Introduction
Among females, ovarian cancer is a frequently
diagnosed cancer type and a leading cause of mortality (1). In China, the incidence rate of ovarian
cancer has increased exponentially over the last decade (2). Inherited germline mutations in
BRCA2 predisposes individuals to a high risk of ovarian
cancer. Therefore, genetic screening of BRCA2 mutations may
be a significant component of clinical practice for individuals
with a family history of ovarian cancer. In females, BRCA2
mutations result in a lifetime risk of 11–18% of developing ovarian
cancer (3,4). Hereditary ovarian cancer is an autosomal
dominant syndrome with incomplete penetrance caused by germline
mutations of BRCA2. Germline mutations in both BRCA1
and BRCA2 are major causes of hereditary ovarian cancer
(5,6).
BRCA1 and BRCA2 serve key roles in the double-strand
break repair system (7). Based on the
Breast Cancer Information Core (BIC) database (http://research.nhgri.nih.gov/bic/),
approximately 3,700 BRCA1/BRCA2 germline mutations have been
associated with ovarian cancer. The present study reports a novel
heterozygous germline insertion mutation in the BRCA2 gene,
c.3195_3196insA. This single nucleotide insertion causes a
frameshift mutation, which results in the formation of a truncated
BRCA2 protein with 1,076 amino acids instead of the wild-type BRCA2
protein with 3,418 amino acids.
Case report
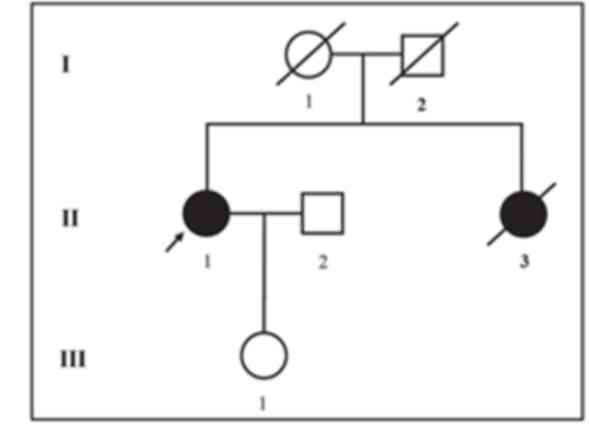
The proband investigated in the current study is a
54-year-old Chinese female clinically diagnosed with ovarian cancer
who belongs to a non-consanguineous Chinese family. The proband's
elder sister passed away at the age of 46 years as a result of
ovarian cancer. The age and cause of mortality for the proband's
parents are unknown (Fig. 1). The
proband had been diagnosed with stage III right ovarian cancer,
along with liver metastasis. The top edge of the liver exhibited a
nodule with mild leukoaraiosis. The magnetic resonance imaging
(MRI) scan demonstrated no metastasis around the brain.
The biopsy sections (thickness, 3–4 µm) were
paraffin embedded, fixed with 4% neutral buffer formaldehyde
solution, at room temperature (25°C) for 6–48 h. Pathological
biopsy was performed at the Fourth Hospital of Hebei Medical
