Introduction
One of the most common solid tumors is ovarian
carcinoma. In the United States, ovarian cancer was the fifth
leading cause of cancer-associated mortality worldwide in 2014
(1). As a result of limited screening
and delayed diagnosis, the majority of patients present with
advanced-stage disease at the time of diagnosis, with a 5-year
survival rate of ~20% (2). The poor
prognosis of ovarian cancer is mainly caused by tumor metastasis or
recurrence (3,4). Therefore, the development of effective
therapeutic strategies and targets for the control of ovarian
cancer metastasis is crucial.
MicroRNAs (miRs/miRNAs) consist of 21–23 endogenous
non-coding nucleotides that control gene expression by binding to
the 3′-untranslated region of their target genes. This binding
results in the suppression of messenger (m)RNA translation or the
degradation of mRNA. Emerging evidence has demonstrated that the
development and progression of ovarian cancer involve the
deregulation of various miRNAs, suggesting that miRNAs could be
novel diagnostic and therapeutic markers (2,5–7). For example, miR-205 has been reported to
induce cell invasion by repressing transcription factor 21 in human
ovarian cancer (8). In addition,
miR-212 serves a tumor-suppressing role in ovarian cancer cells by
inhibiting the expression of heparin-binding EGF-like growth factor
(9).
miR-16 was the first deregulated miRNA reported to
function as a tumor suppressor in a number of types of cancer,
including B-cell chronic lymphocytic leukemia and pituitary
adenomas (10,11). In addition, miR-16 was reported to be
downregulated in numerous tumor types, including chronic
lymphocytic leukemia, prostate cancer and pituitary adenomas
(12). The reduced expression of
miR-16 was also been reported in ovarian cancer tissues and in a
number ovarian cancer cell lines, including OV-202, CP70 and A2780
(13). miR-16 overexpression markedly
inhibits the proliferation and clonal growth of ovarian cancer
cells, indicating the involvement of miR-16 in the carcinogenesis
of ovarian cancer via the deregulation of proliferation (13). However, the role and underlying
mechanism of miR-16 in ovarian cancer cell migration and/or
invasion requires further investigation.
The objective of the present study was to
investigate the functions and underlying mechanisms of miR-16 in
ovarian cancer. Using SKOV3 and OVCAR3 cells as a model, the
effects of miR-16 on proliferation, migration and invasion were
studied, in addition to the epithelial-mesenchymal transition (EMT)
of ovarian cancer in vitro. The present study aimed to
provide novel insights to improve the treatment and prevention of
ovarian cancer.
Materials and methods
Cell lines
Human ovarian cancer cell lines, SKOV3 and OVCAR3,
were obtained from the Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China). Human ovarian epithelial
cells (OECs; cat. no. 7310) were purchased from ScienCell Research
Laboratories, Inc. (San Diego, CA, USA). The cell lines were
cultured in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) containing 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and 1% antibiotic-antimycotic
solution (100 U/ml penicillin and 100 µg/ml streptomycin). The OECs
were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% FBS. All cells were cultured at 37°C with 5%
CO2. SKOV3 and OVCAR3 cells were used in all the
following experiments.
Reagents
CCK-8 solution was obtained from Abcam (Cambridge,
UK). The following antibodies were purchased from Abcam: Rabbit
anti-matrix metallopeptidase (MMP)2, anti-MMP9, anti-cadherin 1
(E-CAD), anti-cadherin 2 (N-CAD), anti-MYC, anti-Cyclin D1,
anti-Vimentin, anti-Slug, anti-snail family transcriptional
repressor 1 (SNAIL), anti-twist family BHLH transcription factor 1
(TWIST), anti-GAPDH, anti-Wnt family member 3A (Wnt3a),
anti-glycogen synthase kinase 3 β (Gsk3β) and anti-β-catenin. Goat
anti-rabbit and anti-mouse antibodies labeled with horseradish
peroxidase were purchased from Santa Cruz Biotechnology, Inc.,
(Dallas, TX, USA). Transwell chambers were purchased from Corning,
Inc. (Corning, NY, USA).
miRNA synthesis, preparation and
storage
The synthetic miR-16 mimic and the mimic negative
control (NC) were purchased from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The synthetic samples were first centrifuged at
12,000 × g for 20 min at 25°C following dilution with 250 µl
diethyl-pyrocarbonate-treated water as the mother liquor per 5 nmol
miRNA. The samples were subpackaged and stored at −20°C until
further use. The miR sequences were as follows: miR-16 mimic,
5′-UAGCAGCACGUAAUAUUGGCGCCAAUAUUUACGUGCUGCUAUU-3′; and miR-16 mimic
NC, 5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′.
Transfection of cells with miRNAs
Cells were inoculated 1 day prior to transfection in
RPMI-1640 medium without penicillin-streptomycin solution in 6-well
plates, (2,000 µl per well) with an increase or decrease in the
proportion of cells per well. Upon reaching a confluence of 30–50%,
the cells were transfected. Opti-MEM (250 µl) was added to dilute
the miRNAs (50 nmol) and Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was subsequently
diluted separately in Opti-MEM (250 µl) by blending gently. This
mixture was incubated for 5 min at room temperature and gently
mixed with the diluted miRNAs prior to further incubation at room
temperature for 20–30 min. The miRNA/Lipofectamine 2000 compound
was added to the 6-well plates, which were subsequently gently
agitated. After 6 h, RPMI-1640 medium containing 10% FBS was added,
and the cells were cultivated at 37°C in a CO2 incubator
for 24–72 h.
Cell Counting Kit (CCK)-8 assay
A CCK-8 assay was used to assess proliferation.
