Introduction
HER2-positive breast cancer accounts for
approximately 15–20% of all breast cancers and has been associated
with poor prognosis and decreased survival (1). The outcome of patients with early or
HER2-positive metastatic breast cancer (MBC) has been greatly
improved following the introduction of trastuzumab, a humanised
monoclonal antibody targeting HER2. Despite the success of
trastuzumab, the median overall survival of patients with
HER2-positive MBC is still limited to 32–42 months (2,3).
Furthermore, a significant number of HER2-positive MBC patients
exhibit de novo or acquired resistance towards trastuzumab
therapy (4). Apart from HER2 itself,
there are currently no clinically established biomarkers
distinguishing patients who will benefit from trastuzumab treatment
from those who will not (2,5–7). With
later generations of HER2-targeted drugs now being available, the
search for prognostic and predictive biomarkers guiding the patient
to the most effective treatment has become increasingly
important.
Trastuzumab mainly exerts its anti-tumoural effects
via the stimulation of antibody-dependent cell-mediated
cytotoxicity (ADCC), the inhibition of ectodomain cleavage, and the
inhibition of ligand-independent HER2 signalling (8–10). The
latter is dependent on the dimerisation of two receptor units,
either homo- or hetero-dimerisation with another HER-receptor
family member, with HER2-HER3 being the most potent dimer (11).
HER2 overexpression may induce downstream activation
of the phosphoinositide 3-kinase (PI3K)/Akt pathway, a key pathway
in carcinogenesis leading to cell proliferation, cell survival, and
upregulated protein synthesis (12).
The activation of PI3K initiates a downstream chain of
phosphorylation events including Akt, mTOR, S6K1 and S6K2, and
ultimately the upregulation of Cyclin D1, which regulates the
G1/S-phase transition through binding to CDK 4/6 and
phosphorylation of Rb (13–15). It is hypothesised that dysregulation
of PI3K/Akt signalling pathway mediators plays a key role in
trastuzumab resistance. PTEN deficiency and/or upregulated Src
signalling have been implicated in de novo and acquired
trastuzumab resistance in vitro (16–18). Src
is activated by upstream receptor tyrosine kinases such as Met or
EGFR, but it is also affected by intracellular protein tyrosine
phosphatases e.g. protein tyrosine phosphatase non-receptor type 1
(PTPN1) and 2 (PTPN2) (19,20). PTPN1 expression status appears to have
no or limited impact in HER2-positive breast cancer or breast
cancer patients undergoing neoadjuvant chemotherapy, whilst PTPN2
has been reported to be frequently lost in breast cancer,
correlating with poor outcome (21–24).
In the present study, we explored the prognostic
values of intra-tumoural biomarkers believed to be involved in
trastuzumab resistance in a long-term follow-up cohort of the first
consecutive patients receiving palliative trastuzumab treatment at
our department.
Materials and methods
Patient material
All patients diagnosed with HER2+ MBC and treated
with trastuzumab at Linköping University Hospital between 2000 and
2007 were included in the cohort. Treatment decisions were based on
HER2-positive disease as determined by IHC and/or FISH diagnostics
performed on primary tumours or, if available, biopsies from
metastatic lesions. Exclusion criteria from the present study were
incomplete key data (i.e., data related to survival, HER2 status,
and/or trastuzumab treatment) and previous anti-HER2 treatment.
Primary tumour material was available for all patients and material
from metastases was available in one-third of the cases.
Formalin-fixed paraffin-embedded (FFPE) tissue specimens, obtained
from surgery, were stored at room temperature until DNA extraction.
Two reviewers (SE and AM) extracted clinical and pathological data
from the medical records. As per clinical routine, results from
metastatic lesions were used for final analysis when available. The
primary endpoint was overall survival (OS), defined as the time
from start of trastuzumab treatment until time of death. The
secondary endpoint was progression-free survival (PFS) as measured
from the start of trastuzumab treatment to time of death, or at the
first sign of radiological and/or clinical progression. Analyses
were performed and reported following the REMARK guidelines
(25).
DNA extraction
Genomic DNA was extracted from FFPE tumour tissue
specimens containing at least 50% tumour cells using the QIAamp DNA
FFPE Tissue kit (Qiagen GmbH, Hilden, Germany). The manufacturer's
protocol was followed except for the paraffin removal procedure,
which was performed in Tissue-Tek Tissue-Clear (Sakura Finetek
Europe B.V., Flemingweg, The Netherlands). DNA concentration was
measured using QuantiFluor® ONE dsDNA Dye kit (Promega
Corporation, Madison, WI, USA) on a Quantus™ Fluorometer (Promega
Corporation). DNA samples were stored at −70°C during long-term
storage and at −20°C for short-term storage.