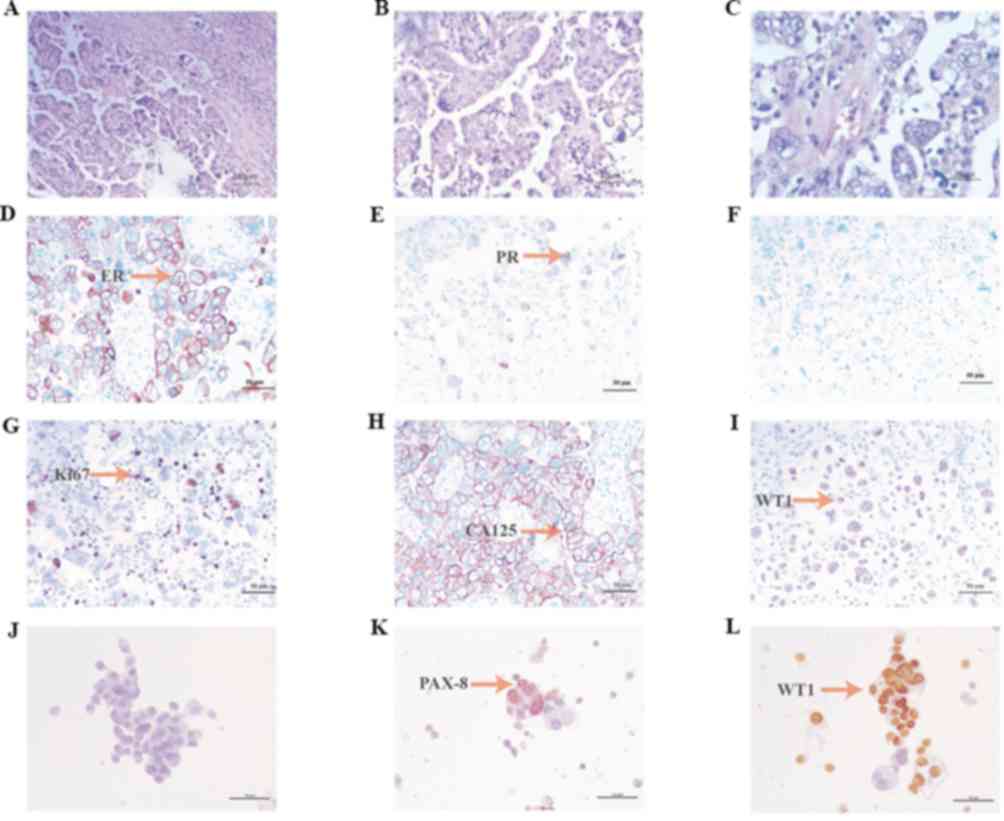
University (Shijiazhuang, China). Hematoxylin and eosin staining of
tumor tissue was performed by optical microscopy (magnification,
×460) and microscopy images were obtained (Fig. 2A-C). Paraffin-embedded slices were
stained with IHC markers (Ventana Benchmark Ultra automatic
immunohistochemical staining platform, primary antibodies: ER:
Anti-estrogen receptor (SP1) rabbit monoclonal primary antibody,
Roche, catalog number: 790–4325; PR: Anti-progesterone receptor
(1E2) rabbit monoclonal primary antibody, Roche, catalog number:
790–4296; P53: Anti-P53 mouse monoclonal primary antibody, Roche,
catalog number: 800–2912; Ki-67: Anti-Ki-67 (30–9) rabbit
monoclonal primary antibody, Roche, catalog number: 790–4286; CEA:
Anti-CEA (COL-1) mouse monoclonal primary antibody, From: MXB
Biotechnologies, catalog number: Kit-0008; CA125: Anti-CA125
(TA347) mouse monoclonal primary antibody, MXB Biotechnologies,
catalog number: MAB-0007; WT1: Anti-WT1 (MX012) mouse monoclonal
primary antibody, MXB Biotechnologies, catalog number: MAB-0678;
PAX-8: Anti-PAX-8 rabbit polyclonal primary antibody, MXB
Biotechnologies, catalog number: RAB-0657; PAX-2: MXB
Biotechnologies, catalog number: RAB-0648. Secondary antibodies:
MaxVision DAB, From: MXB Biotechnologies, catalog number: KIT-5220.
All antibodies were purchased in their ready-to-use format so
dilutions weren't required. IHC experimental procedure according to
the manufacturer's protocols), the percentage of positive staining
was calculated by eye. Immunohistochemistry (IHC) for estrogen
receptors demonstrated a high (70%) expression level in cancer
cells (Fig. 2D). IHC for progesterone
receptors revealed a moderate (30%) expression level in cancer
cells (Fig. 2E). IHC for p53
demonstrated zero expression level of p53 in cancer cells (Fig. 2F). IHC for Ki67 revealed a moderate
(30%) expression level in cancer cells (Fig. 2G). Staining for cancer antigen 125
demonstrated a high expression level (Fig. 2H). IHC for Wilms tumor protein (WT1)
revealed a moderate expression level in cancer cells (Fig. 2I). IHC for paired box gene PAX-2,
PAX-8 and CEA was also performed, revealing zero, positive and
positive expression, respectively (Fig.
