Introduction
Breast cancer is one of the most common types of
tumors among women and the main cause of cancer-related deaths in
women worldwide (1). Some studies
have revealed that the overall morbidity of breast cancer among
Asian women is on the increase (2),
and although the breast cancer treatment methods have improved, the
death toll from breast cancer has not decreased due to the aging
population (3). Due to the high
recurrence rate of tumors and the drug resistance to chemotherapy,
the efficacy of breast cancer treatments and prognosis of breast
cancer have not achieved satisfactory results (4), making breast cancer a hot topic in
clinical research. At present, a number of reports (5–7) have
proven that microRNAs (miRNAS) are involved in the development of
various tumors. miR-221 and miR-489 have been verified to have
abnormal expression (8–10) in colorectal cancer and gastric
cancer; however, miR-221 and miR-489 expression levels in breast
cancer have been rarely reported. Therefore, in the present study
it was speculated that miR-221 and miR-489 have also abnormal
expression in breast cancer, and experimental analysis was carried
out for verification. The aim of the present study was to
investigate whether miR-221 and miR-489 can be used as diagnostic
indicators and disease prediction indices in breast cancer, and to
provide a relevant reference basis and suggestions for the future
clinical treatment of breast cancer.
Subjects and methods
Clinical data
Sixty-two breast cancer patients admitted to the
First Teaching Hospital of Tianjin University of Traditional
Chinese Medicine (Tianjin, China) for tumor surgery, from July 2014
to January 2016, were selected as the research group (RG), and 27
female adults who underwent physical examination during the same
period were selected as the control group (CG). The average age of
all subjects was 48.12±10.36 years. The study was approved by the
Ethics Committee of the First Teaching Hospital of Tianjin
University of Traditional Chinese Medicine (Tianjin, China). All
subjects who participated in this research had complete clinical
data. Signed written informed consents were obtained from the
participants and/or their guardians.
Inclusion and exclusion criteria
All patients in the RG were diagnosed with breast
cancer after biopsy in the Pathology Department of our hospital.
The exclusion criteria were as follows: i) Patients with incomplete
pathological data; ii) patients with other tumors; iii) patients
who had received relevant treatment; iv) patients with other
visceral diseases; v) patients with drug allergy history; vi)
patient referrals.
Methods
Sixty-two breast cancer patients were selected as
the RG and 27 healthy women as the CG. Blood samples were collected
from both groups. miR-221 expression (kit purchased from Guangzhou
Angfeibio Biotech Co., Ltd.; UTS19324) and miR-489 (kit purchased
from Shanghai Yihui Biological Technology Co., Ltd.; HHM0387) were
detected by fluorescence reverse transcription-quantitative PCR
(RT-qPCR). Total RNA was extracted using TRIzol® kit
(Shanghai Mingjing Biology Co., Ltd.; 5003050) according to the
manufacturer's protocol. The extracted total RNA was reverse
transcribed into cDNA according to the manufacturer's instructions
of a reverse transcription kit (Wuhan Chundu Biotech Co., Ltd.;
CD-102539GM). Reverse transcription reaction conditions were 50°C
for 45 min, and the reverse transcriptase was inactivated at 85°C
for 5 min. cDNA was amplified using an amplification kit (Shanghai
Xinyu Biotechnology Co., Ltd.; XY-051021). SYBR Green I (KS26757)
was purchased from Shanghai Keshun Biological Technology Co., Ltd.
The primer sequences used were synthesized by Thermo Fisher
Scientific, Inc. Details are shown in Table I. The following thermocycling
conditions were used for qPCR: Pre-denaturation at 94°C for 30 sec,
denaturation at 94°C for 5 sec, annealing and extension at 60°C for
30 sec for 40 cycles. The experiment was conducted 3 times. U6 was
used as internal reference and 2−∆Cq method was used for
quantification (11).
 | Table I.Primer sequences of miR-221, miR-489
and U6 internal reference gene. |
Table I.
Primer sequences of miR-221, miR-489
and U6 internal reference gene.
| Genes | Upstream primers | Downstream
primers |
|---|
| miR-221 |
5′-GGGAAGCTACTAAGTCTGC-3′ |
5′-GTGCGTGTCGTGGAGTCG-3′ |
| miR-489 |
5′-ACACTCCAGCTGGGGTGACATCACATA-3′ |
5′-TGGTGTCGTGGAGTCG-3′ |
| U6 |
5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
Observation indicators
The expression levels of miR-221 and miR-489 were
observed in the two groups, as well as the diagnostic value of
miR-221 and miR-489 in breast cancer. The patients in the RG were
followed up for 3 years. According to the median values of miR-221
and miR-489 expression levels in the RG, the patients were divided
into miR-221 high- and low-expression groups and into miR-489 high-
and low-expression groups, respectively. The 3-year survival rates
of the high- and low-expression groups were recorded and
compared.
