Introduction
Ovarian cancer is a kind of common malignant
gynecological tumor, whose mortality rate ranks 1st in all kinds of
gynecological tumors (1). Due to the
lack of typical clinical symptoms and specific clinical diagnostic
indexes in the early stage, missed diagnosis easily occur. More
than 80% of patients with ovarian cancer are in the middle-advanced
stage at the initial diagnosis. Despite of the therapeutic regimen
of cytoreductive surgery and postoperative chemotherapy currently,
the 5-year survival rate of patients with ovarian cancer is still
less than 40% (2). Therefore,
searching effective molecular targets has always been a research
hotspot in the treatment of ovarian cancer.
Osteopontin (OPN) is a phosphorylated glycoprotein
that exists as a potential biomarker in the protein matrix of all
body fluids, extracellular matrix (ECM) and mineralized tissues
(3). Some studies suggest that the
OPN levels may increase in tumor tissues and blood of patients, and
there is evidence that the increased OPN may be associated with the
reduced survival rate of patients (4,5). Although
the OPN expression has been widely studied, the role and exact
mechanism of OPN in the occurrence, development and metastasis of
ovarian cancer remain unclear.
In this study, the expression of OPN protein in
ovarian cancer tissues was detected via immunohistochemistry (IHC)
using clinical specimens, and the cytological study was further
conducted, so as to clarify the biological role of OPN in ovarian
cancer and investigate whether OPN can serve as a prognostic index
of patients with ovarian cancer, thus becoming a candidate target
for tumor treatment.
Materials and methods
Clinical data
A total of 81 patients with ovarian cancer without
any previous treatment and treated in the Obstetrics Department of
Shouguang People's Hospital (Weifang, China) from January 2010 to
March 2013 were collected, and they were aged 32–76 years with an
average of 46.8 years. In terms of the pathological type, there
were 35 cases of serous cystadenocarcinoma, 32 cases of mucous
cystadenocarcinoma, 11 cases of endometrioid carcinoma, and 3 cases
of other types. In terms of cytological grading, there were 18
cases of well-differentiated carcinoma, 26 cases of
moderately-differentiated carcinoma, and 37 cases of
poorly-differentiated carcinoma. According to the new pathological
staging criteria of the International Federation of Gynecology and
Obstetrics (FIGO) in 2012, there were 24 cases in stage I, 29 cases
in stage II, 16 cases in stage III, and 12 cases in stage IV.
Inclusion criteria: i) patients pathologically
diagnosed with epithelial ovarian cancer and receiving surgical
treatment, ii) patients without receiving any treatment before
operation, and iii) patients with complete clinical data. All
patients were followed up for 3 years.
This study was approved by the Ethics Committee of
Shouguang People's Hospital, and all patients enrolled in this
study signed the informed consent.
Detection of OPN protein expression
via IHC
Paraffin-embedded specimens of ovarian cancer
tissues in 81 patients were collected from the Pathology
Department, Shouguang People's Hospital, fixed with 40%
formaldehyde at 22°C for 12 h and then cut into 4 µm-thick
sections. Hematoxylin and eosin (H&E) were used for H&E
staining, followed by histopathological observation. Then
pathologically-confirmed ovarian cancer tissues and para-carcinoma
tissues (more than 4 cm) were taken and serially sliced into 4
µm-thick sections, followed by OPN IHC staining. In IHC, two-step
method and universal kit (model: ZLI9610; Beijing Zhongshan Golden
Bridge Biotechnology Co., Ltd., Beijing, China) were used. Tissues
were blocked with 8% milk at 4°C for 2 h. The primary goat
anti-mouse polyclonal antibody of OPN (dilution, 1:800; cat. no.
AF808) and the secondary Donkey anti-goat polyclonal antibody
(dilution, 1:2,000; cat. no. VC004) were purchased from R&D
Systems, Inc. (Minneapolis, MN, USA). The primary antibody was
replaced with phosphate buffered saline (PBS) as the negative
control. IHC results were interpreted independently by two
pathologists. Nuclei or cytoplasm stained dark brown indicated
positive IHC results. Five fields of view were selected randomly
under a high-power microscope (magnification, ×400; Olympus, Tokyo,
Japan), followed by scoring based on the proportion of positive
cells in each field of view and staining intensity: proportion of
positive cells <5% (0 point), 5–30% (1 point), 31–70% (2 points)
and >70% (3 points), and staining intensity: negative (0 point),
weak (1 point), moderate (2 points) and strong (3 points). The
above two scores were added up, and the final score >3 points
indicated positive expression.
