Introduction
Breast cancer (BC) is the most common neoplasm among
women worldwide, according to Global cancer statistics (GLOBOCAN)
there will be about 2.1 million newly diagnosed female breast
cancer cases in 2018, accounting for almost 1 in 4 cancer cases
among women (1). In Mexico, BC is the
most common cancer in women since 2006 (2). This neoplasm comprises a group of
biologically different entities with different pathological and
molecular features involved in their staging and therapeutic
management. Based on standard immunohistochemistry tests (IHT), BC
is classified into three main groups: 1) luminal: Positive for
hormonal receptors; 2) HER2 overexpressed, and 3) Triple-negative
BC (TNBC). TNBC is characterized by the absence of hormone receptor
expression and lack of HER2 amplification (3–5). Targeted
therapies are available for luminal and HER2 amplified categories,
but there is no treatment option for TNBC. Therefore, the precise
classification in clinically relevant subtypes is of particular
importance for therapeutic decision-making. TNBC represents 15–20%
of the BC and is more frequent in young women and individuals of
African and Hispanic heritage (6).
Due to the lack of expression of therapeutic targets, chemotherapy
remains a primary treatment option, along with radiotherapy and
surgery (7,8). While TNBC patients respond better to
chemotherapy than patients with non-TN BC (nTNBC), TNBC patients
who do not respond eventually develop the metastatic form of the
disease. This form is virtually incurable, so TNBC is characterized
by its aggressive clinical course and poor prognosis compared to
other BC subtypes (9).
The absence of therapeutic biomarkers of TNBC
requires determining the molecular profile of TNBC tumors to
propose therapeutic targets. The efforts of the complete genome
sequencing have shown that TNBC presents alterations of TP53
in up to 80% of the cases, followed by a broad set of genes with
lower frequencies, such as PIK3CA and RB (10,11).
Next generation sequencing (NGS) is a powerful
method that allows visualizing the genomic landscape of tumors and
revealing tumor heterogeneity through the detection of genetic
variants that occur in a low percentage (12). NGS has been used to sequence genes
linked to cancer (11,13,14) to
discover mutations that can modulate the repair capacity as well as
the response to chemotherapy (15).
Besides, the increased heterogeneity correlates with poor patient
outcomes to treatment (16,17).
In the present study, we characterize the genetic
alterations of TNBC in fresh tissue biopsies from TNBC patients
from the Northeast of Mexico through NGS, with the aim of
identifying alternative driver mutations, including those
predictive of sensitivity and/or clinical response to chemotherapy
and new molecularly directed drugs.
Materials and methods
Patients and tissue sample
The protocol was approved by the Ethics and Research
Committee of the School of Medicine (Universidad Autonoma de Nuevo
Leon), with the registered number BI11-005. Each participant was
asked to sign an informed consent. Demographic information and
personal data were obtained from the medical records. Tissue
samples were obtained from patients under clinical and radiological
suspicion of locally advanced BC (Tumor size > 2 cm, palpable
ipsilateral lymph nodes, and ulceration) (18) of University Hospital “Dr. José
Eleuterio Gonzalez” between 2011 and 2014. Core biopsies were
obtained using a 12 Fr gauge (Bard®). Each patient
underwent histopathological diagnosis by immunohistochemistry (ER,
PR, HER2 and Ki67 status).
DNA isolation
Genomic DNA was obtained from biopsies with the
DNeasy Blood and Tissue kit (Qiagen, Inc., Valencia, CA, USA)
following the manufacturer's instructions (www.qiagen.com). The tissues were lysed by incubation
with proteinase K at 56°C until the tissues were completely lysed,
followed by purification and elution on a centrifugation column.
The DNA was initially quantified in the Nanodrop 8000
spectrophotometer (ratio 260/280> 1.8; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). A subsequent quantification was done using
Quant-iT Picogreen (Thermo Fisher Scientific, Inc.) following the
instructions from the manufacturer. DNA concentration was then
adjusted to 50 µg/ml.
Sequencing
Libraries were constructed using the TruSeq Amplicon
Cancer Panel (FC-130-1008) (19) a
kit available on https://www.illumina.com/ that has been designed to
cover mutational hotspot of 48 genes associated with cancer that
can generate data for treatment with drugs approved by the US Food
and Drug Administration. 250 ng of DNA was mixed with the pool of
oligonucleotides containing all the primers to generate 212
amplicons (~35 kilobases) from hotspot regions of 48 genes.
Libraries were amplified in the Eppendorf EP Master Faster City
thermal cycler (Eppendorf, Hamburg, Germany). The quality of the
libraries were evaluated in Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc., Santa Clara, CA, USA). Each library was
standardized according to the manufacturer's instructions. Finally,
libraries were adjusted at a concentration of 12 pM. Library pools
were loaded into a MiSeq Reagent Kit v3 cartridge (Illumina, Inc.,
San Diego, CA, USA), and each library pool was sequenced on an
Illumina MiSeq instrument using a 150 paired-end design.
Data analysis
The Human Genome build 19 construct (hg19) was used
as the reference genome. Alignment and the variant calling were
performed with the MiSeq Reporter TruSeq Amplicon (Illumina, Inc.).
Variants were identified using Variant Interpreter (Illumina Inc.).