Statistical analysis
SPSS 22.0 software (IBM Corp.) was used to
statistically analyze the experimental data and generate the
relevant figures. The measurement data were expressed as the mean ±
standard deviation. The comparison of the measurement data between
two groups was conducted using t-test. Counting data were expressed
as percentage [n (%)], and the comparison of counting data between
groups was performed by Chi-square (χ2) test. The
diagnostic value was analyzed by ROC curve analysis and the
survival rates were calculated by Kaplan-Meier method. Log-rank
test was used for the comparison of the survival curves. P<0.05
was considered to indicate a statistically significant
difference.
Results
Comparison of clinicopathological
characteristics
There was no significant difference in age, weight,
family history of breast cancer, marital status, reproductive
history, smoking or drinking between the RG and CG (P>0.05),
proving that the two groups were comparable. Details are presented
in Table II.
 | Table II.Comparison of clinicopathological
characteristics between the two groups. |
Table II.
Comparison of clinicopathological
characteristics between the two groups.
| Clinicopathological
characteristics | Research group
(n=62) | Control group
(n=27) | t or
χ2 | P-value |
|---|
| Age (years) | 41.29±7.35 | 39.71±6.82 | 0.952 | 0.344 |
| Weight (kg) | 60.47±7.56 | 57.42±6.98 | 1.790 | 0.077 |
| Family history of
breast cancer |
|
| 1.208 | 0.272 |
| Yes | 21 (33.87) | 6
(22.22) |
|
|
| No | 41 (66.13) | 21 (77.78) |
|
|
| Marital status |
|
| 1.394 | 0.498 |
|
Unmarried | 21 (33.87) | 9