Droplet digital polymerase chain
reaction (PCR)
Copy numbers of CCND1, ERBB2, MET, PTPN2, and
RP6SKB1 were measured with droplet digital PCR (ddPCR) using
AP3B1 as a reference gene. The amplicon length of the
reference gene was matched with the amplicon length of the gene of
interest; two different primer-probe assays were therefore used for
AP3B1. The choice of reference gene and the protocol were
previously described, with the appropriate annealing temperatures
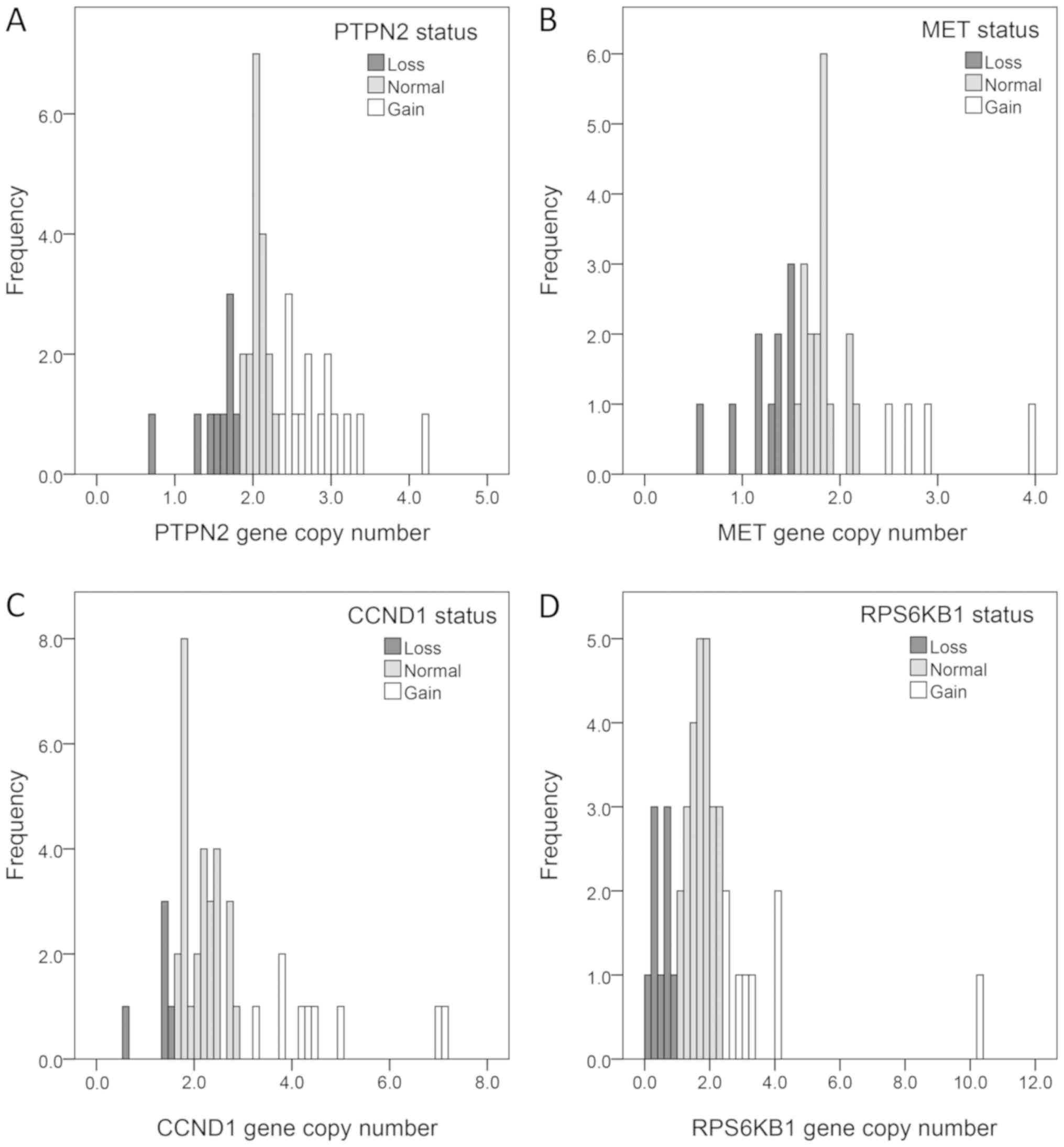
as shown in Table I (26). Gene copy numbers were determined by
analysing the distribution of the ddPCR values, with care taken for
the distribution being skewed towards two gene copies due to any
non-tumour cells in the samples. PTPN2, MET, RPS6KB1, and
CCND1 gene copy number analyses were approached as follows:
less than two gene copies was considered as gene copy loss, two
copies as normal, and three or more was considered as gene copy
gain. The samples were subsequently dichotomised into gain/no gain
(≥3 vs. <3 copies; Fig. 1). As
patients in this cohort were all HER2-positive, ERBB2 gene
copy number was divided into low- and high-grade amplification.