Reading quality Q>90 and reading depth>60 were used. Variants
with an allelic frequency less than 5% were discarded. The clinical
significance of the variants was determined using the ClinVar tool.
Also, the tool Polymorphism Phenotyping v2 (Polyphen-2) was applied
to predict the possible impact of an amino acid substitution on the
function of the proteins.
Results
Patients
A total of 29 frozen tissue biopsies classified as
TNBC were collected. However, there were 19 tissue samples
available for sequencing. The average age of these 19 women was 51
years, with a BMI average of 27.5. All participants were free of
metastases at the time of participating in the study. The main
clinical information of the patients is shown in Table I.
 | Table I.Clinical characteristics of women
participating in this study. The average age at diagnosis was 52
years, with an age range of 41–71 years. The average Body Mass
Index was 27.31. No patient presented metastasis at the time of the
study. The participating women were in clinical stages II and
III. |
Table I.
Clinical characteristics of women
participating in this study. The average age at diagnosis was 52
years, with an age range of 41–71 years. The average Body Mass
Index was 27.31. No patient presented metastasis at the time of the
study. The participating women were in clinical stages II and
III.
| Clinical
characteristics | n=19 | Range | Standard
deviation |
|---|
| Age at diagnosis,
years | 52 | 41–71 | 8.63 |
| BMI,
Kg/m2 | 27.31 | 20.78–37.01 | 4.27 |
| Menopause
status |
|
|
|
|
Pre | 10 | 53% |
|
|
Post | 9 | 47% |
|
| Diabetes
mellitusa |
|
|
|
|
Yes | 3 | 16% |
|
| No | 16 | 84% |
|
| Glucose levels,
mg/dl | 101.8 | 91–116 | 7.69 |
| Number of
childrenb |
|
|
|
|
Nulliparous | 0 | 0% |
|
| 1 to
2 | 6 | 32% |
|
|
>3 | 13 | 68% |
|
| Smoking |
|
|
|
|
Yes | 1 | 5% |
|
| No | 18 | 95% |
|
| TNM |
|
|
|
| T1 | 0 | 0% |
|
| T2 | 9 | 47% |
|
| T3 | 7 | 37% |
|
| T4 | 3 | 16% |
|
| N0 | 0 | 0% |
|
| N1 | 13 | 68% |
|
| N2 | 4 | 21% |
|
| N3 | 2 | 11% |
|
| M0 | 0 | 0% |
|
| Clinical stage |
|
|
|
| I | 0 | 0% |
|
| II | 11 | 58% |
|
|
III | 8 | 42% |
|
TNBC sequencing
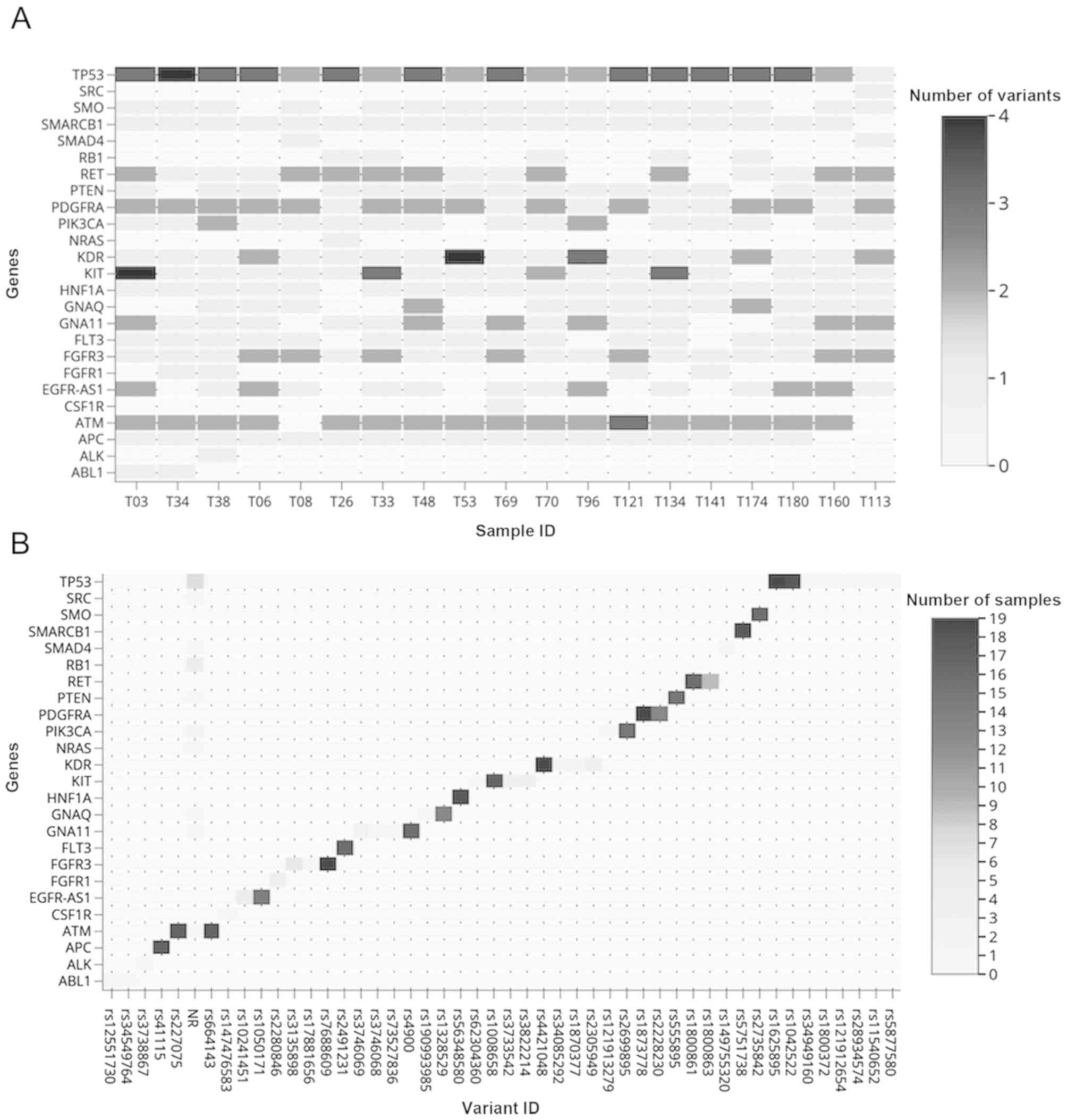
We found 65 variants in 25 of the 48 genes analyzed.