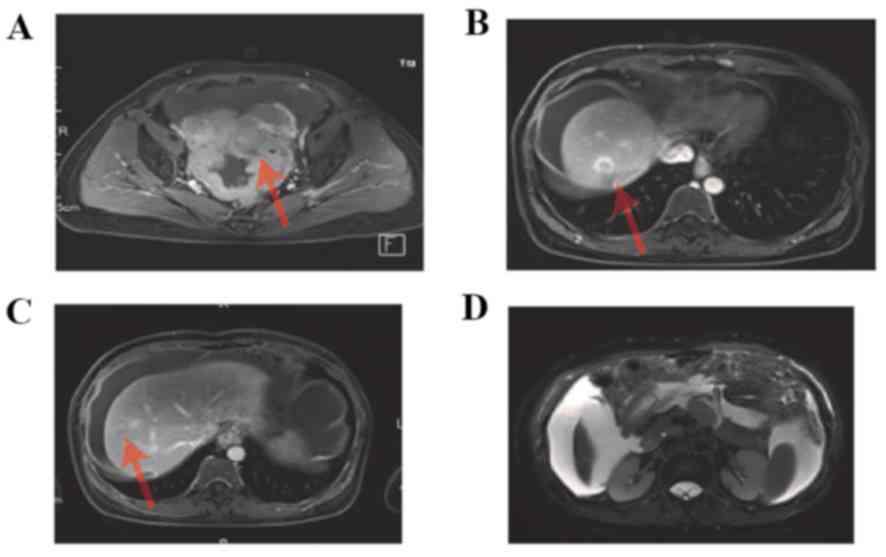
2J-L). An MRI scan was performed, which demonstrated
enhancement of the pelvic tumor (Fig.
3A), enhancement of hepatic metastasis (Fig. 3B,C), and thickening of the omentum
majus and tumor tissue (Fig. 3D).
The patient's treatment regimen consisted of TC
scheme (paclitaxel and carboplatin: Paclitaxel liposome, 240 mg/day
1+ carboplatin, 0.5 g/day 1, intravenous injection) chemotherapy
for two cycles. Following treatment, the patient showed an
improvement, and a gynecological surgeon contacted the patient's
family.
Prior to the experiment, written informed consent
was obtained from each patient. Peripheral blood samples of the
patients were collected and genomic DNA was extracted. Targeted
high-throughput sequencing (DNA polymerase is T4 DNA POLYMERASE,
supplier is ENZYMATICS; Qiagen, Inc., Valencia, CA, USA) was
performed with a panel of a 21 genes (BRCA1, BRCA2, CHEK2,
PALB2, BRIP1, TP53, PTEN, STK11, CDH1, ATM, BARD1, MLH1, MRE11A,
MSH2, MSH6, MUTYH, NBN, PMS1, PMS2, RAD50 and RAD51C).
Roche NimbleGen's (Roche Applied Science, Madison, MI, USA) custom
Sequence Capture Human array was used to designed to capture
targeted sequence, according to the manufacturer's protocols. A
target area of 172,959 bp, a target area coverage of 99.49% and a
target area average depth of 586.13×. Samples were sequenced
simultaneously on Illumina HiSeq 2500 Analyzers (Illumina Inc., San
Diego, CA, USA) for 90 cycles according to the manufacturer's
protocols. Targeted high-throughput sequencing identified a novel
heterozygous mutation, c.3195_3196insA, in BRCA2, which
causes a frameshift mutation that generates a premature stop codon
and results in the formation of a truncated BRCA2 protein. This
mutation was confirmed by Sanger sequencing (Fig. 1R). The mutation was absent in the
Human Gene Mutation database (www.hgmd.cf.ac.uk/) and the Online Mendelian
Inheritance in Man database (www.omim.org).
In addition, the mutation could not be identified in our in-house
databases, which contains ~30,000 samples from Chinese Han
individuals.