Detection of OPN protein expression
via western blotting
After 200 µl mixed solution (1:10) containing
protease inhibitor cocktail and protein lysis buffer RIPA was added
into a 6-well plate, the cell lysis buffer was taken into an EP
tube and centrifuged at 12,000 × g and 4°C for 30 min. The protein
concentration was determined using the bicinchoninic acid (BCA)
protein assay kit (Beyotime, Guangzhou, China). A total of 10 µg
protein was added in each lane. After 40 µg total protein was
isolated via 20% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis, it was transferred onto a polyvinylidene fluoride
(PVDF) membrane. The membrane was blocked with 5% BSA at 20°C for
1.5 h. The primary mouse anti-human antibody of OPN (dilution,
1:2,000; cat. no. 1433-OP-050/CF; R&D Systems, Inc.) and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal
antibody (dilution, 1:1,000; cat. no. 9484; Abcam, Cambridge, UK)
were incubated at 4°C, followed by incubation with the
corresponding horseradish peroxidase-labeled secondary goat
anti-mouse polyclnonal antibody (dilution, 1:2,000; cat. no. A0216;
Beyotime, Shanghai, China) at room temperature for 1 h. Finally,
the membrane was visualized using the enhanced chemiluminescence
(ECL) detection system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) and the gray-scale analysis was performed using a gel
analyzer.
Lentivirus transfection
Cell culture
Human epithelial ovarian cancer cell lines (SKOV-3,
COC1, A2780, HO-8910 and OVCAR-3) were purchased from Bnbio (cat.
nos. ATCC HTB-77, BNCC101678, BNCC100884, BNCC100717 and
BNCC338624, Beijing, China). IOSE80 cell line was purchased from
Wuhan Cell Bank, Chinese Academy of Sciences (Wuhan, China).
Lipofectamine 2000 was from Invitrogen; Thermo Fisher Scientific,
Inc. (Waltham, MA, USA). Roswell Park Memorial Institute
(RPMI)-1640 culture solution and fetal bovine serum (FBS) were from
Gibco; Thermo Fisher Scientific, Inc. All cells were inoculated
into the RPMI-1640 medium containing 10% FBS and cultured in an
incubator with 5% CO2 at 37°C. Cells in the logarithmic
growth phase were taken for subsequent experiments.
Cell transfection
According to the manufacturer's instructions, short
hairpin ribonucleic acid (sh-RNA) solution was mixed with
Lipofectamine 2000 at a volume ratio of 1:1. Cells were divided
into sh-vector group (transfected with negative control sequence)
and sh-OPN group (transfected with targeted OPN sequence). One
targeted OPN sequence (sh-OPN) and one negative control sequence
(sh-vector) were designed and synthesized by Shanghai GenePharma
Co., Ltd. (Shanghai, China). Targeted OPN sequence (sh-OPN): sense
strand: 5′-GGGACUGGAAUACGCUAAUTT-3′, antisense strand:
5′-AUUAGCGUAUUCCAGUCCCTT-3′. Negative control (sh-vector) sequence:
sense strand: 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense strand:
5′-ACGUGACACGUUCGGAGAATT-3′. After successful transfection, cells
continued to be incubated in the incubator, and western blotting
was used to identify the interference effect after 48 h (not shown
here).
Detection of cell proliferation via
cell counting kit 8 (CCK8) assay
Cells in the logarithmic growth phase were taken,
inoculated into a 96-well plate (1×103/well) and
cultured, followed by CCK-8 assay at 6, 12, 24, 48 and 72 h. CCK-8
solution (100 µl) diluted with RPMI-1640 medium was added into each
well, followed by incubation in a dark place at 37°C for 2 h. Then
the optical density was read at 450 nm using a microplate reader
(Bio-Rad Laboratories, Inc.). Three repeated wells were set in each
group. The growth curve was drawn after measurement for 7
consecutive days.
Detection of apoptosis level via flow
cytometry
The apoptosis level in each group of cells was
detected using the apoptosis kit (Harlingen; BD Biosciences,
Franklin Lakes, NJ, USA). After cells were treated with different
extract liquids for 48 h, the cell culture fluid was taken and
retained, and then cells were centrifuged at 800 × g for 5 min,
washed with PBS, centrifuged again twice, and resuspended in 100 µl
1X binding buffer. Then 5 µl propidium iodide (PI) and 5 µl Annexin
V were added, followed by incubation in the dark at room
temperature for 15 min. Then the mixture was sent to the scientific
research center for on-machine detection within 1 h. Apoptotic rate
= early apoptotic rate + late apoptotic rate.