(33.33) |
|
|
|
Married | 38 (61.29) | 18 (66.67) |
|
|
|
Widowed | 3
(4.84) | 0
(00.00) |
|
|
| Childbearing
history |
|
| 0.048 | 0.826 |
| Having
children | 36 (58.06) | 15 (55.56) |
|
|
| Not
having children | 26 (41.94) | 12 (44.44) |
|
|
| Smoking |
|
| 0.185 | 0.667 |
| Yes | 14 (22.58) | 5
(18.52) |
|
|
| No | 48 (77.42) | 22 (81.48) |
|
|
| Drinking |
|
| 0.203 | 0.653 |
| Yes | 19 (30.65) | 7
(25.93) |
|
|
| No | 43 (69.35) | 20 (74.07) |
|
|
| Type of
carcinoma |
|
Infiltrating carcinoma | 49 (79.03) |
|
|
|
|
Non-infiltrating
carcinoma | 13 (20.97) |
|
|
|
| Degree of
differentiation |
|
Moderately and poorly
differentiated | 50 (80.65) |
|
|
|
| Highly
differentiated | 12 (19.35) |
|
|
|
| Pathological
staging |
| Stages
I–II | 27 (43.55) |
|
|
|
| Stages
III–IV | 35 (56.45) |
|
|
|
| Distant
metastasis |
|
Yes | 25 (40.32) |
|
|
|
| No | 37 (59.68) |
|
|
|
Comparison of miR-221 and miR-489
expression levels
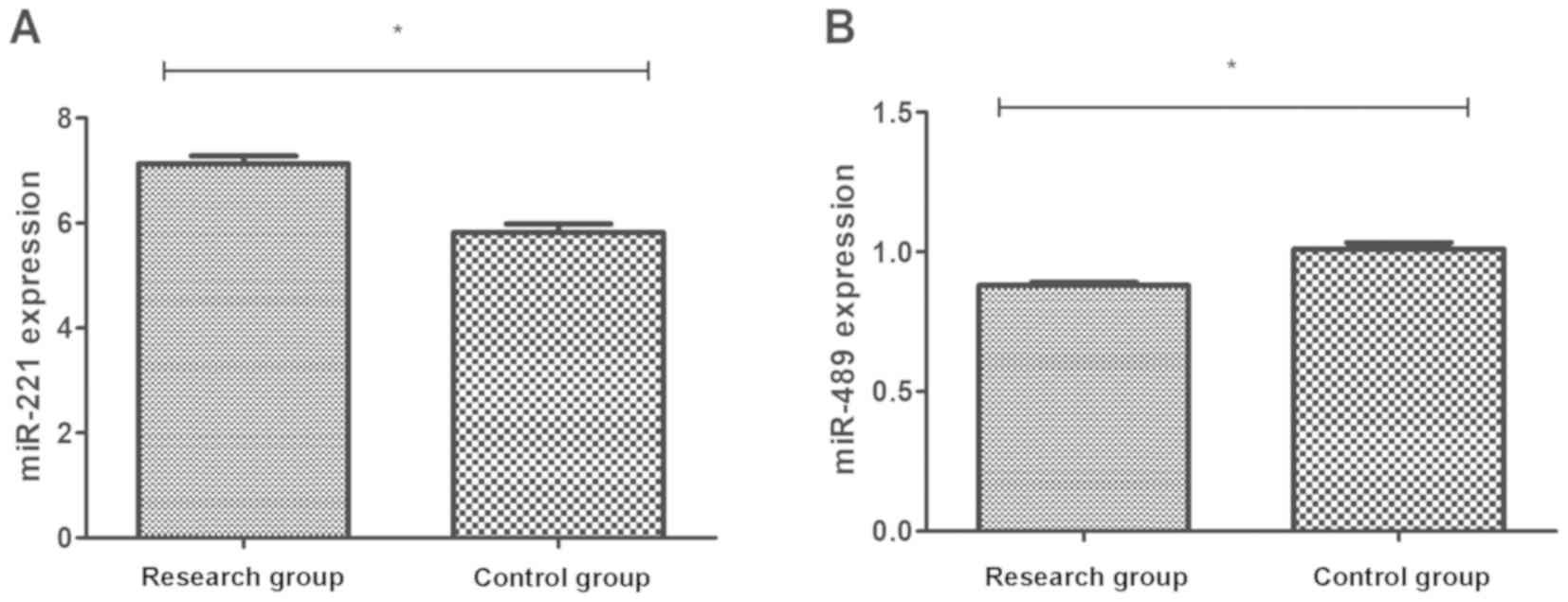
Blood samples were collected from the RG and CG, and
the expression levels of miR-221 and miR-489 were detected. The
results revealed that the expression level of miR-221 in the RG was
7.13±1.19, significantly higher than that in the CG (5.82±0.84)
(P<0.01), whereas, the expression level of miR-489 in the RG was
0.88±0.09, significantly lower than that in the CG (1.01±0.12)
(P<0.01) (Fig. 1).
Diagnostic value of miR-221 and
miR-489 expression levels in breast cancer
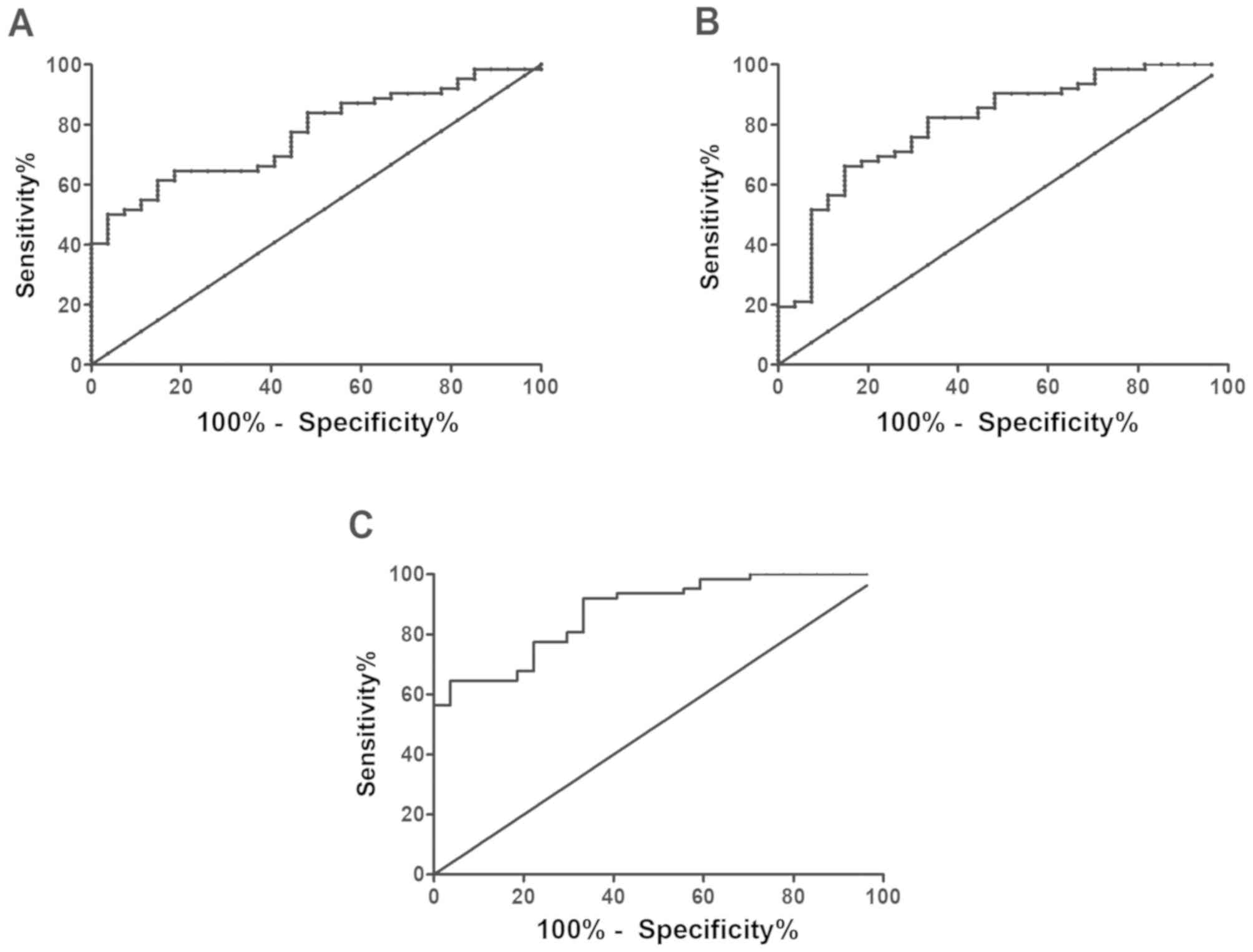
ROC curve analysis revealed that the area under the
miR-221 curve was 0.769, the sensitivity was 61.29%, the