Cells were seeded into 96-well plates (Corning, Inc.) and
transfected with either the miR-16 mimic or the NC as
aforementioned. CCK-8 (10 µg) solution (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was added into 100 µl
RPMI-1640 medium at 24, 48 and 72 h post-transfection. The cells
were diluted with CCK-8 in RPMI-1640 medium for 2 h, lysed in
radioimmunoprecipitation assay (RIPA) buffer and centrifuged at
13,000 × g for 10 min at 4°C. The absorbance of the supernatants
was measured at a wavelength of 450 nm on an iMark microplate
reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA samples
were qualified by using a UV spectrophotometer, and optical density
(OD)260/OD280 was limited to 1.8–2.0. Total
RNA was reverse transcribed to cDNA using a reverse transcription
system (cat. no. 4368814; Fermentas, Thermo Fisher Scientific,
Inc.). In brief, total RNA was incubated at 70°C for 10 min and
centrifuged at 1,000 × g for 20 min at room temperature. cDNA was
synthesized by Avian Myeloblastosis Virus Reverse Transcriptase
(Takara Bio, Inc., Otsu, Japan), according to the manufacturer's
protocols. RT-qPCR with SYBR Green qPCR dye (Toyobo Life Science,
Osaka, Japan) was conducted, according to the manufacturer's
protocol, to study the quantitative expression of miR-16. The
thermocycling conditions were as follows: 45°C for 5 min for
reverse transcription, followed by 94°C for 30 sec and then 40
cycles of 94°C for 5 sec and 60°C for 30 sec. Three replicates were
included in the RT-qPCR analysis. The relative expression levels of
miR-16 were normalized to β-actin and calculated using the
2−∆∆Cq method (14). The
sequences of the primers used are: miR-16,
5′-TAGCAGCACGTAAATATTGGCG-3′ (forward) and 5′-TGCGTGTCGTGGAGTC-3′
(reverse); and GAPDH, 5′-AGAAGGCTGGGGCTCATTTG-3′ (forward) and
5′-AGGGGCCATCCACAGTCTTC-3′ (reverse).
Scratch assay
Overall, ~1×105 cells were seeded into
12-well plates (Costar; Corning, Inc.) and incubated at 37°C until
reaching ≥90% confluency. The tip of a sterile 10-µl pipette was
used to create a scratch at time 0 h for each experiment. After 48
h, an inverted microscope was used to image the cells. The
migration distance was calculated as follows: Distance=(gap width
at 0 h-gap width at 48 h)/gap width at 0 h.
Invasion assay
For the invasion assays, 24-well Transwell chamber
inserts with an 8-µm pore size (BD Biosciences, Franklin Lakes, NJ,
USA) were used. In total, ~1×105 cells/well were
resuspended in 200 µl FBS-free medium and seeded on the upper
chamber, which featured a Matrigel-coated membrane. Additionally,
500 µl RPMI-1640 medium supplemented with 10% FBS was added to the
lower chamber, and the plates were incubated for 24 h at 37°C with
5% CO2. Finally, the membranes were stained with 0.1%
crystal violet for 10 min at room temperature. An inverted
microscope was used to count the number of cells on each membrane.
Each assay was repeated at least three times.
Western blot analysis
RIPA lysis buffer was applied to the protein
extracts from the SKOV3 and OVCAR3 cells, according to the
manufacturer's protocol. Bovine serum albumin was used as the
standard for determining the protein concentration by the Bradford
protein assay (Bio-Rad Laboratories, Inc.). The expression levels
of E-CAD, N-CAD, Vimentin, Slug, Twist, SNAIL, GAPDH, MMP2, MMP9,
Wnt3a, Gsk3β, β-catenin, MYC proto-oncogene, BHLH transcription
factor (c-MYC) and cyclin D1 were determined by western blot
analysis. Equal amounts (20 µg) of lysate were subjected to 10%
SDS-PAGE, and the proteins were subsequently transferred to a
polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA,
USA) using a semidry transfer method. The PVDF membrane was blocked
with 5% skimmed milk in Tween 20-BST buffer for 2 h at room
temperature. Following blocking, the membrane was incubated
overnight with the primary antibodies at 4°C followed by incubation
for 1 h with a suitable secondary antibody at room temperature. An
Enhanced Chemiluminescence Detection kit (Thermo Fisher Scientific,
Inc.) was used for the detection of proteins, according to the
manufacturer's protocols. Protein bands were subsequently detected
using a Bio-Rad imaging system (Bio-Rad Laboratories, Inc.), and
band intensity was quantified by densitometry analysis using
Image-Pro Plus 4.5 software (Media Cybernetics, Inc., Rockville,
MD, USA). The primary antibodies against MMP9 (cat. no. ab38898;
1:1,000), MMP2 (cat. no. ab37150; 1:1,000), GAPDH (cat. no. ab8245;
1:10,000), TWIST (cat. no. ab50581; 1:1,000), Vimentin (cat. no.
ab8978; 1:1,000), SNAIL (cat. no. ab53519; 1:1,000), Slug (cat. no.
ab27568; 1:1,000), E-CAD (cat. no. ab40772; 1:1,000), N-CAD (cat.
no. ab18203; 1:1,000), β-catenin (cat. no. ab16051; 1:1,000), Gsk3β
(cat. no. ab32391; 1:1,000), Wnt3a (cat. no. ab219412; 1:1,000),
c-MYC (cat. no. ab32072; 1:1,000) and cyclin D1 (cat. no. ab16663;
1:1,000) were purchased Abcam. Goat anti-rabbit (cat. no. sc-2004;
1:10,000) and anti-mouse antibodies (cat. no. sc-2005; 1:10,000)
labeled with horseradish peroxidase were purchased from Santa Cruz
Biotechnology, Inc.