Statistical analysis
GraphPad Prism software (version 5.01, GraphPad
Software, Santiago, Chile) was used for analysis. Chi-square test
was used for the correlation of OPN expression with
clinicopathological data of patients. The disease-free survival
(DFS) and overall survival (OS) curves of patients in high/low OPN
expression groups were drawn using the Kaplan-Meier method. The
survival difference was compared between the two groups of patients
using the log-rank test, and the differences in indexes were
compared between the two groups via independent t-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
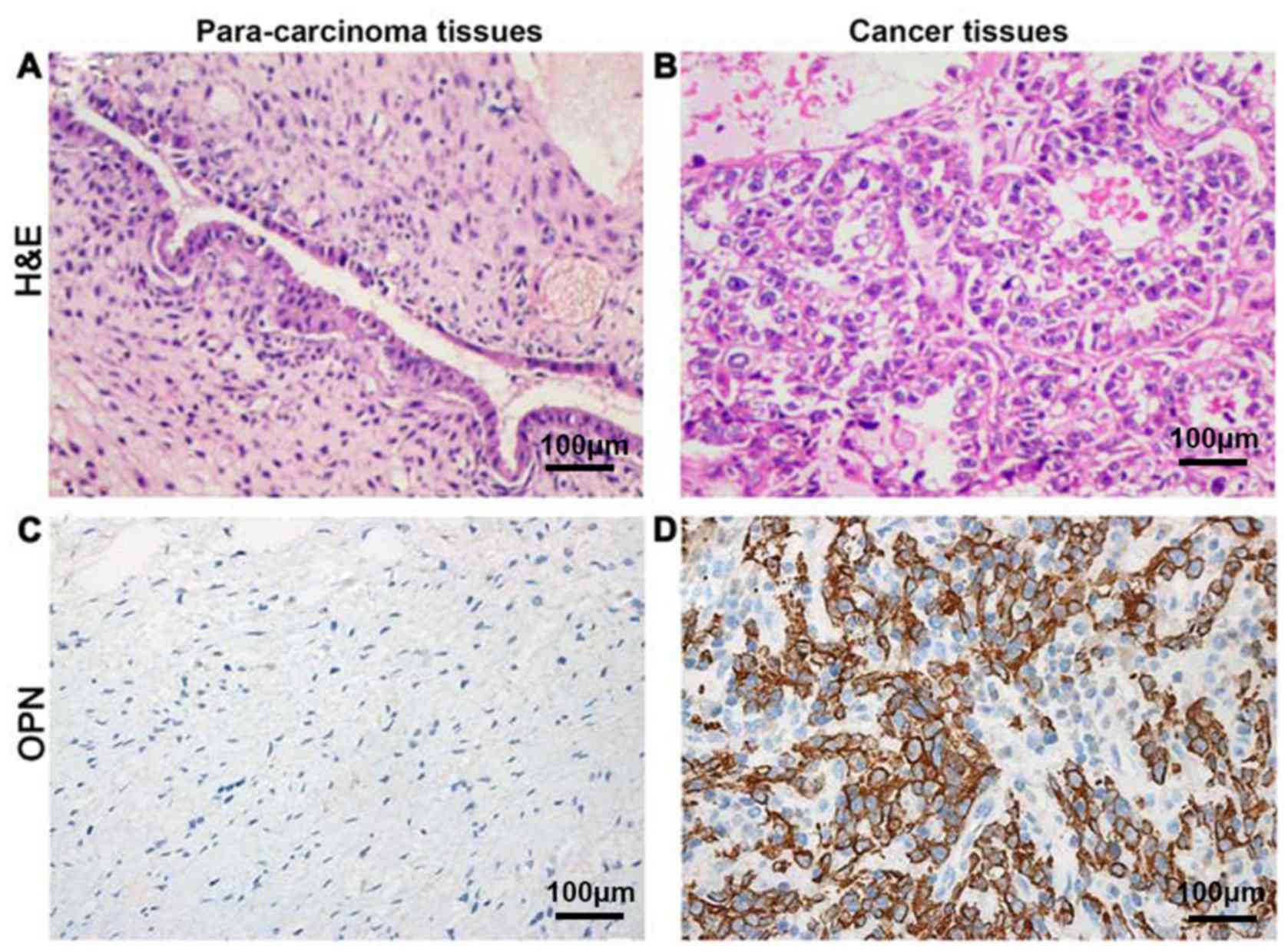
Detection of OPN expression levels in
cancer and para-carcinoma tissues via IHC
OPN was located in the cytoplasm
The positive expression rate of OPN protein in tumor
tissues was 77.8% (63/81), which was significantly higher than that
in para-carcinoma tissues 11.1% (9/81), showing a statistically
significant difference (P<0.05) (Fig.
1).
Correlation of OPN protein expression
in ovarian cancer tissues with clinicopathological factors of
patients
According to the IHC score, 63 patients were defined
as OPN positive and 18 patients as OPN negative. Results of
Chi-square test revealed that there were no significant differences
in the positive rate of OPN protein in ovarian cancer tissues among
patients with different age, sex and tumor progression (P>0.05).
Compared with that in patients with early ovarian cancer (stage
I+II), the positive expression rate of OPN in patients with
middle-advanced ovarian cancer (stage III+IV) was significantly
increased (89.3 vs. 71.7%, χ2=6.95, P=0.019). Besides,
the positive expression rates of OPN in ovarian cancer tissues in
well-differentiated group, moderately-differentiated group and
poorly-differentiated group were 66.7, 69.3 and 89.2%,
respectively. The positive expression rate of OPN was gradually
increased with the decreased degree of cell differentiation, and
differences were statistically significant (P<0.05). The
positive rate of OPN in metastatic ovarian cancer tissues was
obviously higher than that in non-metastatic ovarian cancer tissues
(83.9 vs. 74.0%, χ2=7.86, P=0.009) (Table I).
 | Table I.Correlation of OPN protein level in
ovarian cancer tissues with pathological indexes of patients. |
Table I.