specificity was 85.19%, the cut-off value was 6.806, and the 95% CI
was 66.27–95.81%. The area under the miR-489 curve was 0.805, the
sensitivity was 66.13%, the specificity was 85.19%, the cut-off
value was 0.903, and the 95% CI was 66.27–95.81%. In addition, the
area under the curve of the combined detection of the two was 0.88,
the sensitivity was 64.52%, the specificity was 96.3%, the cut-off
value was 0.165, and the 95% CI was 81.03–99.91% (Fig. 2).
Prognosis and survival of
patients
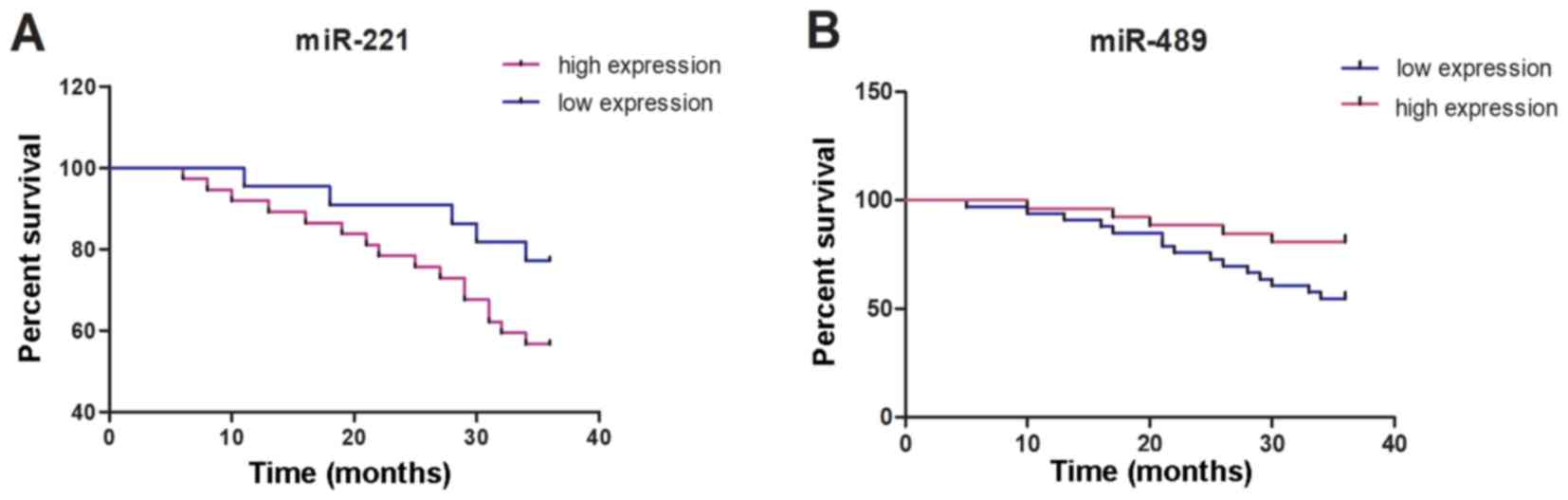
According to the median values of the expression
levels of miR-221 and miR-489, the patients in the RG were divided
into 37 cases of high miR-221 expression (≥7.13), 22 cases of low
miR-221 expression (<7.13), 24 cases of high miR-489 expression
(≥0.88) and 35 cases of low miR-489 expression (<0.88). By March
2019, 59 patients of the RG were successfully followed up through
telephones, hospital re-examination and dropping-in follow-up, with
a follow-up success rate of 95.16%. The 1-, 2- and 3-year survival
rates in the miR-221 high-expression group were 91.89, 78.38 and
56.76%, respectively, whereas those in the miR-221 low-expression
group were 95.45, 90.91 and 81.82%, respectively, which were
significantly higher than those in the miR-221 high-expression
group (P=0.049). The 1-, 2- and 3-year survival rates in the
miR-489 low-expression group were 93.94, 75.76 and 54.55%,
respectively, whereas those in the miR-489 high-expression group
were 96.15, 88.46 and 80.77%, respectively, which were
significantly higher than those in the miR-489 low-expression group