Statistical analysis
The SPSS 17.0 software (SPSS, Inc., Chicago, IL,
USA) was used for the conduction of all statistical tests. The data
were compared by one-way analysis of variance followed by Dunnett's
multiple comparison test. All data are expressed as the mean ±
standard deviation. All the experiments were repeated in
triplicate. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of miR-16 in human ovarian
cancer cell lines
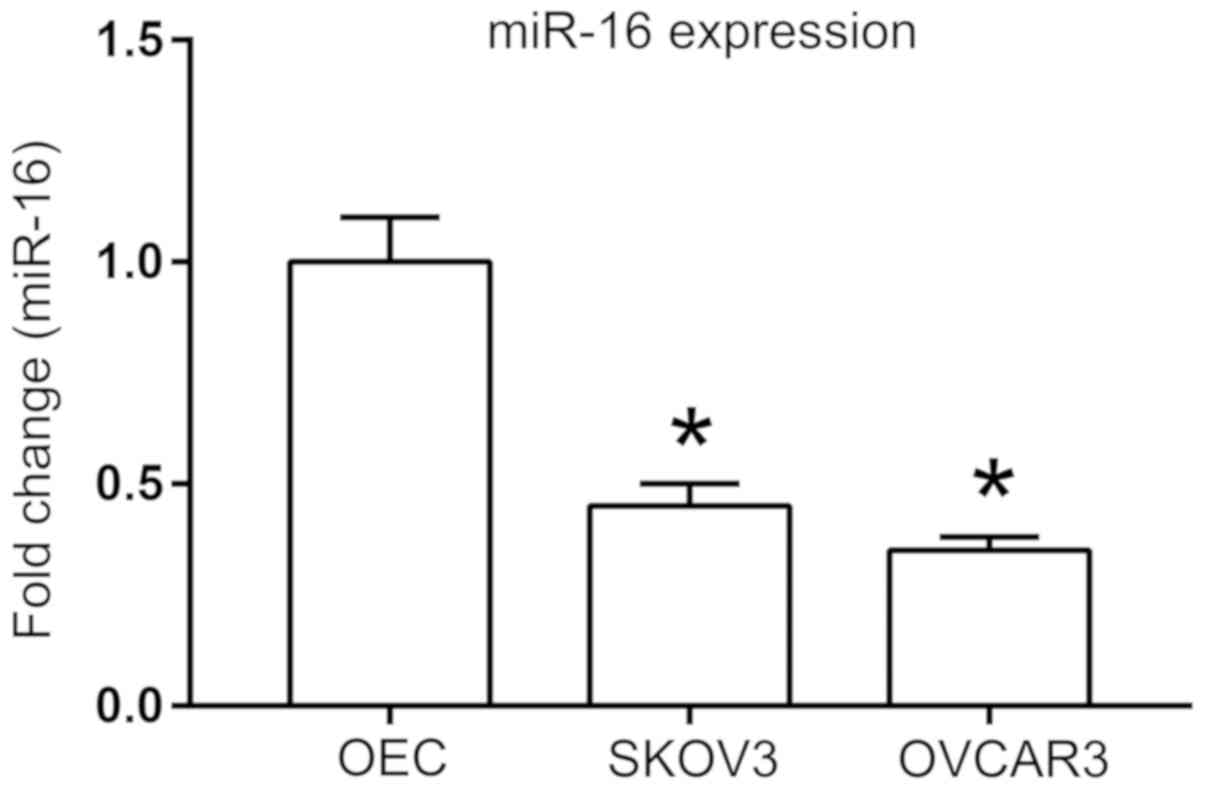
A previous study identified that miR-16 expression
is downregulated in a number of ovarian cancer cell lines,
including OV-167, CP-70, A2780 and OV-202 (13). To further investigate the role of
miR-16 in other ovarian cancer cell lines, SKOV3 and OVCAR3, the
expression of miR-16 was first examined with RT-qPCR. Significant
downregulation of miR-16 expression was observed in the SKOV3 and
OVCAR3 cells compared with that in the ovarian epithelial cells
(P<0.05; Fig. 1).
Effect of miR-16 on the proliferation
of human ovarian cancer cells
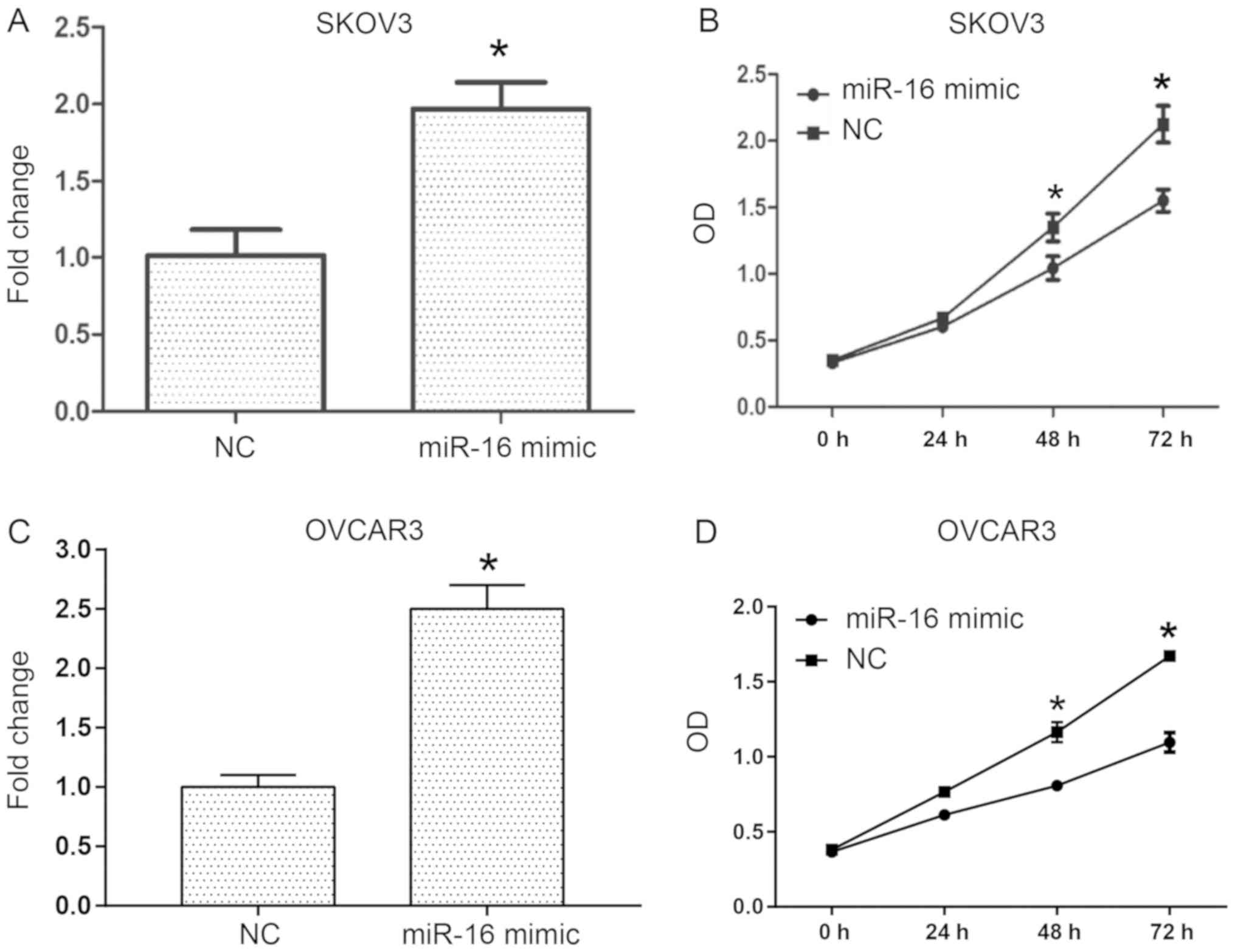
To investigate the role of miR-16 in the
proliferation of human ovarian cancer cells, transient transfection
of miR-16 mimic and NC was conducted into SKOV3 and OVCAR3 cells
(Fig. 2). RT-qPCR was performed to
examine the miR-16 expression level in the SKOV3 and OVCAR3 cells
(Fig. 2A and C), and the
proliferative ability of the cells was assessed with a CCK-8 assay.