Since the proband's elder sister passed away at an
age of 46 years due to ovarian cancer, it can be hypothesized that
she carried the same mutation in BRCA2. In addition, the
proband's father or mother may carry the same mutation. However, as
the proband's elder sister and parents had passed away, it was not
possible to investigate this. The proband's children did not
volunteer genetic testing. Therefore, only the following
suggestions can be made. Firstly, it could be hypothesized that the
proband and her sister carried the same mutation on the
BRCA2 gene, which was inherited from their father or mother.
By contrast, it could be hypothesized that the mutation carried by
the proband is a de novo mutation. In this case, her parents
and sister would not have carried the same mutation. With the
knowledge that the proband's sister passed away due to ovarian
cancer, we speculate that the first hypothesis is more likely. The
Ethical Committee of The Fourth Hospital of Hebei Medical
University (Shijiazhuang, China) approved the current study
protocol, which was in compliance with the Declaration of
Helsinki.
Discussion
A c.3195_3196insA frameshift mutation in exon 10 of
the BRCA2 gene, which has not previously been reported in
the BIC database, was detected in a 54 year-old female Chinese
patient diagnosed with ovarian cancer. Not all genetic variants are
harmful; BRCA1/2 variations can be classified into the five
following categories: ‘Pathogenic’, ‘likely pathogenic’, ‘uncertain
significance’, ‘likely benign’ and ‘benign’, according to American
College of Medical Genetics’ guidelines (8). ‘Pathogenic’ or ‘likely pathogenic’
indicates that a mutation was identified in a specific gene, which
increases the lifetime risk of developing certain cancer types, but
does not indicate that cancer will definitely develop. Germline
BRCA1/2 deleterious mutations lead to an increased risk of
early onset hereditary breast and ovarian cancer (HBOC)-associated
tumors, including breast, ovarian and pancreatic cancer in females,
and to a lesser extent, breast and prostate cancer in males
(9,10).
Genetic sequencing techniques have developed
radically during the past few decades; therefore, novel testing
techniques have been widely used in the medical sector (11–15). For
example, the enrichment step, a preparation technique used prior to
next-generation sequencing (NGS), can significantly improve the
accuracy and efficiency of medical screening (16). This technique was used in the current
study to identify the largest possible number of mutations that may
be associated with HBOC. Due to technology limitations, there are
still many challenges in detecting copy-number variants by NGS
(17), certain patients with a
negative NGS testing result may also require multiplex
ligation-dependent probe application, particularly patients with a
family history of cancer.
BRCA1 and BRCA2 were not the only
genes analyzed in the current study; multiple gene analysis was
performed. Numerous genes should be involved in sequencing tests as
the mutational spectra of these genes can be accurately detected
with NGS technology, which provides additional data that can
support clinical diagnoses. However, involving multiple genes in
sequencing tests may lead to the detection of a higher number of
variants of uncertain significance (VUS) (18). Clinical recommendations may not be
standardized for the presence of VUS mutations in certain cancer
susceptibility genes, which may complicate results. These complex
results are why suggestions from medical geneticists or certified
genetic counselors are important. Since a number of NGS-based
techniques are used in genetic testing and different genetic
companies perform different techniques, customers should have
access to the sensitivity and specificity of any results, as well
as comparisons of various techniques (19). In our opinion, positive results should
be validated using standard Sanger sequencing, which should also be
available to relatives who are concerned about the risk of
cancer.
Acknowledgements
Not applicable.
Funding
The current study was supported by the Special
Foundation for High-level Talents of Guangdong (grant no.
2016TX03R171).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
SB, DJ, ZP designed and coordinated the study. DJ,
YC, NZ, JH, FL, SW, QD, QZ and HJ assessed the clinical findings of
the cases. YW, HH, JW, KL, WC, WL and JX performed the molecular
genetic studies and analyzed the data. YW wrote the draft of the
manuscript with input from the other co-authors. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All study participants provided written informed
consent and the study design was approved by an Institutional
Ethics Review Board of the Department of Internal Medicine, The
Fourth Hospital of Hebei Medical University (Shijiazhuang,
China).