(P=0.035) (Fig. 3).
Relationship between the miR-221 and
miR-489 expression levels and the clinicopathological
characteristics of the patient in the RG
The expression levels of miR-221 and miR-489 in the
RG were related to age, family history of breast cancer, type of
carcinoma, degree of differentiation, pathological staging and
distant metastasis (P<0.05). Details are presented in Tables III and IV.
 | Table III.Relationship between miR-221
expression level and the clinicopathological characteristics of
patients in the RG. |
Table III.
Relationship between miR-221
expression level and the clinicopathological characteristics of
patients in the RG.
| Clinicopathological
characteristics | n | Expression
level | t | P-value |
|---|
| Age (years) |
|
| 2.119 | 0.038 |
|
≥40 | 28 | 7.33±2.12 |
|
|
|
<40 | 34 | 6.41±1.26 |
|
|
| Family history of
breast cancer |
|
| 2.379 | 0.021 |
|
Yes | 21 | 7.62±2.47 |
|
|
| No | 41 | 6.53±1.15 |
|
|
| Type of
carcinoma |
|
| 2.163 | 0.035 |
|
Infiltrating carcinoma | 49 | 7.59±1.31 |
|
|
|
Non-infiltrating
carcinoma | 13 | 6.73±1.12 |
|
|
| Degree of
differentiation |
|
| 2.284 | 0.026 |
|
Moderately and poorly
differentiated | 50 | 7.63±1.25 |
|
|
| Highly
differentiated | 12 | 6.72±1.19 |
|
|
| Pathological
staging |
|
| 2.056 | 0.044 |
| Stages
I–II | 27 | 7.09±1.26 |
|
|
| Stages
III–IV | 35 | 7.88±1.66 |
|
|
| Distant
metastasis |
|
| 2.066 | 0.043 |
|
Yes | 25 | 7.89±1.86 |
|
|
| No | 37 | 7.02±1.45 |
|
|
 | Table IV.Relationship between miR-489
expression level and the clinicopathological characteristics of
patients in the RG. |
Table IV.