The results indicated that following transfection for 3 days,
miR-16 overexpression in the SKOV3 and OVCAR3 cells significantly
decreased the OD values compared with the NC (Fig. 2B and D).
Effect of miR-16 on the migration and
invasion of SKOV3 cells
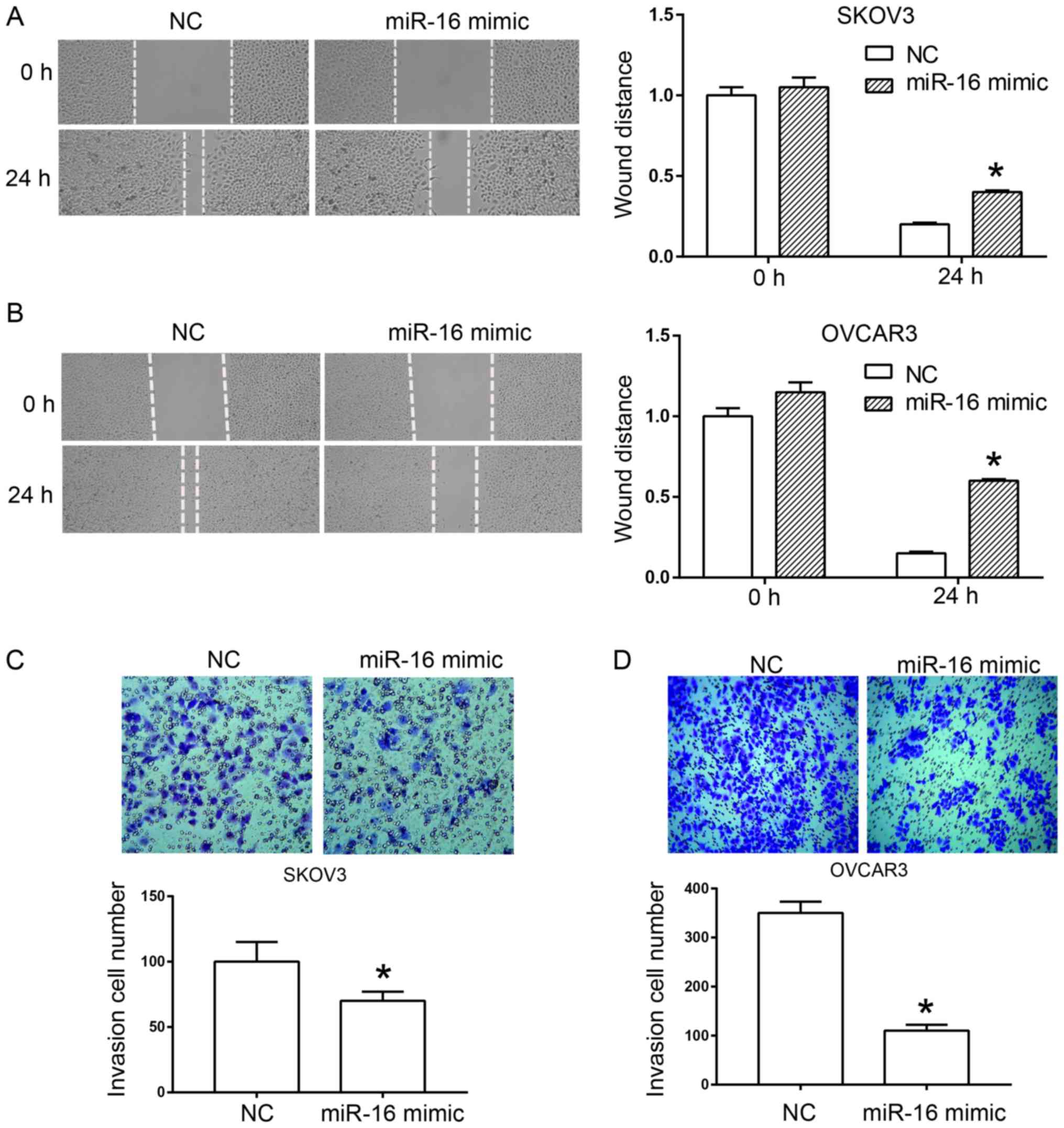
A scratch test and a Transwell assay were used to
determine the effect of miR-16 on the migration and invasion of
human ovarian cancer cells, respectively. In the scratch test,
miR-16 overexpression significantly inhibited wound healing of the
SKOV3 and OVCAR3 cells compared with the NC (Fig. 3A and B). In the Transwell assay, a
significant reduction was observed in the invasion rate of SKOV3
and OVCAR3 cells compared with the NC due to the overexpression of
miR-16 (Fig. 3C and D).
Effect of miR-16 on MMP expression in
human ovarian cancer cells
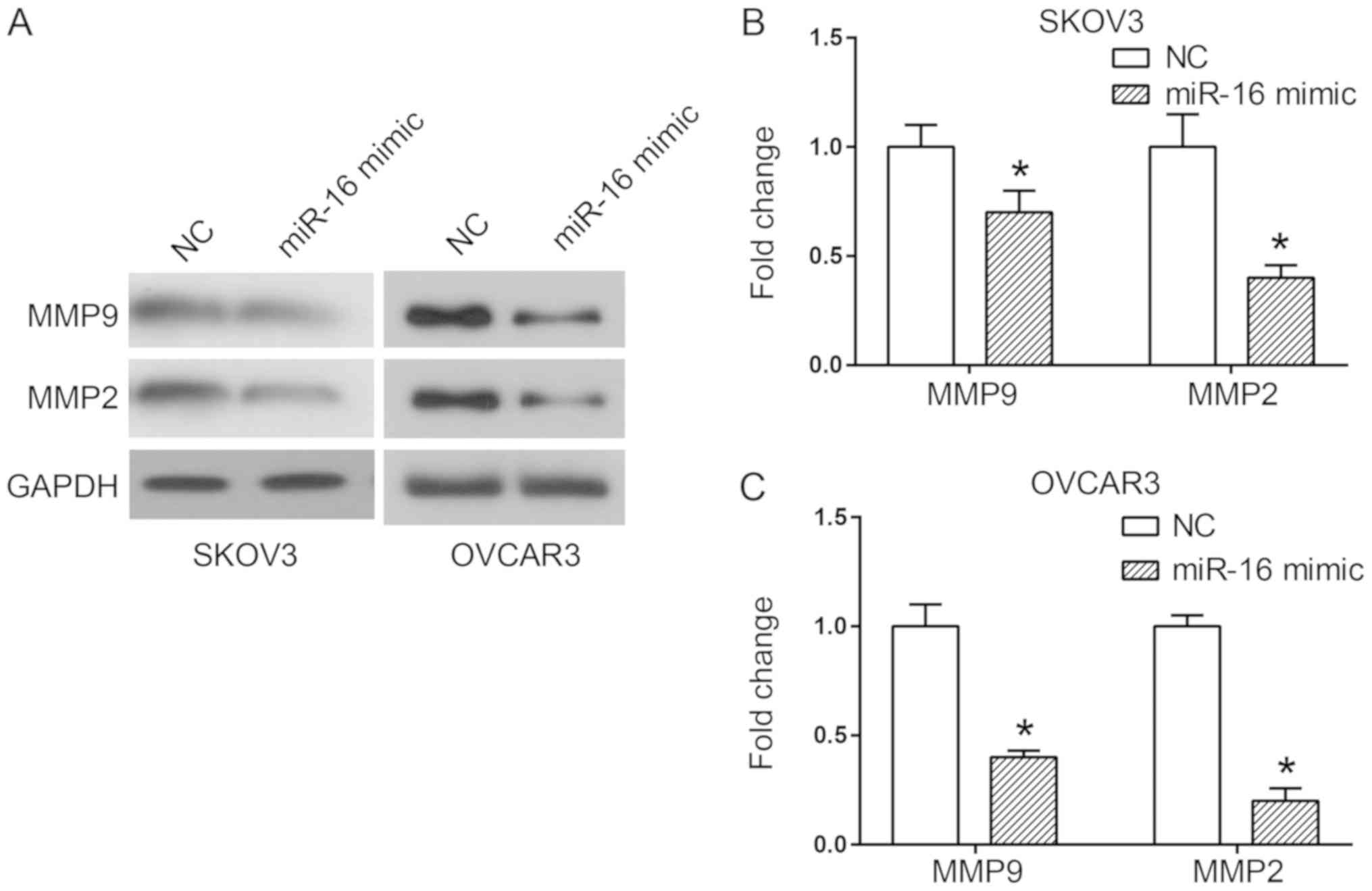
Two critical proteins involved in cell migration and
invasion are MMP2 and MMP9 (15). A
western blot analysis was used to detect the expression of MMP
proteins when miR-16 was overexpressed, in order to observe the
association between these proteins and the invasion and migration