Of these genes, TP53, PIK3CA and FLT3 genes presented
nonsense, missense, stop-gained variants, or variants in the
splicing region with pathogenic significance. In the rest of the
genes, intronic and variants that have been classified as benign
were observed. Fig. 1A shows the
distribution of each gene variants. All the samples presented
between 18 and 26 genetic variants. Fig.
1B shows the distribution of the variants of each gene
analyzed, including the non-reported variants found in ATM,
GNA11, GNAQ, NRAS, PIK3CA, PTEN, RB1, SMAD4, SRC and
TP53. The highest number of variants were found in the
intronic regions (from 10 to 14 per sample) and 49% were in exonic
regions (32/65). An average of 5 synonymous variants were
identified regarding the type of variant, and 2 to 5 missense
variants were found per sample (Fig.
2). TP53 was mutated in all the samples and presented 15
variants, of which 7 (46%) were missense variants. We found four
genetic variants in the PIK3CA gene, including two missense
variants. FLT3 presented one variant in the splicing region
between exons 14 and 15 (Table
II).
 | Table II.Genes with missense, stop gained
variants or splicing region variants. TP53, PIK3CA, and FLT3 were
the genes that presented variants with transcriptional
consequences, in addition to intronic variants. The gene with the
highest number of variants was TP53 with 15 variants; PIK3CA
presented 4 variants and FLT3 presented a variant. |
Table II.
Genes with missense, stop gained
variants or splicing region variants. TP53, PIK3CA, and FLT3 were
the genes that presented variants with transcriptional
consequences, in addition to intronic variants. The gene with the
highest number of variants was TP53 with 15 variants; PIK3CA
presented 4 variants and FLT3 presented a variant.
| Gene
(variants) | Variant | Samples | Exon | HGVSc/HGVSp | Consequence | dbSNP | ClinVar |
|---|
| TP53 (15) | T>C | 19 | − | c.672+62A>G | Intron variant | rs1625895 | Benign |
|
| G>C | 18 | 004/11 | p.Pro72Arg | Missense
variant | rs1042522 | Drug
response |
|
| T>C | 1 | − | c.672+31A>G | Intron variant | rs34949160 | Benign |
|
| T>C | 1 | 006/11 |
c.639A>G(p.=) | Synonymous
variant | rs1800372 | Benign |
|
| G>C | 1 | 008/11 | p.Arg282Gly | Missense
variant | rs28934574 |
Pathogenic/likely |
|
|
|
|
|
|
|
|
pathogenic |
|
| T>C | 1 | 007/11 | p.tyr234cys | Missense
variant |
rs5877580 | b |
|
| C>A | 1 | 005/11 | p.Val157Phe | Missense
variant |
rs121912654 | b |