Correlation of OPN protein level in
ovarian cancer tissues with pathological indexes of patients.
|
|
| OPN |
|
|
|---|
|
|
|
|
|
|
|---|
| Parameters | n=81 | High expression
(n=63) | Low expression
(n=18) | χ2
value | P-value |
|---|
| Sex |
|
|
| 1.74 | 0.271 |
| Male | 47 | 35 (74.5) | 12 (25.5) |
|
|
|
Female | 34 | 28 (82.3) | 6
(17.7) |
|
|
| Age (years) |
|
|
| 1.15 | 0.364 |
| ≤45 | 38 | 28 (73.7) | 10 (26.3) |
|
|
|
>45 | 43 | 35 (81.4) | 8
(18.6) |
|
|
| Pathological
type |
|
|
| 2.23 | 0.156 |
|
Serous | 20 | 11 (55.0) | 9
(45.0) |
|
|
|
Non-serous | 62 | 52 (83.9) | 10 (16.1) |
|
|
| FIGO staging |
|
|
| 6.95 | 0.019 |
| I+II | 53 | 38 (71.7) | 15 (28.3) |
|
|
|
III+IV | 28 | 25 (89.3) | 3
(10.7) |
|
|
| Cytological
differentiation |
|
|
| 6.43 | 0.025 |
| G1 | 18 | 12 (66.7) | 6
(33.3) |
|
|
| G2 | 26 | 18 (69.3) | 8
(30.7) |
|
|
| G3 | 37 | 33 (89.2) | 4
(10.8) |
|
|
| Tumor metastasis |
|
|
| 7.86 | 0.009 |
|
Negative | 50 | 37 (74.0) | 13 (26.0) |
|
|
|
Positive | 31 | 26 (83.9) | 5
(16.1) |
|
|
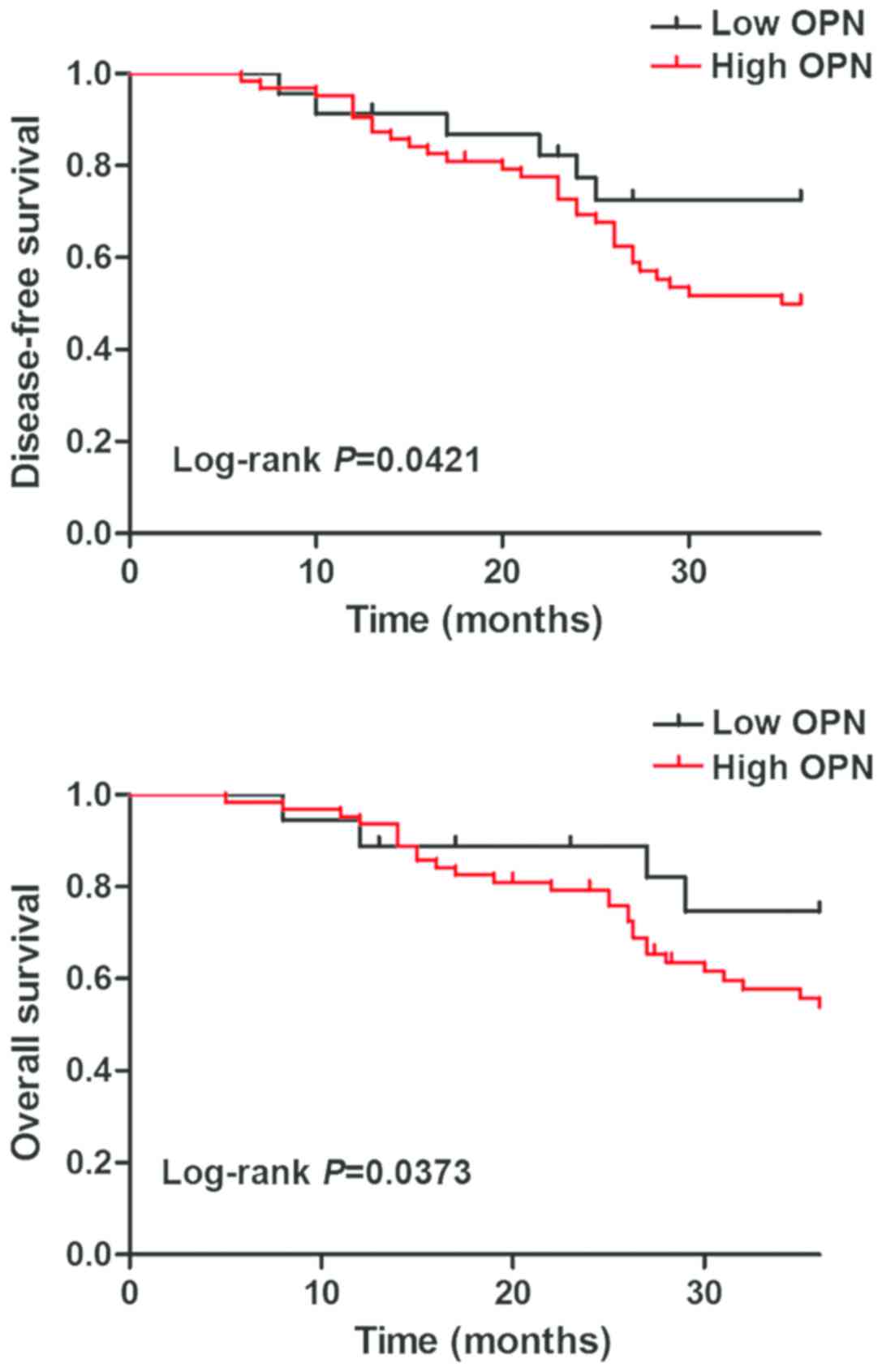
Correlation of OPN expression with
prognosis of patients with ovarian cancer
Survival analysis was performed using the
Kaplan-Meier method, and DFS and OS were compared via the log-rank
test. Both DFS and OS of patients in OPN negative group were
superior to those in OPN positive group, and there were
statistically significant differences (P<0.05) (Fig. 2).