of miR-16-regulated cells. The results indicated a significant
decrease in MMP2 and MMP9 expression levels in the cells
transfected with the miR-16 mimic in comparison with the cells
transfected with the NC (P<0.05; Fig.
4).
Effect of miR-16 on the expression of
EMT-associated proteins in human ovarian cancer cells
EMT is an important process by which epithelial
cells gain migratory and invasive capabilities by transforming into
mesenchymal stem cells (16,17). Western blot analysis was utilized to
observe the expression of different EMT markers upon miR-16
overexpression, in order to investigate whether EMT mediated the
effect of miR-16 on migration and invasion. Overexpression of
miR-16 upregulated the expression of the epithelial marker, E-CAD,
and downregulated the expression of the mesenchymal markers,
including N-CAD, Slug, SNAIL, Vimentin and Twist in the SKOV3 and
OVCAR3 cells (Fig. 5).
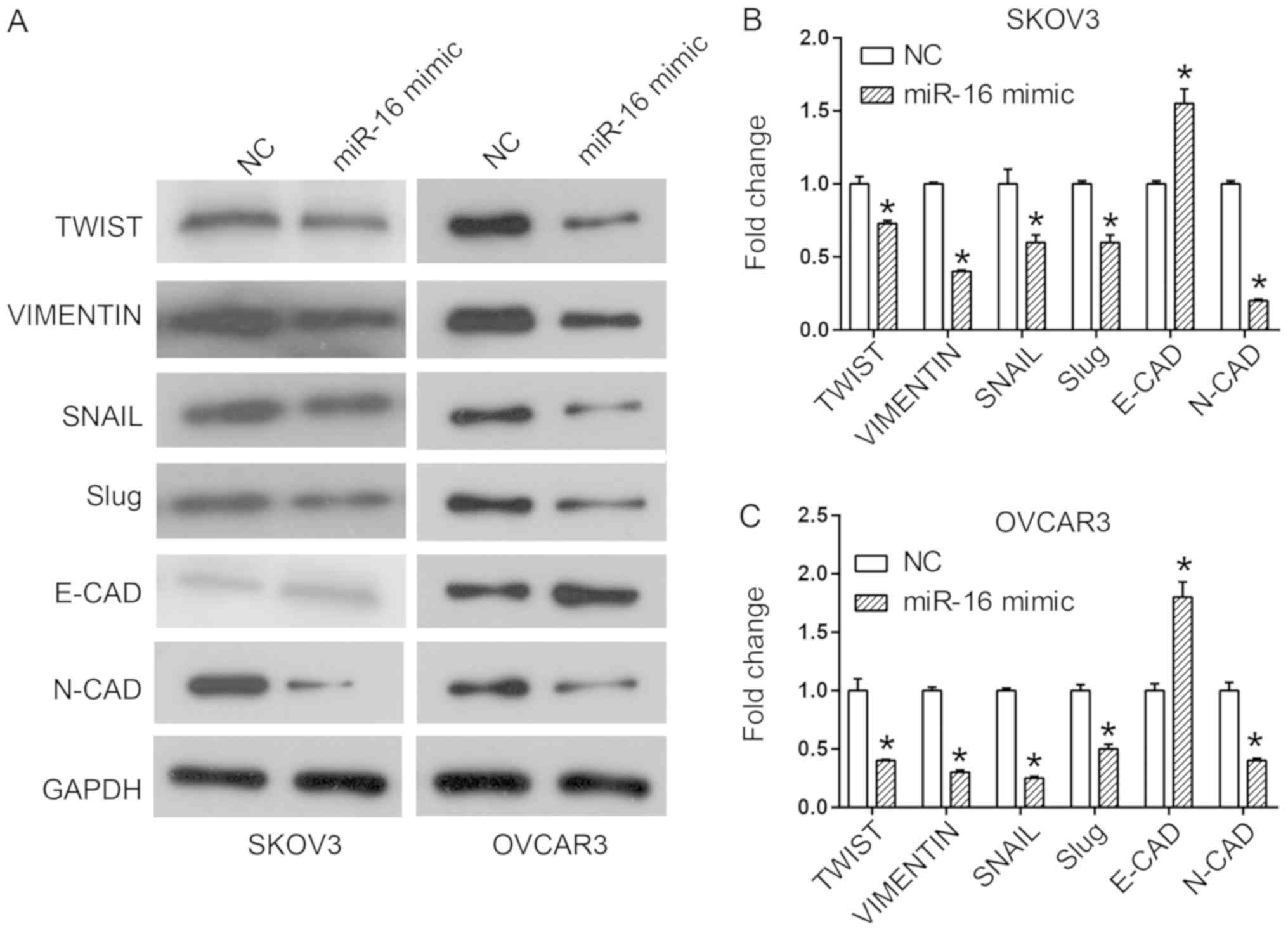 | Figure 5.Effect of miR-16 on the expression
level of EMT-associated proteins in SKOV3 and OVCAR3 cells. (A)
Expression of EMT-associated proteins, including Twist, Vimentin,
SNAIL, Slug, E-CAD and N-CAD in SKOV3 and OVCAR3 cells.