Relationship between miR-489
expression level and the clinicopathological characteristics of
patients in the RG.
| Clinicopathological
characteristics | n | Expression
level | t | P-value |
|---|
| Age (years) |
|
| 2.186 | 0.033 |
|
≥40 | 28 | 0.82±0.06 |
|
|
|
<40 | 34 | 0.86±0.08 |
|
|
| Family history of
breast cancer |
|
| 2.236 | 0.029 |
|
Yes | 21 | 0.80±0.05 |
|
|
| No | 41 | 0.83±0.05 |
|
|
| Type of
carcinoma |
|
| 2.458 | 0.017 |
|
Infiltrating carcinoma | 49 | 0.78±0.05 |
|
|
|
Non-infiltrating
carcinoma | 13 | 0.82±0.06 |
|
|
| Degree of
differentiation |
|
| 4.362 | <0.001 |
|
Moderately and poorly
differentiated | 50 | 0.78±0.05 |
|
|
| Highly
differentiated | 12 | 0.83±0.04 |
|
|
| Pathological
staging |
|
| 2.240 | 0.029 |
| Stages
I–II | 27 | 0.86±0.06 |
|
|
| Stages
III–IV | 35 | 0.83±0.04 |
|
|
| Distant
metastasis |
|
| 2.278 | 0.026 |
|
Yes | 25 | 0.86±0.07 |
|
|
| No | 37 | 0.91±0.06 |
|
|
Discussion
Breast cancer is one of the most common malignant
tumors among women worldwide. Drug resistance and distant organ
metastasis are the main causes of most breast cancer-related deaths
(12). In recent years, the
mortality of breast cancer has decreased due to the promotion of
disease screening; however, the mortality rate remains high
(13,14). Early breast cancer is usually treated
by adjuvant systemic therapy, including chemotherapy, radiotherapy
and endocrine therapy (15).
However, due to the toxic effects of these treatment methods, the
benefits for the breast cancer patients are not obvious (16). At present, as the pathogenesis of
breast cancer has not been completely elucidated and there is no
effective reference index for the diagnosis and prognosis,
biological indicators and new targets for the diagnosis and
treatment of breast cancer have become the research focus in
clinical practice (17). In recent
years, a number of studies have been reported on miRNA as a new
indicator for cancer diagnosis and treatment (18–20).
miRNA is a small non-coding region of 20–22 nucleotide RNA. Under
normal circumstances, miRNAs play a role in feedback mechanisms by
protecting proliferation, differentiation, apoptosis and other
processes of cells, and are key links in a number of biological
processes (21,22). Previous studies have proven that
miRNAs can act as oncogenes or tumor suppressor genes according to
the functions of their target genes (23). Among them, some studies have
confirmed that miR-221 stimulates different types of tumors and
downregulates some tumor suppressor genes. Upregulation of miR-221
is linked to the occurrence of various hematological diseases and
solid malignant tumors (24).
However, miR-489 can play a part in proliferation, survival and
invasion of cells by regulating genes (25). With the deepening of research, it is
gradually considered that miR-221 and miR-489 may be closely
related to the occurrence and development of breast cancer
(26,27).
The results of the present study revealed that the
expression level of miR-221 in patients of the RG was significantly
higher than that in the CG, and the expression level of miR-489 in
the RG was significantly lower than that in the CG, suggesting that
miR-221 and miR-489 might participate in the occurrence and
development of breast cancer, in agreement with the research
results of Patel et al, Pan et al and Ye et al
(28–30). ROC curve analysis showed that when
the cut-off value was 6.806, the sensitivity and specificity of
miR-221 in the diagnosis of breast cancer were 61.29 and 85.19%,
respectively; when the cut-off value was 0.903, the sensitivity and
specificity of miR-489 were 66.13 and 85.19%, respectively; and
when the cut-off value was 0.165, the combined detection of the two
had higher specificity than the single detection of miR-221 and
miR-489. The relationship between the expression levels of miR-221
and miR-489 and the clinicopathological characteristics of breast
cancer patients was further analyzed. miR-221 and miR-489 were
significantly associated with age, family history of breast cancer,
pathological type of carsinoma, degree of differentiation,
pathological staging and distant metastasis, suggesting that
miR-221 and miR-489 are closely associated to differentiation and
proliferation of breast cancer cells. The prognosis of the miR-221
and miR-489 high- and low-expression groups were compared for 3
years, respectively. The survival rate of the miR-221
high-expression group was significantly lower than that of the
miR-221 low-expression group, and the survival rate of the miR-489
low-expression group was significantly lower than that of the
miR-489 high-expression group, suggesting that miR-221 and miR-489
are relevant to the prognosis of breast cancer patients. The aim of
improving prognosis could be achieved by detecting the expression
levels of miR-221 and miR-489 in breast cancer patients.