|
| C>T | 1 | 007/11 | p.Arg248Gln | Missense
variant |
rs11540652 | b |
|
|
A>ACG | 1 | 008/11 |
p.Cys275PhefsTer71 | Frameshift
variant, feature elongation | a | b |
|
| G>A | 1 | 006/11 | p.Arg213Ter | Stop
gained | a | b |
|
| A>C | 1 | 005/11 | p.His179Gln | Missense
variant | a | b |
|
| G>A | 1 | 006/11 | p.Arg196Ter | Stop
gained | a | b |
|
| G>C | 1 | 008/11 | p.Ser269Arg | Missense
variant | a | b |
|
| GT>G | 1 | 008/11 |
p.Asn268ThrfsTer77 | Frameshift variant,
feature truncation | a | b |
|
| CA>C | 1 | 005/11 |
p.Gys135AlafsTer35 | Frameshift
variant, feature truncation | a | b |
| FLT3 (1) | A>G | 16 | − | − | Splice region
variant, intron variant |
rs2491231 | b |
| PIK3CA (4) | A>G | 1 | − |
c.1252-27A>G | Intron
variant | a | b |
|
| G>C | 1 | 21/21 | p.Glu1012Gln | Missense
variant | a | b |
|
| A>G | 1 | 21/21 | p.His1047Arg | Missense
variant | rs121913279 |
Pathogenic/likely pathogenic |
|
| C>A | 15 | − |
c.1059+62C>A | Intron variant |
rs2699895 | b |
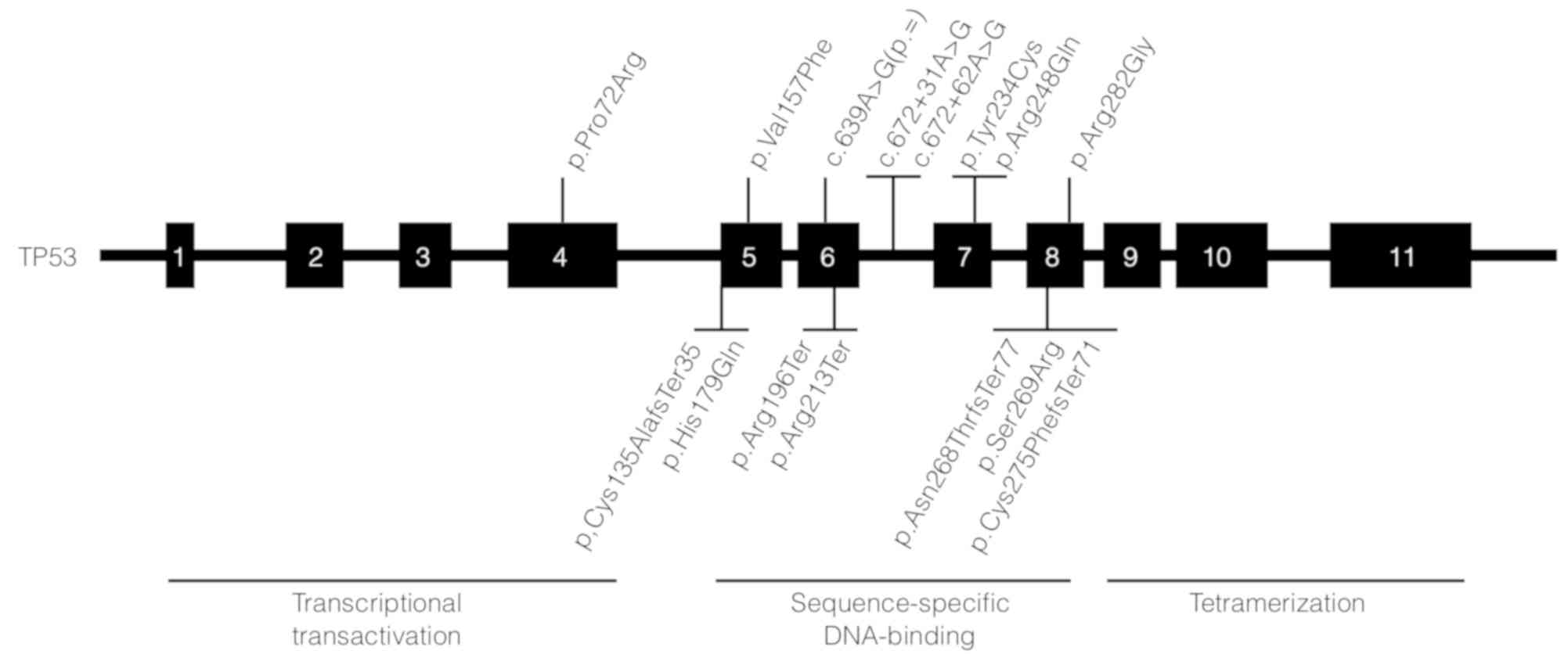
TP53 variants
When comparing the variants found in each sample, we
observed the exonic variant rs1042522 located in the TP53
gene in 94% of the TNBC biopsies (18/19). In addition, we found
rs1625895, rs34949160, rs1800372, rs5877580, rs121912654,
rs28934574 and rs11540652 variants of TP53. The first three
variants, located in intron 6 of the gene, are classified as benign
according to the ClinVar database. The rs5877580, rs121912654, and
rs11540652 variants are probably damaging variants based on
PolyPhen-2 with score s of 0.993, 0.998 and 0.992 respectively. The
rs28934574 is a variant with a possibly damaging score (0.583). We
found 7 non-reported exonic variants. Five of these variants are
SNVs: p.Arg213Ter, p.His179Gln, p.Arg196Ter and p.Ser269Arg. Three
variants correspond to insertion or deletion of a nucleotide:
p.Asn268ThrfsTer77, p.Gys135AlafsTer35 and p.Cys275PhefsTer71. All
these variants affect part of the DNA binding domain of p53.
FLT3
The rs2491231 variant of the FLT3 gene, which
has not been reported before for TNBC, was found in 84% (16/19) of
the samples. This variant is located in the splicing region between
exons 14 and 15.
PIK3CA
Two missense variants were observed in
PIK3CA: The exonic variant rs121913279 of the PIK3CA
gene was detected in one sample. This variant has been classified
as Pathogenic/Likely pathogenic in ClinVar and the exonic variant,
p.Glu1012Gln, that has not yet been reported.