After the level of ERBB2 amplification was assessed, tumours
were partitioned into tertiles, with the lower tertile showing less
than six gene copies and the upper tertile showing more than 16
gene copies. This was later dichotomised and the cut-off for
high-grade ERBB2 amplification was set to six or more gene
copies.
 | Table I.Gene copy number assay overview. |
Table I.
Gene copy number assay overview.
| Gene | Chromosome | ddPCR assay | Company | Annealing
temperature (°C) | Amplicon length
(nt) |
|---|
| MET | 7q31 | dHsaCP2500321 | Bio-Rad
Laboratories, Inc. | 60 | 62 |
| ERBB2 | 17q12 | FP:
5′-GGTCCTGGAAGGCCACAAGG-3′ | Sigma-Aldrich; | 61 | 80 |
|
|
| RP:
5′-GGTTTTCCCACCACATCCTCt-3′ | Merck KGaA |
|
|
|
|
| Probe:
5′ACACAACACATCCCCCTCCTTGACTA TCAA-3′ | Applied Biosystems;
Thermo Fisher Scientific, Inc. |
|
|
| CCND1 | 11q13 | dHsaCP2500371 | Bio-Rad
Laboratories, Inc. | 55 | 64 |
| PTPN2 | 18p11 | FP:
5′-AAGCCCACTCCGGAAACTAAA-3′ | Sigma-Aldrich; | 64.2 | 65 |
|
|
| RP:
5′-AAACAAACAACTGTGAGGCAATCTA-3′ | Merck KGaA |
|
|
|
|
| Probe:
5′-TGAGGCTCGCTAACC-3′ | Applied Biosystems;
Thermo Fisher Scientific, Inc. |
|
|
| RPS6KB1 | 17q23.1a | Hs04469680_cn | Thermo Fisher
Scientific, Inc. | 60 | 87 |
| AP3B1 | 5q14.1 | dHsaCP2500348 | Bio-Rad
Laboratories, Inc. |
| 60 |
|
|
| dHsaCP1000001 | Bio-Rad
Laboratories, Inc. |
| 85 |
Tissue microarray
Sections cut from tumour donor blocks were stained
with haematoxylin and eosin, and morphologically representative
regions were selected in each tumour sample. Three tissue cores
with a diameter of 0.8 mm were taken from these regions and mounted
in recipient blocks, forming tissue microarrays (TMAs) created with
a manual arrayer (Beecher Instruments Inc., Sun Prairie, WI, USA).
The TMA blocks were cut into 5 µm sections and transferred to
frost-coated glass slides for immunohistochemistry (IHC).
Immunohistochemistry
Deparaffinisation, rehydration, and antigen
retrieval were performed on the TMA sections using DAKO PT link
(Dako; Agilent Technologies GmbH, Waldbronn, Germany) with either
high or low pH buffer (EnVision FLEX target retrieval solution
low/high pH; Dako; Agilent Technologies GmbH) in most cases.
Otherwise, these steps were performed in xylene, serial dilutions
of ethanol, and in 10 mM citrate buffer (pH 6) in a pressure cooker
(Digital Decloaking Chamber, Biocare Medical, Concord, CA, USA),
respectively. The TMA sections were blocked in serum-free protein
block (Spring Bioscience, Fremont, CA, USA) for 10–60 min followed
by overnight incubation at 4°C with the primary antibody. The
sections were then incubated for 30 min at room temperature with
the appropriate secondary antibody (EnVision+System-HRP; Dako;
Agilent Technologies GmbH), stained with 3′-diaminobenzidine
tetrahydrochloride (DAB/H2O2) solution,
counterstained with Mayer's Haematoxylin (Fluka Analytical;
Honeywell Specialty Chemicals Seelze GmbH, Seelze, Germany), and
dehydrated using a series of increasing ethanol dilutions.
Scoring was done by two independent investigators
per antigen, without any knowledge of clinical data. The expression
levels were based on staining intensity (negative, weak, moderate,
and strong) and percentage of positive cells. Staining intensity
was evaluated in the nucleus, the cell membrane, and the cytoplasm
depending on the localisation of the respective protein. If
discordant scorings were found, the slide was re-examined in tandem
and a joint score was made. The antibodies, the conditions used,
and how the scoring was performed are summarised in Table II. Phospho-specificity of the
pAkt-S473, p4EBP1-S65, pMet-Y1349, and pS6K-T389 antibodies was
previously validated in our lab (26–29).
 | Table II.Overview of the antibodies used for
immunohistochemistry. |
Table II.