Quantification of expression levels of EMT-associated proteins in
(B) SKOV3 and (C) OVCAR3 cells *P<0.05 compared with the NC.
Slug, Snail family transcriptional repressor 2; SNAIL, snail family
transcriptional repressor 1; Twist, twist family BHLH transcription
factor 1, E-CAD, cadherin 1; N-CAD, cadherin 2; miR, microRNA; NC,
negative control; EMT, epithelial-mesenchymal transition. |
Effect of miR-16 on the Wnt/β-catenin
signaling pathway in human ovarian cancer cells
The results of previous studies have demonstrated
the important role of Wnt/β-catenin signaling in the acquisition of
the EMT phenotype, and in cancer metastasis (18,19).
Therefore, the expression of proteins involved in the Wnt/β-catenin
pathway was examined in miR-16-regulated SKOV3 and OVCAR3 cells by
western blot analysis. The results of the western blot analysis
revealed an increase in Gsk3β expression, but a decrease in Wnt3a
and β-catenin expression in the miR-16 mimic-transfected cells
(Fig. 6). In addition, the downstream
members of the Wnt/β-catenin pathway, c-MYC and cyclin D1, were
additionally downregulated by the miR-16 mimic (Fig. 6).
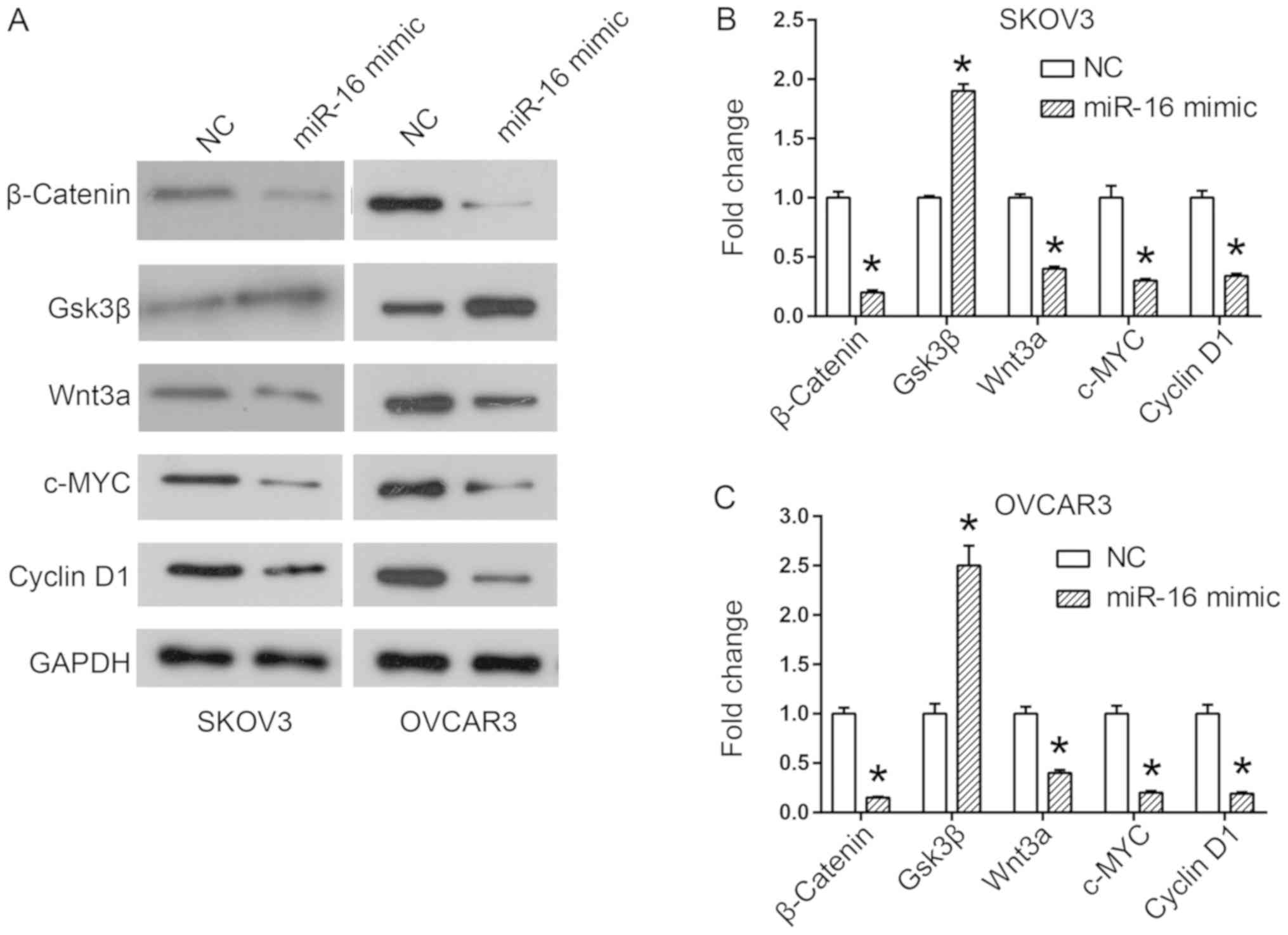 | Figure 6.Effect of miR-16 on the expression of
Wnt/β-catenin signaling pathway-associated proteins in SKOV3 and
OVCAR3 cells. (A) Expression of Wnt/β-catenin signaling pathway
components, including β-catenin, Gsk3β, Wnt3a, c-MYC and cyclin D1
in SKOV3 and OVCAR3 cells. Quantification of expression levels of
Wnt/β-catenin signaling pathway components in (B) SKOV3 and (C)
OVCAR3 cells. *P<0.05 compared with the NC. Gsk3β, glycogen
synthase kinase 3 beta; c-MYC, MYC proto-oncogene, BHLH
transcription factor; Wnt3a, Wnt family member 3A; miR, microRNA;
NC, negative control. |
Discussion
An miR-15a/miR-16-1 cluster was identified as a
tumor suppressor in chronic lymphocytic leukemia, providing the
first evidence linking miRNAs to human cancer (13). Following this pioneering study, there
has been increasing evidence of the significant role of miRNAs in
the early diagnosis, prognosis, prevention and therapeutic
evaluation of cancer (2,20–22).