Discussion
NGS is a very useful tool in disease
characterization of multigenic origin such as cancer, where the
accumulation of a series of mutations in several genes is the key
to tumor development. The ability of NGS to evaluate the mutational
status of a relevant set of oncogenes and tumor suppressor genes in
a single test, such as those evaluated in this work could be
helpful to identify TNBC mutation drivers to design better
diagnostic and therapeutic strategies. TruSeq Amplicon Cancer Panel
was validated in 2015 (19), with the
purpose of detecting somatic mutations through hundreds of
mutational hotspots of essential genes related to cancer, including
PIK3CA, TP53, and EGFR. Mutations in these genes are
related to cancer and are involved in many cellular pathways.
Previously, a difference in the pattern of somatic
mutations among the intrinsic subtypes of BC has been observed
(11). We found variants in 25 genes,
of which variants in TP53, PIK3CA and FLT3 showed
missense and non-sense variants, or variants in the splicing
region. Tumors with a triple negative phenotype have a high
prevalence of mutations in TP53 (80%) vs. luminal breast
cancer (12%). In this work, we analyze the triple negative
phenotype, observing that the gene with a high prevalence of exonic
variants was TP53, as previously indicated (20,21).
Although in 2012 it was reported that most of the TP53
variants in basal tumors were non-sense and frameshift (11), Shah and Weisman et al (10,22),
independently reported missense mutations in TP53 on TNBC
whereas we found that 7 of 15 variants were missense type.
TP53 is a tumor suppressor gene that encodes for a
multifunctional DNA binding protein that regulates the
transcription of hundreds of genes related to cell cycle
regulation, differentiation, and apoptosis (23–25). The
rs1042522 of TP53 corresponds to an arginine (CGC) by a
proline (CCC) change in codon 72 of exon 4. The proteins p53Arg72
and p53Pro72 do not differ in their ability to bind to DNA in a
sequence-specific manner, but differ in other ways (26): p53Arg72 protein induces faster
apoptosis and suppresses transformation more efficiently than the
variant p53Pro72 (27,28). This variant was found in 18 of the 19
analyzed samples, and it is classified in ClinVar as drug response
variant. TNBC, unlike the other BC subtypes, responds better to
chemotherapy (9). Our results suggest
that when p53Arg72 is present, apoptosis is more efficiently
activated. However, the association of this TP53 variant
with the risk of various types of cancer, including BC, remains
controversial (29,30). The missense TP53 variants
rs121912654 and rs28934574 have been previously associated with
hepatocellular carcinoma (31) and
osteosarcoma (32), respectively. The
rs121912654 located in exon 5 causes the substitution of valine by
phenylalanine. The rs28934574 is located in exon 8, and the final
product is a substitution of tryptophan by arginine. The rs28934574
variant meets the criteria published in 2013 by the American
College of Medical Genetics (ACMG) (33) as a variant that is recommended to
inform the patient.
We found seven unreported variants in TP53
(p.Arg213Ter, p.His179Gln, p.Arg196Ter, p.Ser269Arg,
p.Asn268ThrfsTer77, p.Gys135AlafsTer35 and p.Cys275PhefsTer71) in
exons 5, 6, 7 and 8, which affect the DNA binding domain of the
protein. These results match with those reported in 2014 by
Silwal-Pandit et al (34).
Their results conclude that more than 80% of mutations in BC are
grouped in exons 5–8 (the rest was eliminated). They observed that
the p.Arg213Ter variant was located in a hotspot area of basal
tumors. We found this variant in 1 of 9 patients. It would be
important to analyze whether these changes affect the function of
p53.
PIK3CA codes for the catalytic subunit p110
alpha (p110α) of the phosphatidylinositol 3-kinase enzyme (PI3K)
(35). The PI3K signaling pathway is
essential for several cellular processes, including cell growth,
proliferation, migration, and survival (36). Some works have shown gene
amplifications, deletions and, more recently, missense mutations in
the PIK3CA gene in human cancers, including colon, liver,
stomach, brain, lung and breast cancers (37). We found four variants of the
PIK3CA gene, two of them already identified as rs2699895 and
rs121913279. The first is a variant located in the intron 5. The
second is a missense variant. The ancestral allele is an adenine
changed by guanine, which represents a change of histidine by
arginine in position 1047 of the protein. In ClinVar, it is
classified as probably pathogenic since it has been associated with
breast and colorectal cancer, melanoma and non-small cell lung
cancer. We also found two not previously reported variants,
c.1252-27A>G, and p.Glu1012Gln. The first one is located in an
intronic region and the second one is located in exon 21, therefore
a change from glutamic acid to glutamine is observed. In previous
works, it has been reported that PIK3CA is the second most
frequently mutated gene in TNBC (10,38).
PIK3CA presents at least one variant in 15 of the 19 samples
analyzed, and two samples have an exonic gene variant. It has been
reported that the frequency of mutations in PIK3CA in BC is
10–30% (39), what is similar to our
data. However, most of the mutations reported are located in exons
9 and 20 (39). These variants
produce a gain of function and transformation capacity in the
PIK3CA protein, thus it is relevant to investigate the role
of the variants that we found in this work. Lips and colleagues
reported that mutations in PIK3CA are associated with
mutations in BRCA1 (40). In
this work, we do not investigate the BRCA1 gene, so it would
be interesting to analyze this theory in these patients.