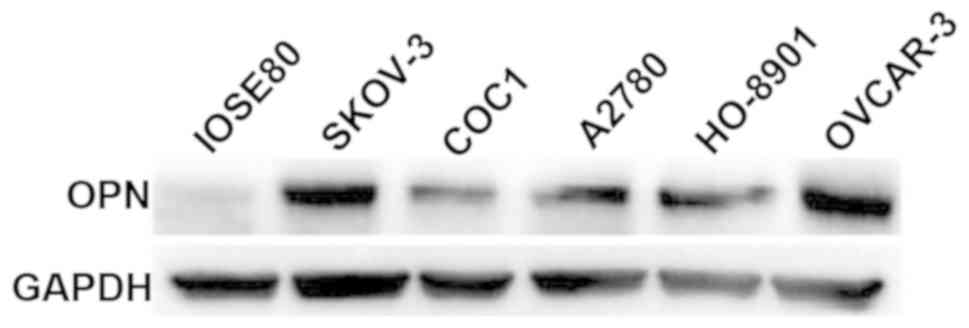
Detection of OPN expression levels in
normal ovarian epithelial and cancer cells via western
blotting
The expression levels of OPN in normal ovarian
epithelial IOSE80 cells and 5 ovarian cancer cell lines (SKOV-3,
COC1, A2780, HO-8910 and OVCAR-3) were detected via western
blotting. Results revealed that OPN was weakly expressed in IOSE80
cells, whereas it was highly expressed in 5 ovarian cancer cell
lines, among which the OPN protein expression levels were
relatively higher in SKOV-3 and OVCAR-3 cell lines (Fig. 3).
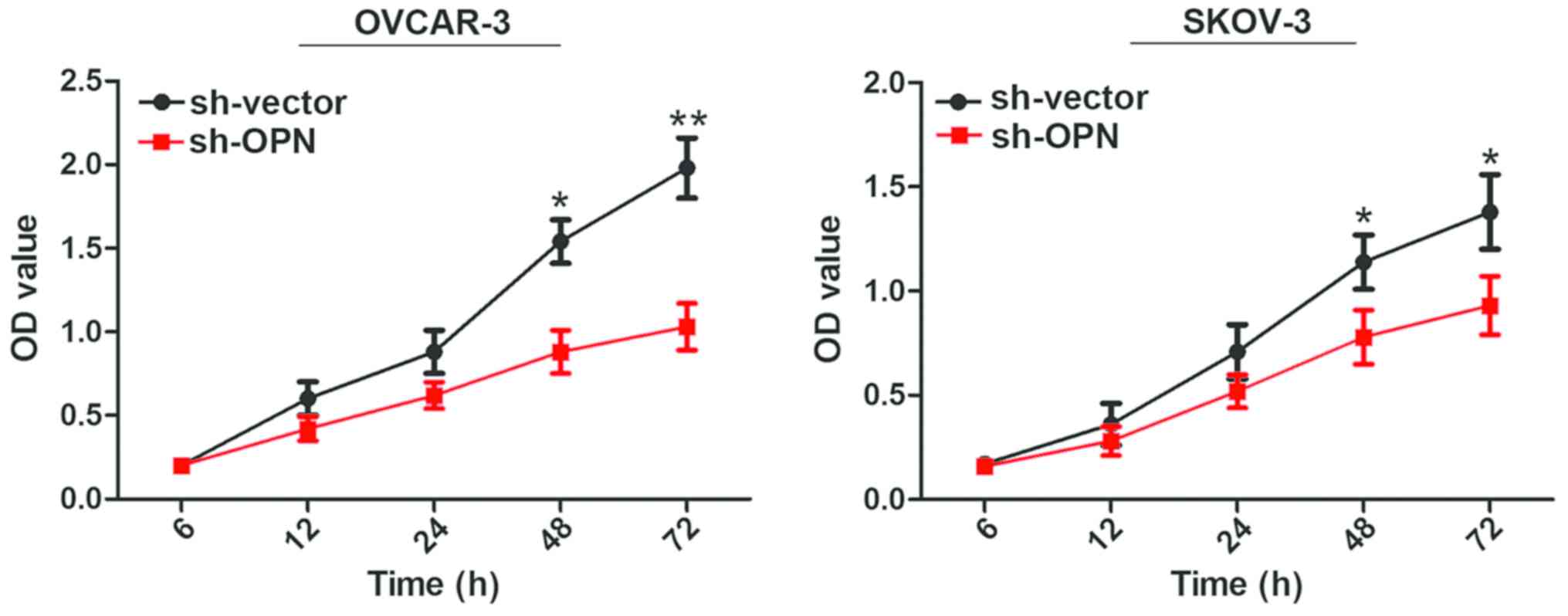
Effect of lentivirus interference in
OPN on cell proliferation capacity
Results of CCK8 assay demonstrated that the cell
proliferation rates of OVCAR-3 and SKOV-3 cells were decreased
remarkably from 48 h after OPN gene knockout with sh-OPN, and
differences were statistically significant (P<0.05) (Fig. 4).