Identification of alterations in specific miRNAs, in addition to
understanding their functions in each type of cancer will improve
the accuracy of diagnosis and prognosis of numerous types of
cancer. Previous studies have indicated that certain miRNAs,
including the members of the miR-200 and let-7 families, are
involved in the progression and prognosis of ovarian cancer, as
well as the response of ovarian cancer to chemotherapy (23). Bhattacharya et al (13) reported that miR-16 is downregulated in
ovarian cancer cell lines and patient samples, whereas miR-16
upregulation inhibits the proliferation of ovarian carcinoma cells
(13). The specific roles and
underlying mechanisms of miR-16 in ovarian tumor metastasis remain
unclear.
In the present study, downregulation of miR-16
expression was observed in two other ovarian cancer cell lines,
SKOV3 and OVCAR3, providing evidence of the role of miR-16 in the
process of proliferation. The results of the present study are
consistent with those in the study by Bhattacharya et al
(13), where miR-16 was downregulated
in ovarian cancer cell lines and tumor tissues. Additionally, the
present study indicated that overexpression of miR-16 inhibited the
migration and invasion of SKOV3 and OVCAR3 cells. Previous studies
have also examined the roles of miR-16 in tumor metastasis. Li
et al (24) reported that
miR-16-1 functions as a negative regulator of cell migration and
invasion in glioma. In addition, Wu et al (25) revealed the suppressive effects of
miR-16 on the metastasis of hepatocellular carcinoma.
In addition, the results of the present study
indicate that the overexpression of miR-16 is able to decrease MMP2
and MMP9 expression in human ovarian cancer cells. The MMPs are a
group of proteases involved in the degradation of the extracellular
matrix, as these proteases also regulate invasion and tumor
metastasis (15,17,18). MMP2
expression levels have been reported to be increased in patients
with ovarian cancer with advanced tumors and metastasis (26). Therefore, the roles of miR-16 in
ovarian cancer invasion and migration may be associated with the
effect of MMPs on the extracellular matrix.
In addition, the effect of miR-16 on EMT may be
another factor that regulates tumor metastasis. During EMT,
epithelial cells lose their apical-basal polarity and cell-cell
adhesion, allowing their conversion to motile mesenchymal cells, as
this conversion promotes invasion and metastasis in cancer
(16,17,27,28).
Decreased expression of the intercellular adhesion molecule E-CAD
has been reported as an EMT marker, along with increased expression
of a series of mesenchymal markers, including Slug, SNAIL and
Vimentin (27,29). In a previous study, Wang et al
(30) reported that miR-16 may
inhibit the expression of EMT-associated proteins in human glioma.
Wang et al (31) reported that
miR-16 mimics inhibited transforming growth factor-β1-induced EMT
via activation of autophagy in non-small cell lung carcinoma cells.
In the present study, it was determined that overexpression of
miR-16 in SKOV3 and OVCAR3 cells inhibited the expression of Slug,
SNAIL, Vimentin and Twist, but downregulated the expression of
E-CAD.
It was also revealed that miR-16 mimics led to a
significant increase in the expression of Gsk3β and decreased the
expression levels of Wnt3a and β-catenin in SKOV3 and OVCAR3 cells.
The Wnt/β-catenin signaling pathway has a regulatory function in a
number of cellular processes, including development,
differentiation, proliferation and adult tissue homeostasis.
Therefore, aberrant Wnt/β-catenin signaling has been indicated to
be involved in the pathogenesis of multiple tumors (18,19,32,33).
Additional evidence indicates that the activation of the
Wnt/β-catenin signaling pathway promotes the EMT process and the
secretion of MMP2 in cancer cells, providing them with an increased
ability for survival and metastasis (18,34–36).
However, the present study included a number of
limitations. Firstly, the study was conducted to examine the
effects of miR-16 on SKOV3 and OVCAR3 cell proliferation, invasion
and metastasis, and to determine the involved signaling pathways.
However, the target genes of miR-16 in these processes remain
unknown. Further experiments are required. Secondly, this study is
an in vitro study; therefore, in vivo studies should
be conducted in the future to confirm the findings.
In conclusion, the present study demonstrated that
miR-16 served a negative role in the proliferation, invasion and
metastasis of SKOV3 and OVCAR3 cells. Furthermore, overexpression
of miR-16 inhibited the process of EMT by inactivating the
Wnt/β-catenin signaling pathway. The aforementioned results suggest
that miR-16 may be a promising therapeutic target for ovarian
cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW contributed to the study design, data acquisition
and analysis, and drafted the manuscript. NL was involved in the
study design, data acquisition and revision of the manuscript. LY
and YS assisted in the performance of the statistical analysis. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Prahm KP, Novotny GW, Høgdall C and
Høgdall E: Current status on microRNAs as biomarkers for ovarian
cancer. APMIS. 124:337–355. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yeung TL, Leung CS, Yip KP, Au Yeung CL,
Wong ST and Mok SC: Cellular and molecular processes in ovarian
cancer metastasis. A review in the theme: Cell and molecular
processes in cancer metastasis. Am J Physiol Cell Physiol.
309:C444–C456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Kim S and Kim IM: Regulation of
metastasis by microRNAs in ovarian cancer. Front Oncol. 4:1432014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang L, Nadeem L, Connor K and Xu G:
Mechanisms and therapeutic targets of microRNA-associated
chemoresistance in epithelial ovarian cancer. Curr Cancer Drug
Targets. 16:429–441. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang ZH and Xu CJ: Research progress of
MicroRNA in early detection of ovarian cancer. Chin Med J (Engl).
128:3363–3370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Langhe R: microRNA and ovarian cancer. Adv
Exp Med Biol. 889:119–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wei J, Zhang L, Li J, Zhu S, Tai M, Mason
CW, Chapman JA, Reynolds EA, Weiner CP and Zhou HH: MicroRNA-205
promotes cell invasion by repressing TCF21 in human ovarian cancer.