In Mexico, Vaca-Paniagua et al (41) reported TP53 and RB1 as
the most frequently mutated genes in TNBC by complete exome
sequencing. Differences in the results found in our work with
respect to RB1 could be attributed to the design of the
study. In the work of Vaca-Paniagua, the whole exome was sequenced
in paraffin embedded tissues (n=12), while we sequenced the hotspot
regions of the 48 genes in frozen tissue biopsies. Although they
obtained a greater coverage of the gene, we have a greater depth of
reading. To properly compare both studies not only an
intrapopulation analysis is needed, but also an interpopulation
comparison is required. Finally, differences also could be
explained by the clinical criteria of selection, geographic origin
of the patient and his ancestors, analytical methods, sample size,
exposure to environmental risk factors and dietary, among others
factors.
In addition, we found the rs2491231 variant of
FLT3, which corresponds to an SNV type change for which
there is no evaluation in the ClinVar database. This variant is
found in a region of splicing between exons 14 and 15 that is part
of the region that codes for the cytoplasmic domain of the protein.
The FLT3 gene encodes a tyrosine kinase receptor class III
regulating hematopoiesis. When this receptor is activated, it
phosphorylates and activates multiple cytoplasmic effector
molecules in pathways involved in apoptosis, proliferation, and
differentiation of hematopoietic cells in the bone marrow. The most
common reported mutations are located in exons 14 or 20 and result
in constitutive activation of this receptor, which are observed in
acute myeloid leukemia and acute lymphoblastic leukemia (42,43). It
would be essential to evaluate the effect of the FLT3
variant on TNBC.
It would be interesting to determine the pathogenic
significance of the genetic variants reported in this work in TNBC,
because these genes are potential therapeutic targets. Currently,
new compounds with different specificity and potency are being
developed, targeting different components of the PI3K/AKT/mTOR
pathway (44), small molecule
compounds that specifically target the mutant p53 (45,46) and
compounds that inhibit tyrosine kinase enzymes, such as FLT3
(47). Mutations in DNA are not the
only form of gene regulation; it is important to consider some
other molecular events, including Copy Number Variation,
chromosomal and epigenetic alterations, as well as the role that
play micro RNAs (miRNAs) and non-coding RNA (ncRNA) (48).
In the present study, the diagnosis of TNBC was the
only criterion for the variant analysis, other factors, such as
age, ethnicity as well as the mutational signature will be
considered in a future study, including damage to DNA and its
repair components (49). Although for
this study we do not have healthy tissue and our sample number is
small, we can draw some conclusions. First, the mutation spectrum
remains diverse even in a carefully selected and untreated group of
patients with TNBC. All samples were from the same institution and
the laboratory procedures were carefully monitored. Our results
strongly suggest that each tumor has its unique molecular
composition. However, it is observed that the total of the biopsies
studied have at least two variants in the TP53 gene. The
rs1042522 drug responsive variant is the most representative, since
it was found in 94% of the samples analyzed. We also found seven
previously unreported variants with probable deleterious
characteristics of the p53 tumor suppressor protein. The variant
rs2491231 of the FLT3 gene was identified in 84% (16/19) of
the samples, which has not been reported before for TNBC.
In conclusion, we found intron, missense, stop
gained and splicing variants in TP53, PIK3CA, and
FLT3 genes. Some of these variants have not been reported.
Studies should be carried out to elucidate if they have a role in
the development of TNBC y and their possible role as therapeutic
targets. It is important to validate the presence of these variants
in a large cohort that includes healthy tissue and non TNBC tissue
as well as in cell culture to evaluate their impact on diagnosis,
prognosis and management of such aggressive TBNC.
Acknowledgements
The authors would like to thank Mrs Patricia
Villarreal of Unidad de Genomica, Centro de Investigacion y
Desarrollo en Ciencias de la Salud, Universidad Autonoma de Nuevo
Leon for her technical assistance.
Funding
The present study was partially supported by the
SS/IMSS/ISSSTE-CONACYT-2011-162301. GIUP and SKSF had scholarships
from CONACYT (CVU nos. 369773 and 217104, respectively).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
GIUP conducted the experiments, acquired, analyzed
and interpreted the data, and drafted the manuscript. SKSF designed
the study, conducted the experiments, analyzed and interpreted the
data, and critically revised the manuscript. CNSD made substantial
contributions to the conception of the study, and drafted and
critically revised the manuscript. SCH, GMM, JFGG and JVG as
clinicians, selected the patients, performed the biopsies to obtain
the tissue samples, collected the clinical information from medical
records and contributed to the design of the study. PRF and ARM as
geneticists selected the patients and participated in the
interpretation of data. JRBW and LEOR assisted in technical support
during the experimental work and made substantial contributions in
the analysis and intepretation of the data. GSGM, ABQ, OBQ and RGG
as pathologists, conducted the histopathological diagnosis of the
patients. ROL made substantial contributions to the conception and
design of study, and drafted and critically revised the
manuscript.