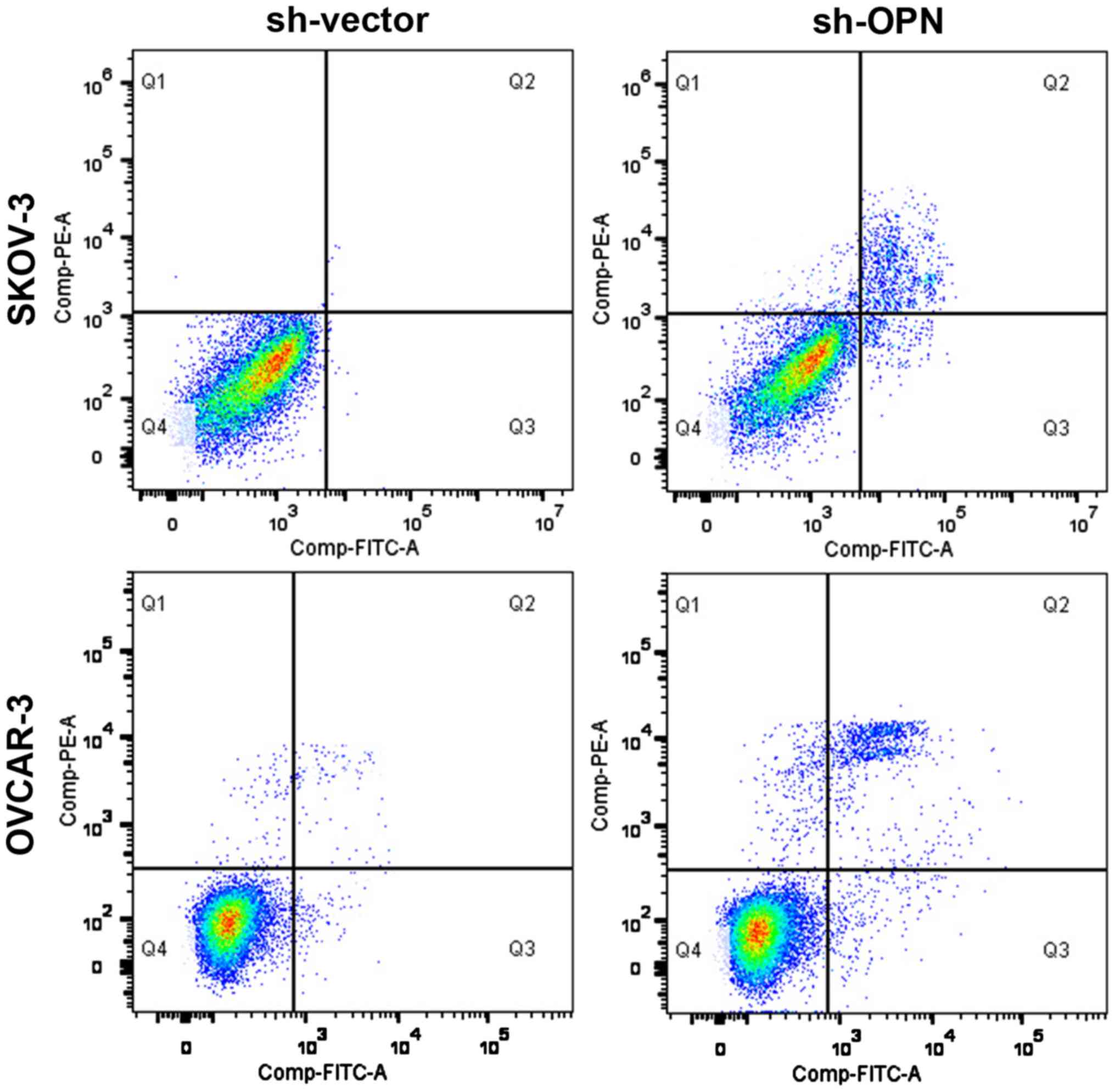
Effect of OPN knockout on apoptosis
level
After OPN gene knockout with sh-OPN, the apoptosis
levels of OVCAR-3 and SKOV-3 cells were obviously increased (28.2
vs. 1.3% and 25.3 vs. 3.2%), and there were statistically
significant differences (P<0.05) (Fig.
5).
Discussion
OPN protein was described for the first time as a
marker of epithelial cell transformation, and there has been
considerable interest in the role of OPN in human tumorigenesis
subsequently. OPN is both a marker of malignant tumors and a
candidate for detection as a prognostic factor (6). The OPN expression is detected via IHC in
tumor tissues of lung cancer, breast cancer, prostate cancer,
gastric cancer, esophagus cancer, glioma and other human cancers
(7,8).