At present, biopsy is still the only definite way to
diagnose breast cancer, although it causes great damage to the
body. Therefore, a convenient and effective biomarker is urgently
needed in clinic for the diagnosis and treatment of breast cancer
diseases. As miRNA can be detected from serum, plasma, body fluids
and tissues, it has become a hot spot in the research of tumor
diseases. In the present study, the expression levels of miR-221
and miR-489 in the blood of breast cancer patients were analyzed,
and it was shown that miR-221 and miR-489 can be effective
indicators for the diagnosis and treatment of breast cancer
diseases. However, there are still some limitations in this study,
such as the small number of experimental subjects, the short
follow-up time, and the lack of basic experiments. The mechanism of
miR-221 and miR-489 on breast cancer is not yet clear. Further
research is needed to continuously improve our experiments in order
to obtain the best and most accurate experimental results.
In conclusion, miR-221 expression is upregulated and
miR-489 expression is downregulated in the blood of breast cancer
patients, which has a certain impact on prognosis. In the future,
miR-221 can be used as an effective indicator for the diagnosis,
treatment and prognosis of breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FL conceived and designed the study, performed the
experiments and the statistical analysis, and wrote the manuscript.
The author read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Teaching Hospital of Tianjin University of Traditional
Chinese Medicine (Tianjin, China). All subjects who participated in
this research had complete clinical data. Signed written informed
consents were obtained from the participants and/or their
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ghislain I, Zikos E, Coens C, Quinten C,
Balta V, Tryfonidis K, Piccart M, Zardavas D, Nagele E,
Bjelic-Radisic V, et al European Organisation for Research and
Treatment of Cancer (EORTC) Quality of Life Group and Breast Cancer
Group; EORTC Headquarters, : Health-related quality of life in
locally advanced and metastatic breast cancer: Methodological and
clinical issues in randomised controlled trials. Lancet Oncol.
17:e294–e304. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Ma J, Goding Sauer A, Newman
LA and Jemal A: Breast cancer statistics, 2017, racial disparity in
mortality by state. CA Cancer J Clin. 67:439–448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Malvezzi M, Carioli G, Bertuccio P,
Boffetta P, Levi F, La Vecchia C and Negri E: European cancer
mortality predictions for the year 2019 with focus on breast
cancer. Ann Oncol. 30:781–787. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gao C, Li H, Zhuang J, Zhang H, Wang K,
Yang J, Liu C, Liu L, Zhou C and Sun C: The construction and
analysis of ceRNA networks in invasive breast cancer: A study based
on The Cancer Genome Atlas. Cancer Manag Res. 11:1–11. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng X, Müller V, Milde-Langosch K,
Trillsch F, Pantel K and Schwarzenbach H: Diagnostic and prognostic
relevance of circulating exosomal miR-373, miR-200a, miR-200b and
miR-200c in patients with epithelial ovarian cancer. Oncotarget.
7:16923–16935. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu Y, Nangia-Makker P, Farhana L, G
Rajendra S, Levi E and Majumdar AP: miR-21 and miR-145 cooperation
in regulation of colon cancer stem cells. Mol Cancer. 14:982015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ying X, Wu Q, Wu X, Zhu Q and Wang X,
Jiang L, Chen X and Wang X: Epithelial ovarian cancer-secreted
exosomal miR-222-3p induces polarization of tumor-associated
macrophages. Oncotarget. 7:43076–43087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mukohyama J, Shimono Y, Dalerba P, Isobe
T, Hu Q, Sahoo D, Shibuya N, Minami H, Mimori K, Kakeji Y, et al:
Epigenetic regulation of colorectal cancer stem cells by the
miR-221/QKI5 axis. Cancer Res. 78:4882018.
|
|
9
|
Fang Z, Zhong M, Wang Y, Yuan X, Guo H,
Yao Y, Feng M, Chen J, Xiong J and Xiang X: miR-381 and miR-489
suppress cell proliferation and invasion by targeting CUL4B via the
Wnt/β-catenin pathway in gastric cancer. Int J Oncol. 54:733–743.
2019.PubMed/NCBI
|
|
10
|
Xu D, Liu R, Meng L, Zhang Y, Lu G and Ma
P: Long non-coding RNA ENST01108 promotes carcinogenesis of glioma
by acting as a molecular sponge to modulate miR-489. Biomed
Pharmacother. 100:20–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vergara-Ortega DN, Sevilla-Reyes EE,
Herrera-Ortiz A, Torres-Ibarra L, Salmerón J, Lazcano-Ponce E and
Sánchez- Alemán MA: Real time PCR to evaluate HSV-2 shedding from
anal and genital samples among men who have sex with men, living
with HIV. J Med Virol. 90:745–752. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Singh SK, Singh S, Lillard JW Jr and Singh
R: Drug delivery approaches for breast cancer. Int J Nanomedicine.