Ethics approval and consent to
participate
The protocol and informed consent was approved by
the Ethics and Research Committee of the Faculty of Medicine
(Universidad Autonoma de Nuevo Leon), with registration number
BI11-005.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soto-Perez-de-Celis E and Chavarri-Guerra
Y: National and regional breast cancer incidence and mortality
trends in Mexico 2001–2011: Analysis of a population-based
database. Cancer Epidemiol. 41:24–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Spitale A, Mazzola P, Soldini D,
Mazzucchelli L and Bordoni A: Breast cancer classification
according to immunohistochemical markers: Clinicopathologic
features and short-term survival analysis in a population-based
study from the South of Switzerland. Ann Oncol. 20:628–635. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sotiriou C and Pusztai L: Gene-expression
signatures in breast cancer. N Engl J Med. 360:790–800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Weigelt B, Baehner FL and Reis-Filho JS:
The contribution of gene expression profiling to breast cancer
classification, prognostication and prediction: A retrospective of
the last decade. J Pathol. 220:263–280. 2010.PubMed/NCBI
|
|
6
|
Dietze EC, Sistrunk C, Miranda-Carboni G,
O'Regan R and Seewaldt VL: Triple-negative breast cancer in
African-American women: Disparities versus biology. Nat Rev Cancer.
15:248–254. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carey LA, Dees EC, Sawyer L, Gatti L,
Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML and Perou
CM: The triple negative paradox: Primary tumor chemosensitivity of
breast cancer subtypes. Clin Cancer Res. 13:2329–2334. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, et al: Response to neoadjuvant therapy and long-term
survival in patients with triple-negative breast cancer. J Clin
Oncol. 26:1275–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shah SP, Roth A, Goya R, Oloumi A, Ha G,
Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al: The clonal
and mutational evolution spectrum of primary triple-negative breast
cancers. Nature. 486:395–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Metzker ML: Sequencing technologies-the
next generation. Nat Rev Genet. 11:31–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kan Z, Jaiswal BS, Stinson J, Janakiraman
V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al:
Diverse somatic mutation patterns and pathway alterations in human
cancers. Nature. 466:869–873. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wagle N, Emery C, Berger MF, Davis MJ,
Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE,
Hahn WC, et al: Dissecting therapeutic resistance to RAF inhibition
in melanoma by tumor genomic profiling. J Clin Oncol. 29:3085–3096.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hrstka R, Coates PJ and Vojtesek B:
Polymorphisms in p53 and the p53 pathway: Roles in cancer
susceptibility and response to treatment. J Cell Mol Med.
13:440–453. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mroz EA, Tward AD, Pickering CR, Myers JN,
Ferris RL and Rocco JW: High intratumor genetic heterogeneity is
related to worse outcome in patients with head and neck squamous
cell carcinoma. Cancer. 119:3034–3042. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gerlinger M, Rowan AJ, Horswell S, Math M,
Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N,
Stewart A, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Macdonald SM, Harris EE, Arthur DW, Bailey
L, Bellon JR, Carey L, Goyal S, Halyard MY, Horst KC, Moran MS and
Haffty BG: ACR appropriateness criteria® locally
advanced breast cancer. Breast J. 17:579–585. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Simen BB, Yin L, Goswami CP, Davis KO,
Bajaj R, Gong JZ, Peiper SC, Johnson ES and Wang ZX: Validation of
a next-generation-sequencing cancer panel for use in the clinical
laboratory. Arch Pathol Lab Med. 139:508–517. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oren M and Rotter V: Mutant p53
gain-of-function in cancer. Cold Spring Harb Perspect Biol.
2:a0011072010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Muller PA and Vousden KH: p53 mutations in
cancer. Nat Cell Biol. 15:2–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weisman PS, Ng CK, Brogi E, Eisenberg RE,
Won HH, Piscuoglio S, De Filippo MR, Ioris R, Akram M, Norton L, et
al: Genetic alterations of triple negative breast cancer by
targeted next-generation sequencing and correlation with tumor
morphology. Mod Pathol. 29:476–488. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu H and el-Gewely MR: P53-responsive
genes and the potential for cancer diagnostics and therapeutics
development. Biotechnol Annu Rev. 7:131–164. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Levine AJ: p53, the cellular gatekeeper
for growth and division. Cell. 88:323–331. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wallace-Brodeur RR and Lowe SW: Clinical
implications of p53 mutations. Cell Mol Life Sci. 55:64–75. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jones JS, Chi X, Gu X, Lynch PM, Amos CI
and Frazier ML: p53 polymorphism and age of onset of hereditary
nonpolyposis colorectal cancer in a Caucasian population. Clin
Cancer Res. 10:5845–5849. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thomas M, Kalita A, Labrecque S, Pim D,
Banks L and Matlashewski G: Two polymorphic variants of wild-type
p53 differ biochemically and biologically. Mol Cell Biol.