In a study of 25 cases of lung tumor specimens, it was found that
both OPN mRNA and protein levels are elevated compared with those
in normal lung tissues, and the positive rate of OPN protein has
statistical significance in the survival of patients (9). In a recent study, tissue specimens of 68
patients with primary breast cancer, metastatic tumor and
fibroadenoma, and normal subjects were studied, and results
manifested that OPN mRNA and protein levels are increased in
malignant tissues compared with those in benign/normal tissues, and
the positive tumor cell immunity is associated with local
recurrence and metastasis of tumors formed in the liver and bone
(10). However, there is still a lack
of studies on the expression and biological effects of OPN protein
in ovarian cancer.
In this study, the expression levels of OPN protein
in 81 pairs of ovarian cancer tissues and para-carcinoma tissues
were detected via IHC. Based on the immunostaining score of 3
points, 63 cases were defined as OPN positive and 18 cases as OPN
negative. Results of Chi-square test revealed that there were no
significant differences in the positive rate of OPN protein in
ovarian cancer tissues among patients with different age, sex and
tumor progression (P>0.05). Compared with that in patients with
early ovarian cancer (stage I+II), the positive expression rate of
OPN in patients with middle-advanced ovarian cancer (stage III+IV)
was significantly increased (89.3 vs. 71.7%, χ2=6.95,
P=0.019). Besides, the positive expression rates of OPN in ovarian
cancer tissues in well-differentiated group,
moderately-differentiated group and poorly-differentiated group
were 66.7, 69.3 and 89.2%, respectively. The positive expression
rate of OPN was gradually increased with the decreased degree of
cell differentiation, and differences were statistically
significant (P<0.05). The positive rate of OPN in metastatic
ovarian cancer tissues was obviously higher than that in
non-metastatic ovarian cancer tissues (83.9 vs. 74.0%,
χ2=7.86, P=0.009). The above results are consistent with
the research results of Yeatman and Chambers on colon cancer
(11).
In addition, all the patients enrolled were followed
up for 3 years, and DFS and OS curves were drawn using Kaplan-Meier
method. Results manifested that there were negative correlations of
baseline OPN level with DFS and OS, and the increased OPN level had
a significant correlation with the mortality risk over time. Such a
result is similar to that of Donati et al who found that the
positive rate of OPN is significantly associated with the survival
of patients with lung cancer (12).
Moreover, in a study involving 163 patients with prostate cancer
who were followed up for at least 6 years, it was demonstrated that
the high expression level of OPN is an extremely adverse prognostic
factor for OS and DFS, and the expression of OPN is proved to be
remarkably associated with OS and DFS in univariate and
multivariate analyses (13).
OPN is expressed and secreted by a variety of cells
(osteoclasts, fibroblasts, macrophages and tumor cells), which
possess a variety of functions, and play an important role in cell
adhesion, chemotaxis, prevention of apoptosis, invasion, migration,
and anchorage-independent growth of tumor cells (14,15).
Previous studies mostly focused on the correlations of OPN with
invasion and metastasis of tumor cells. Studies have found that the
signal transduction after OPN activates the cell surface receptors
can increase the expression of proteolytic enzymes that can degrade
ECM proteins, especially matrix metalloproteinases (MMPs) and
urokinase plasminogen activator (UPA), thus enhancing invasiveness
(16,17). In addition, OPN selectively induces
CD44-dependent chemotaxis, the latter of which contributes to tumor
cell migration and invasion (18).
Recent studies have displayed that after treatment of colon cancer
cells with OPN, the cell proliferation capacity is significantly
enhanced, and OPN can protect suspension-cultured liver cancer
cells from stress-induced apoptosis (19). Gong et al added OPN into
gastric cancer cell lines to resist ultraviolet-induced apoptosis,
which has been proved to be mediated through binding to the αvβ3
integrin receptor, thereby, in turn, activating the transcription
factor nuclear factor κ-B (NFκ-B) to exert biological effects
(20).
In this study, the expression levels of OPN in
normal ovarian epithelial IOSE80 cells and 5 ovarian cancer cell
lines (SKOV-3, COC1, A2780, HO-8910 and OVCAR-3) were detected via
western blotting. Results revealed that the expression levels of
OPN in 5 ovarian cancer cell lines were obviously higher than that
in normal ovarian epithelial cells. Furthermore, the OPN genes in
SKOV-3 and OVCAR-3 cell lines with higher OPN protein expression
were knocked out using the lentivirus. Subsequently, CCK8 assay and
flow cytometry demonstrated that OPN knockdown significantly
inhibited the cell proliferation level, whose mechanism might be
mediated through inducing apoptosis.