J Ovarian Res. 10:332017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wei LQ, Liang HT, Qin DC, Jin HF, Zhao Y
and She MC: MiR-212 exerts suppressive effect on SKOV3 ovarian
cancer cells through targeting HBEGF. Tumour Biol. 35:12427–12434.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hanlon K, Rudin CE and Harries LW:
Investigating the targets of MIR-15a and MIR-16-1 in patients with
chronic lymphocytic leukemia (CLL). PLoS One. 4:e71692009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bottoni A, Piccin D, Tagliati F, Luchin A,
Zatelli MC and degli Uberti EC: miR-15a and miR-16-1
down-regulation in pituitary adenomas. J Cell Physiol. 204:280–285.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Di Leva G and Croce CM: The role of
microRNAs in the tumorigenesis of ovarian cancer. Front Oncol.
3:1532013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bhattacharya R, Nicoloso M, Arvizo R, Wang
E, Cortez A, Rossi S, Calin GA and Mukherjee P: MiR-15a and MiR-16
control Bmi-1 expression in ovarian cancer. Cancer Res.
69:9090–9095. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pietruszewska W, Bojanowska-Poźniak K and
Kobos J: Matrix metalloproteinases MMP1, MMP2, MMP9 and their
tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: An
immunohistochemical study. Otolaryngol Pol. 70:32–43. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deng J, Wang L, Chen H, Hao J, Ni J, Chang
L, Duan W, Graham P and Li Y: Targeting epithelial-mesenchymal
transition and cancer stem cells for chemoresistant ovarian cancer.
Oncotarget. 7:55771–55788. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou XM, Zhang H and Han X: Role of
epithelial to mesenchymal transition proteins in gynecological
cancers: Pathological and therapeutic perspectives. Tumour Biol.
35:9523–9530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McCubrey JA, Fitzgerald TL, Yang LV,
Lertpiriyapong K, Steelman LS, Abrams SL, Montalto G, Cervello M,
Neri LM, Cocco L, et al: Roles of GSK-3 and microRNAs on epithelial
mesenchymal transition and cancer stem cells. Oncotarget.
8:14221–14250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ghahhari NM and Babashah S: Interplay
between microRNAs and WNT/β-catenin signalling pathway regulates
epithelial-mesenchymal transition in cancer. Eur J Cancer.
51:1638–1649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu L and Fang J: The structure and
clinical roles of MicroRNA in colorectal cancer. Gastroenterol Res
Pract. 2016:13603482016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shukla KK, Misra S, Pareek P, Mishra V,
Singhal B and Sharma P: Recent scenario of microRNA as diagnostic
and prognostic biomarkers of prostate cancer. Urol Oncol.
35:92–101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shah MY, Ferrajoli A, Sood AK,
Lopez-Berestein G and Calin GA: microRNA therapeutics in cancer-an
emerging concept. EBioMedicine. 12:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pal MK, Jaiswar SP, Dwivedi VN, Tripathi
AK, Dwivedi A and Sankhwar P: MicroRNA: A new and promising
potential biomarker for diagnosis and prognosis of ovarian cancer.
Cancer Biol Med. 12:328–341. 2015.PubMed/NCBI
|
|
24
|
Li X, Ling N, Bai Y, Dong W, Hui GZ, Liu
D, Zhao J and Hu J: MiR-16-1 plays a role in reducing migration and
invasion of glioma cells. Anat Rec (Hoboken). 296:427–432. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu WL, Wang WY, Yao WQ and Li GD:
Suppressive effects of microRNA-16 on the proliferation, invasion
and metastasis of hepatocellular carcinoma cells. Int J Mol Med.
36:1713–1719. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ekinci T, Ozbay PO, Yiğit S, Yavuzcan A,
Uysal S and Soylu F: The correlation between immunohistochemical
expression of MMP-2 and the prognosis of epithelial ovarian cancer.
Ginekol Pol. 85:121–130. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan H, He Q, Gong G, Wang Y, Li J, Wang J,
Zhu D and Wu X: miR-382 inhibits migration and invasion by
targeting ROR1 through regulating EMT in ovarian cancer. Int J
Oncol. 48:181–190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Amankwah EK, Lin HY, Tyrer JP, Lawrenson
K, Dennis J, Chornokur G, Aben KK, Anton-Culver H, Antonenkova N,
Bruinsma F, et al: Epithelial-mesenchymal transition (EMT) gene
variants and epithelial ovarian cancer (EOC) risk. Genet Epidemiol.
39:689–697. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takai M, Terai Y, Kawaguchi H, Ashihara K,
Fujiwara S, Tanaka T, Tsunetoh S, Tanaka Y, Sasaki H, Kanemura M,
et al: The EMT (epithelial-mesenchymal-transition)-related protein
expression indicates the metastatic status and prognosis in
patients with ovarian cancer. J Ovarian Res. 7:762014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Q, Li X, Zhu Y and Yang P:
MicroRNA-16 suppresses epithelial-mesenchymal transition-related
gene expression in human glioma. Mol Med Rep. 10:3310–3314. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang H, Zhang Y, Wu Q, Wang Y-B and Wang
W: miR-16 mimics inhibit TGF-β1-induced epithelial-to-mesenchymal
transition via activation of autophagy in non-small cell lung
carcinoma cells. Oncol Rep. 39:247–254. 2018.PubMed/NCBI
|
|
32
|
Bodnar L, Stanczak A, Cierniak S, Smoter
M, Cichowicz M, Kozlowski W, Szczylik C, Wieczorek M and
Lamparska-Przybysz M: Wnt/β-catenin pathway as a potential
prognostic and predictive marker in patients with advanced ovarian
cancer. J Ovarian Res. 7:162014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Arend RC, Londoño-Joshi AI, Straughn JM
Jr..Buchsbaum DJ: The Wnt/β-catenin pathway in ovarian cancer: A
review. Gynecol Oncol. 131:772–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Y, Ginther C, Kim J, Mosher N, Chung S,
Slamon D and Vadgama JV: Expression of Wnt3 activates Wnt/β-catenin
pathway and promotes EMT-like phenotype in trastuzumab-resistant
HER2-overexpressing breast cancer cells. Mol Cancer Res.
10:1597–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kwon YJ, Baek HS, Ye DJ, Shin S, Kim D and
Chun YJ: CYP1B1 enhances cell proliferation and metastasis through
induction of EMT and activation of Wnt/β-catenin signaling via Sp1
upregulation. PLoS One. 11:e01515982016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sonderegger S, Haslinger P, Sabri A,
Leisser C, Otten JV, Fiala C and Knöfler M: Wingless (Wnt)-3A
induces trophoblast migration and matrix metalloproteinase-2
secretion through canonical Wnt signaling and protein kinase B/AKT
activation. Endocrinology. 151:211–220. 2010. View Article : Google Scholar : PubMed/NCBI
|