Patient consent for publication
A written consent was obtained from the patient for
the publications of the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tielent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen S and Parmigiani G: Meta-analysis of
BRCA1 and BRCA2 penetrance. J Clin Oncol. 25:1329–1333. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mavaddat N, Peock S, Frost D, Ellis S,
Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, et al:
Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from
prospective analysis of EMBRACE. J Natl Cancer Inst. 105:812–822.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Claus EB, Risch N and Thompson WD:
Autosomal dominant inheritance of earlyonset breast cancer.
Implications for risk prediction. Cancer. 73:643–651. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Petrucelli N, Daly MB and Pal T: BRCA1-and
BRCA2-associated hereditary breast and ovarian cancer. Adam MP,
Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K and Amemiya
A: University of Washington; Seattle: 2016
|
|
7
|
Narod SA and Foulkes WD: BRCA1 and BRCA2:
1994 and beyond. Nat Rev Cancer. 4:665–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the American college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song H, Cicek MS, Dicks E, Harrington P,
Ramus SJ, Cunningham JM, Fridley BL, Tyrer JP, Alsop J, et al: The
contribution of deleterious germline mutations in BRCA1, BRCA2 and
the mismatch repair genes to ovarian cancer in the population. Hum
Mol Genet. 23:4703–4709. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim YC, Zhao L, Zhang H, Huang Y, Cui J,
Xiao F, Downs B and Wang SM: Prevalence and spectrum of BRCA
germline variants in mainland Chinese familial breast and ovarian
cancer patients. Oncotarget. 7:9600–9612. 2016.PubMed/NCBI
|
|
11
|
Lifton RP: Individual genomes on the
horizon. N Engl J Med. 362:1235–1236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goossens D, Moens LN, Nelis E, Lenaerts
AS, Glassee W, Kalbe A, Frey B, Kopal G, De Jonghe P, De Rijk P and
Del-Favero J: Simultaneous mutation and copy number variation (CNV)
detection by multiplex PCR-based GS-FLX sequencing. Hum Mutat.
30:472–476. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Daiger SP, Sullivan LS, Bowne SJ, Birch
DG, Heckenlively JR, Pierce EA and Weinstock GM: Targeted
high-throughput DNA sequencing for gene discovery in retinitis
pigmentosa. Adv Exp Med Biol. 664:325–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoischen A, Gilissen C, Arts P, Wieskamp
N, van der Vliet W, Vermeer S, Steehouwer M, de Vries P, Meijer R,
Seiqueros J, et al: Massively parallel sequencing of ataxia genes
after array-based enrichment. Hum Mutat. 31:494–499. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chou LS, Liu CS, Boese B, Zhang X and Mao
R: DNA sequence capture and enrichment by microarray followed by
next-generation sequencing for targeted resequencing:
Neurofibromatosis type 1 gene as a model. Clin Chem. 56:62–72.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mamanova L, Coffey AJ, Scott CE, Kozarewa
I, Turner EH, Kumar A, Howard E, Shendure J and Turner DJ:
Target-enrichment strategies for next-generation sequencing. Nat
Methods. 7:111–118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Teo SM, Pawitan Y, Ku CS, Chia KS and
Salim A: Statistical challenges associated with detecting copy
number variations with next-generation sequencing. Bioinformatics.
28:2711–2718. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kapoor NS, Curcio LD, Blakemore CA,
Bremner AK, McFarland RE, West JG and Banks KC: Multigene panel
testing detects equal rates of pathogenic BRCA1/2 mutations and has
a higher diagnostic yield compared to limited BRCA1/2 analysis
alone in patients at risk for hereditary breast cancer. Ann Surg
Oncol. 22:3282–3288. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mattocks CJ, Morris MA, Matthijs G,
Swinnen E, Corveleyn A, Dequeker E, Müller CR, Pratt V and Wallace
A; EuroGentest Validation Group, : A standardized framework for the
validation and verification of clinical molecular genetic tests.
Eur J Hum Genet. 18:1276–1288. 2010. View Article : Google Scholar : PubMed/NCBI
|