In conclusion, OPN is highly expressed in ovarian
cancer tissues, and overexpressed OPN enhances ovarian cancer cell
proliferation, which is an adverse factor for the survival and
prognosis of patients with ovarian cancer. Targeting OPN has
potential significance in providing new treatment opportunity for
patients with ovarian cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HH wrote the manuscript and was responsible for cell
culture. ZL performed cell transfection. CL contributed to CCK8
assay. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Shouguang People's Hospital (Weifang, China) and informed consents
were signed by the patients or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Patch AM, Christie EL, Etemadmoghadam D,
Garsed DW, George J, Fereday S, Nones K, Cowin P, Alsop K, Bailey
PJ, et al: Australian Ovarian Cancer Study Group: Whole-genome
characterization of chemoresistant ovarian cancer. Nature.
521:489–494. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj
A, Burnell M, Kalsi JK, Amso NN, Apostolidou S, Benjamin E,
Cruickshank D, et al: Ovarian cancer screening and mortality in the
UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): A
randomised controlled trial. Lancet. 387:945–956. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Anborgh PH, Mutrie JC, Tuck AB and
Chambers AF: Pre- and post-translational regulation of osteopontin
in cancer. J Cell Commun Signal. 5:111–122. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ue T, Yokozaki H, Kitadai Y, Yamamoto S,
Yasui W, Ishikawa T and Tahara E: Co-expression of osteopontin and
CD44v9 in gastric cancer. Int J Cancer. 79:127–132. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Agrawal D, Chen T, Irby R, Quackenbush J,
Chambers AF, Szabo M, Cantor A, Coppola D and Yeatman TJ:
Osteopontin identified as lead marker of colon cancer progression,
using pooled sample expression profiling. J Natl Cancer Inst.
94:513–521. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weber GF: The metastasis gene osteopontin:
A candidate target for cancer therapy. Biochim Biophys Acta.
1552:61–85. 2001.PubMed/NCBI
|
|
7
|
Rudland PS, Platt-Higgins A, El-Tanani M,
De Silva Rudland S, Barraclough R, Winstanley JH, Howitt R and West
CR: Prognostic significance of the metastasis-associated protein
osteopontin in human breast cancer. Cancer Res. 62:3417–3427.
2002.PubMed/NCBI
|
|
8
|
Thalmann GN, Sikes RA, Devoll RE, Kiefer
JA, Markwalder R, Klima I, Farach-Carson CM, Studer UE and Chung
LW: Osteopontin: Possible role in prostate cancer progression. Clin
Cancer Res. 5:2271–2277. 1999.PubMed/NCBI
|
|
9
|
Chambers AF, Wilson SM, Kerkvliet N,
O'Malley FP, Harris JF and Casson AG: Osteopontin expression in
lung cancer. Lung Cancer. 15:311–323. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rodrigues LR, Teixeira JA, Schmitt FL,
Paulsson M and Lindmark-Mänsson H: The role of osteopontin in tumor
progression and metastasis in breast cancer. Cancer Epidemiol
Biomarkers Prev. 16:1087–1097. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yeatman TJ and Chambers AF: Osteopontin
and colon cancer progression. Clin Exp Metastasis. 20:85–90. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Donati V, Boldrini L, Dell'Omodarme M,
Prati MC, Faviana P, Camacci T, Lucchi M, Mussi A, Santoro M,
Basolo F, et al: Osteopontin expression and prognostic significance
in non-small cell lung cancer. Clin Cancer Res. 11:6459–6465. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khodavirdi AC, Song Z, Yang S, Zhong C,
Wang S, Wu H, Pritchard C, Nelson PS and Roy-Burman P: Increased
expression of osteopontin contributes to the progression of
prostate cancer. Cancer Res. 66:883–888. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He B, Mirza M and Weber GF: An osteopontin
splice variant induces anchorage independence in human breast
cancer cells. Oncogene. 25:2192–2202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Katagiri YU, Sleeman J, Fujii H, Herrlich
P, Hotta H, Tanaka K, Chikuma S, Yagita H, Okumura K, Murakami M,
et al: CD44 variants but not CD44s cooperate with beta1-containing
integrins to permit cells to bind to osteopontin independently of
arginine-glycine-aspartic acid, thereby stimulating cell motility
and chemotaxis. Cancer Res. 59:219–226. 1999.PubMed/NCBI
|
|
16
|
Liu H, Chen A, Guo F and Yuan L: A
short-hairpin RNA targeting osteopontin downregulates MMP-2 and
MMP-9 expressions in prostate cancer PC-3 cells. Cancer Lett.
295:27–37. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Buommino E, De Filippis A, Gaudiello F,
Balato A, Balato N, Tufano MA and Ayala F: Modification of
osteopontin and MMP-9 levels in patients with psoriasis on
anti-TNF-α therapy. Arch Dermatol Res. 304:481–485. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Desai B, Rogers MJ and Chellaiah MA:
Mechanisms of osteopontin and CD44 as metastatic principles in
prostate cancer cells. Mol Cancer. 6:182007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gotoh M, Sakamoto M, Kanetaka K, Chuuma M
and Hirohashi S: Overexpression of osteopontin in hepatocellular
carcinoma. Pathol Int. 52:19–24. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gong M, Lu Z, Fang G, Bi J and Xue X: A
small interfering RNA targeting osteopontin as gastric cancer
therapeutics. Cancer Lett. 272:148–159. 2008. View Article : Google Scholar : PubMed/NCBI
|