19:1092–1100. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dumont P, Leu JI, Della Pietra AC III,
George DL and Murphy M: The codon 72 polymorphic variants of p53
have markedly different apoptotic potential. Nat Genet. 33:357–365.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soleimani A, Rahmani Y, Farshchian N,
Delpisheh A, Khassi K, Shahmohammadi A and Amirifard N: The
evaluation of p53 polymorphism at codon 72 and association with
breast cancer in iran: A systematic review and meta-analysis. J
Cancer Prev. 21:288–293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bansal A, Das P, Kannan S, Mahantshetty U
and Mulherkar R: Effect of p53 codon 72 polymorphism on the
survival outcome in advanced stage cervical cancer patients in
India. Indian J Med Res. 144:359–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bressac B, Kew M, Wands J and Ozturk M:
Selective G to T mutations of p53 gene in hepatocellular carcinoma
from southern Africa. Nature. 350:429–431. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Smith-Sørensen B, Gebhardt MC, Kloen P,
McIntyre J, Aguilar F, Cerutti P and Børresen AL: Screening for
TP53 mutations in osteosarcomas using constant denaturant gel
electrophoresis (CDGE). Hum Mutat. 2:274–285. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Green RC, Berg JS, Grody WW, Kalia SS,
Korf BR, Martin CL, McGuire AL, Nussbaum RL, O'Daniel JM, Ormond
KE, et al: CORRIGENDUM: ACMG recommendations for reporting of
incidental findings in clinical exome and genome sequencing. Genet
Med. 19:6062017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Silwal-Pandit L, Vollan HK, Chin SF, Rueda
OM, McKinney S, Osako T, Quigley DA, Kristensen VN, Aparicio S,
Børresen-Dale AL, et al: TP53 mutation spectrum in breast cancer is
subtype specific and has distinct prognostic relevance. Clin Cancer
Res. 20:3569–3580. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Volinia S, Hiles I, Ormondroyd E, Nizetic
D, Antonacci R, Rocchi M and Waterfield MD: Molecular cloning, cDNA
sequence, and chromosomal localization of the human
phosphatidylinositol 3-kinase p110 alpha (PIK3CA) gene. Genomics.
24:472–477. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Katso R, Okkenhaug K, Ahmadi K, White S,
Timms J and Waterfield MD: Cellular function of phosphoinositide
3-kinases: Implications for development, homeostasis, and cancer.
Annu Rev Cell Dev Biol. 17:615–675. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karakas B, Bachman KE and Park BH:
Mutation of the PIK3CA oncogene in human cancers. Br J Cancer.
94:455–459. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cancer Genome Atlas Network. Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Samuels Y, Wang Z, Bardelli A, Silliman N,
Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al:
High frequency of mutations of the PIK3CA gene in human cancers.
Science. 304:5542004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lips EH, Michaut M, Hoogstraat M, Mulder
L, Besselink NJ, Koudijs MJ, Cuppen E, Voest EE, Bernards R,
Nederlof PM, et al: Next generation sequencing of triple negative
breast cancer to find predictors for chemotherapy response. Breast
Cancer Res. 17:1342015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vaca-Paniagua F, Alvarez-Gomez RM,
Maldonado-Martinez HA, Pérez-Plasencia C, Fragoso-Ontiveros V,
Lasa-Gonsebatt F, Herrera LA, Cantú D, Bargallo-Rocha E, Mohar A,
et al: Revealing the molecular portrait of triple negative breast
tumors in an understudied population through omics analysis of
formalin-fixed and paraffin-embedded tissues. PLoS One.
10:e01267622015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Uras IZ, Walter GJ, Scheicher R, Bellutti
F, Prchal-Murphy M, Tigan AS, Valent P, Heidel FH, Kubicek S,
Scholl C, et al: Palbociclib treatment of FLT3-ITD+ AML cells
uncovers a kinase-dependent transcriptional regulation of FLT3 and
PIM1 by CDK6. Blood. 127:2890–2902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Adamia S, Bar-Natan M, Haibe-Kains B,
Pilarski PM, Bach C, Pevzner S, Calimeri T, Avet-Loiseau H, Lode L,
Verselis S, et al: NOTCH2 and FLT3 gene mis-splicings are common
events in patients with acute myeloid leukemia (AML): New potential
targets in AML. Blood. 123:2816–2825. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Costa RLB, Han HS and Gradishar WJ:
Targeting the PI3K/AKT/mTOR pathway in triple-negative breast
cancer: A review. Breast Cancer Res Treat. 169:397–406. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Parrales A and Iwakuma T: Targeting
oncogenic mutant p53 for cancer therapy. Front Oncol. 5:2882015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Synnott NC, Murray A, McGowan PM, Kiely M,
Kiely PA, O'Donovan N, O'Connor DP, Gallagher WM, Crown J and Duffy
MJ: Mutant p53: A novel target for the treatment of patients with
triple-negative breast cancer? Int J Cancer. 140:234–246. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Stone RM, Mandrekar SJ, Sanford BL,
Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Döhner K,
Marcucci G, et al: Midostaurin plus chemotherapy for acute myeloid
leukemia with a FLT3 mutation. N Engl J Med. 377:454–464. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bautista RR, Gómez AO, Miranda AH, Dehesa
AZ, Villarreal-Garza C, Ávila-Moreno F and Arrieta O: Correction
to: Long non-coding RNAs: Implications in targeted diagnoses,
prognosis, and improved therapeutic strategies in human non- and
triple-negative breast cancer. Clin Epigenetics. 10:1062018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nik-Zainal S, Van Loo P, Wedge DC,
Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J,
Ramakrishna M, et al: The life history of 21 breast cancers. Cell.
149:994–1007. 2012. View Article : Google Scholar : PubMed/NCBI
|