2015 World Health Organization Classification of
Tumors of the Pleura: Advances since the 2004 classification. J
Thorac Oncol. 11:142–154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hjerpe A, Ascoli V, Bedrossian C, Boon M,
Creaney J, Davidson B, Dejmek A, Dobra K, Fassina A, Field A, et
al: Guidelines for cytopathologic diagnosis of epithelioid and
mixed type malignant mesothelioma. Complementary statement from the
International Mesothelioma Interest Group, also endorsed by the
International Academy of Cytology and the Papanicolaou Society of
Cytopathology. Cytojournal. 12:262015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bruno R, Alì G and Fontanini G: Molecular
markers and new diagnostic methods to differentiate malignant from
benign mesothelial pleural proliferations: A literature review. J
Thorac Dis. 10 (Suppl 2):S342–S352. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Churg A, Sheffield BS and Galateau-Salle
F: New markers for separating benign from malignant mesothelial
proliferations: Are we there yet? Arch Pathol Lab Med. 140:318–321.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ladanyi M: Implications of P16/CDKN2A
deletion in pleural mesotheliomas. Lung Cancer. 49 (Suppl
1):S95–S98. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Illei PB, Ladanyi M, Rusch VW and Zakowski
MF: The use of CDKN2A deletion as a diagnostic marker for malignant
mesothelioma in body cavity effusions. Cancer. 99:51–56. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dacic S, Kothmaier H, Land S, Shuai Y,
Halbwedl I, Morbini P, Murer B, Comin C, Galateau-Salle F, Demirag
F, et al: Prognostic significance of p16/cdkn2a loss in pleural
malignant mesotheliomas. Virchows Arch. 453:627–635. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chung CT, Santos GD, Hwang DM, Ludkovski
O, Pintilie M, Squire JA and Tsao MS: FISH assay development for
the detection of p16/CDKN2A deletion in malignant pleural
mesothelioma. J Clin Pathol. 63:630–634. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sheffield BS, Hwang HC, Lee AF, Thompson
K, Rodriguez S, Tse CH, Gown AM and Churg A: BAP1
immunohistochemistry and p16 FISH to separate benign from malignant
mesothelial proliferations. Am J Surg Pathol. 39:977–982. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bruno R, Alì G, Giannini R, Proietti A,
Lucchi M, Chella A, Melfi F, Mussi A and Fontanini G: Malignant
pleural mesothelioma and mesothelial hyperplasia: A new molecular
tool for the differential diagnosis. Oncotarget. 8:2758–2770. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Travis WD, Brambilla E, Burke AP, Marx A
and Nicholson AG: Introduction to the 2015 World Health
Organization Classification of tumors of the lung, pleura, thymus,
and heart. J Thorac Oncol. 10:1240–1242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cigognetti M, Lonardi S, Fisogni S,
Balzarini P, Pellegrini V, Tironi A, Bercich L, Bugatti M, Rossi G,
Murer B, et al: BAP1 (BRCA1-associated protein 1) is a highly
specific marker for differentiating mesothelioma from reactive
mesothelial proliferations. Mod Pathol. 28:1043–1057. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McGregor SM, Dunning R, Hyjek E,
Vigneswaran W, Husain AN and Krausz T: BAP1 facilitates diagnostic
objectivity, classification, and prognostication in malignant
pleural mesothelioma. Hum Pathol. 46:1670–1678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McGregor SM, McElherne J, Minor A,
Keller-Ramey J, Dunning R, Husain AN, Vigneswaran W, Fitzpatrick C
and Krausz T: BAP1 immunohistochemistry has limited prognostic
utility as a complement of CDKN2A (p16) fluorescence in situ
hybridization in malignant pleural mesothelioma. Hum Pathol.
60:86–94. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiosea S, Krasinskas A, Cagle PT,
Mitchell KA, Zander DS and Dacic S: Diagnostic importance of 9p21
homozygous deletion in malignant mesotheliomas. Mod Pathol.
21:742–747. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Monaco SE, Shuai Y, Bansal M, Krasinskas
AM and Dacic S: The diagnostic utility of p16 FISH and GLUT-1
immunohistochemical analysis in mesothelial proliferations. Am J
Clin Pathol. 135:619–627. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Churg A, Colby TV, Cagle P, Corson J,
Gibbs AR, Gilks B, Grimes M, Hammar S, Roggli V and Travis WD: The
separation of benign and malignant mesothelial proliferations. Am J
Surg Pathol. 24:1183–1200. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Husain AN: Mesothelial proliferations:
Useful marker is not the same as a diagnostic one. Am J Clin
Pathol. 141:152–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nasu M, Emi M, Pastorino S, Tanji M,
Powers A, Luk H, Baumann F, Zhang YA, Gazdar A, Kanodia S, et al:
High Incidence of Somatic BAP1 alterations in sporadic malignant
mesothelioma. J Thorac Oncol. 10:565–576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hwang HC, Sheffield BS, Rodriguez S,
Thompson K, Tse CH, Gown AM and Churg A: Utility of BAP1
immunohistochemistry and p16 (CDKN2A) FISH in the diagnosis of
malignant mesothelioma in effusion cytology specimens. Am J Surg
Pathol. 40:120–126. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsang HF, Xue VW, Koh SP, Chiu YM, Ng LP
and Wong SC: NanoString, a novel digital color-coded barcode
technology: Current and future applications in molecular
diagnostics. Expert Rev Mol Diagn. 17:95–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Linton A, Cheng YY, Griggs K, Schedlich L,
Kirschner MB, Gattani S, Srikaran S, Chuan-Hao Kao S, McCaughan BC,
Klebe S, et al: An RNAi-based screen reveals PLK1, CDK1 and NDC80
as potential therapeutic targets in malignant pleural mesothelioma.
Br J Cancer. 110:510–519. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kato T, Lee D, Wu L, Patel P, Young AJ,
Wada H, Hu HP, Ujiie H, Kaji M, Kano S, et al: SORORIN and PLK1 as
potential therapeutic targets in malignant pleural mesothelioma.
Int J Oncol. 49:2411–2420. 2016. View Article : Google Scholar : PubMed/NCBI
|