12:6205–6218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Welch HG, Prorok PC, O'Malley AJ and
Kramer BS: Breast-cancer tumor size, overdiagnosis, and mammography
screening effectiveness. N Engl J Med. 375:1438–1447. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Burwinkel B, Cuk K, Zucknick M and
Madhavan D: Circulating miRNAs as markers for breast cancer. US
Patent 10,316,367. Filed June 21, 2013; issued June 11, 2019.
|
|
15
|
Sparano JA, Gray RJ, Makower DF, Pritchard
KI, Albain KS, Hayes DF, Geyer CE Jr, Dees EC, Goetz MP, Olson JA
Jr, et al: Adjuvant chemotherapy guided by a 21-gene expression
assay in breast cancer. N Engl J Med. 379:111–121. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cardoso F, van't Veer LJ, Bogaerts J,
Slaets L, Viale G, Delaloge S, Pierga JY, Brain E, Causeret S,
DeLorenzi M, et al MINDACT Investigators, : 70-gene signature as an
aid to treatment decisions in early-stage breast cancer. N Engl J
Med. 375:717–729. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Malih S, Saidijam M and Malih N: A brief
review on long noncoding RNAs: A new paradigm in breast cancer
pathogenesis, diagnosis and therapy. Tumour Biol. 37:1479–1485.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shah MY, Ferrajoli A, Sood AK,
Lopez-Berestein G and Calin GA: MicroRNA therapeutics in cancer -
an emerging concept. EBioMedicine. 12:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Acunzo M, Romano G, Wernicke D and Croce
CM: MicroRNA and cancer - a brief overview. Adv Biol Regul. 57:1–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reddy KB: MicroRNA (miRNA) in cancer.
Cancer Cell Int. 15:382015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lou W, Liu J, Gao Y, Zhong G, Chen D, Shen
J, Bao C, Xu L, Pan J, Cheng J, et al: MicroRNAs in cancer
metastasis and angiogenesis. Oncotarget. 8:115787–115802. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Santolla MF, Lappano R, Cirillo F,
Rigiracciolo DC, Sebastiani A, Abonante S, Tassone P, Tagliaferri
P, Di Martino MT, Maggiolini M, et al: miR-221 stimulates breast
cancer cells and cancer-associated fibroblasts (CAFs) through
selective interference with the A20/c-Rel/CTGF signaling. J Exp
Clin Cancer Res. 37:942018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao S, Liu H, Hou S, Wu L, Yang Z, Shen J,
Zhou L, Zheng SS and Jiang B: miR-489 suppresses tumor growth and
invasion by targeting HDAC7 in colorectal cancer. Clin Transl
Oncol. 20:703–712. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Wang YW, Xing AY, Xiang S, Shi DB,
Liu L, Li YX and Gao P: Suppression of SPIN1-mediated PI3K-Akt
pathway by miR-489 increases chemosensitivity in breast cancer. J
Pathol. 239:459–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Roscigno G, Quintavalle C, Donnarumma E,
Puoti I, Diaz-Lagares A, Iaboni M, Fiore D, Russo V, Todaro M,
Romano G, et al: miR-221 promotes stemness of breast cancer cells
by targeting DNMT3b. Oncotarget. 7:580–592. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patel Y, Shah N, Lee JS, Markoutsa E, Jie
C, Liu S, Botbyl R, Reisman D, Xu P and Chen H: A novel
double-negative feedback loop between miR-489 and the
HER2-SHP2-MAPK signaling axis regulates breast cancer cell
proliferation and tumor growth. Oncotarget. 7:18295–18308. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pan Y, Li J, Zhang Y, Wang N, Liang H, Liu
Y, Zhang CY, Zen K and Gu H: Slug-upregulated miR-221 promotes
breast cancer progression through suppressing E-cadherin
expression. Sci Rep. 6:257982016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ye Z, Hao R, Cai Y, Wang X and Huang G:
Knockdown of miR-221 promotes the cisplatin-inducing apoptosis by
targeting the BIM-Bax/Bak axis in breast cancer. Tumour Biol.
37:4509–4515. 2016. View Article : Google Scholar : PubMed/NCBI
|