Overview of the antibodies used for
immunohistochemistry.
| Protein | Antibody | Company | Antigen
retrieval | Blocking time | Antibody
dilution | Scoring
nuclear | Scoring
cytoplasm | Scoring
membrane |
|---|
| p4EBP1-S65 | Phospho-4E-BP1 | Cell | 10 mM citrate
buffer, | 10 min | 1:100 | Low: Weak-moderate
in >50% | Low: Weak | N/A |
|
| (Ser65)
(174A9) | signaling | pH6 pressure
cooker |
|
| High: Strong in
>50% | High: Moderate in
>50%-strong in >50% |
|
| pAkt-T308 | Phospho-Akt
(Thr308) (244F9) | Cell signaling | 10 mM citrate
buffer, pH6 pressure cooker | 10 min | 1:25 | Low: Negative High:
Positive | Low: Negative High:
Weak/moderate/strong | N/A |
| pAkt-S473 | Phopsho-Akt
(Ser473) High: Weak/moderate/strong | Cell signaling | 10 mM citrate
buffer, pH6 pressure cooker | 10 min | 1:50 | Low: Negative High:
Weak/strong | Low: Negative High:
Weak/moderate/strong | N/A |
| Cyclin D1 | Cyclin D1 SP4 | ThermoFisher
scientific | PT-link, high
pH | 10 min | 1:100 | Low: Negative/weak
High: Moderate/strong | Low: Negative High:
Positive | N/A |
| HER3 | erbB3/Her3 clone
5A12 | Nano tools | PT-link, high
pH | 60 min | 1:20 | N/A | Low:
Negative/moderate High: Strong | Low: Negative High:
Weak/moderate-strong |
| HER4 | ERBB4 polyclonal
antibody | Abnova | PT-link, low
pH | 60 min | 1:20 | N/A | Low: Negative/weak
High: Moderate/strong | Low: Negative-weak
High: Moderate/strong |
| Met | Met (D1C2)
XP® | Cell signaling | PT-link, high
pH | 60 min | 1:100 | N/A | Low: Negative/weak
High: Moderate/strong | Low: <10% High:
>10% |
| pMet-Y1349 | Anti-Met (c-Met)
(phospho Y1349) | Abcam | PT-link, high
pH | 60 min | 1:25 | N/A | Low: Negative High:
Weak/moderate-strong | Low: <10% High:
>10% |
| PTEN | PTEN (138G6) | Cell signaling | 10 mM citrate
buffer, pH6 pressure cooker | 10 min | 1:50 | N/A | Low: Negative/weak
High: Normal/strong | N/A |
| PTPN2 | PTPN2 polyclonal
antibody | Proteintech | PT-link, low
pH | 60 min | 1:400 | N/A | Low: Negative/weak
High: Moderate/strong | N/A |
| S6K1 | P70 S6 kinase
(49D7) | Cell signaling | PT-link, low
pH | 10 min | 1:100 | Low: Weak-moderate
in >50% High: Strong in >50% | Low: Weak High:
Moderate or strong in >50% | N/A |
| pS6K1-T389 | Phospho-p70 S6
kinase (Thr389) (1A5) | Cell signaling | PT-link, low
pH | 10 min | 1:100 | Low: 0–25% High:
26–100% | Low: Negative High:
Positive | N/A |
Statistics
Statistical analyses were carried out using IBM SPSS
Statistics v.23 (IBM Corp., Armonk, NY, USA). Univariable hazard
ratios (HR) were calculated using Cox regression for all biomarkers
and clinicopathological prognostic factors. Parameters found to be
significant in the univariable analyses were further analysed with
multiple Cox regression analysis. Comparisons of categorical
outcomes were analysed using Fisher's exact test. Kaplan-Meier
curves were used to visualise survival and progression-free
survival and differences between groups were estimated with
Mantel-Cox. No imputations for missing data were used and P<0.05
was considered to indicate a statistically significant
difference.
Results
Fifty patients with HER2-positive MBC patients who
received de novo-treatment with trastuzumab at our
institution between 2000 and 2007 were identified and 46 of these
were included in this study. Three patients were excluded due to
incomplete clinical data and one patient was excluded due to
HER2-negative disease. Patient follow-up was performed in 2016 and
the results are summarised in Table
III. The age of the included patients ranged between 35 and 79
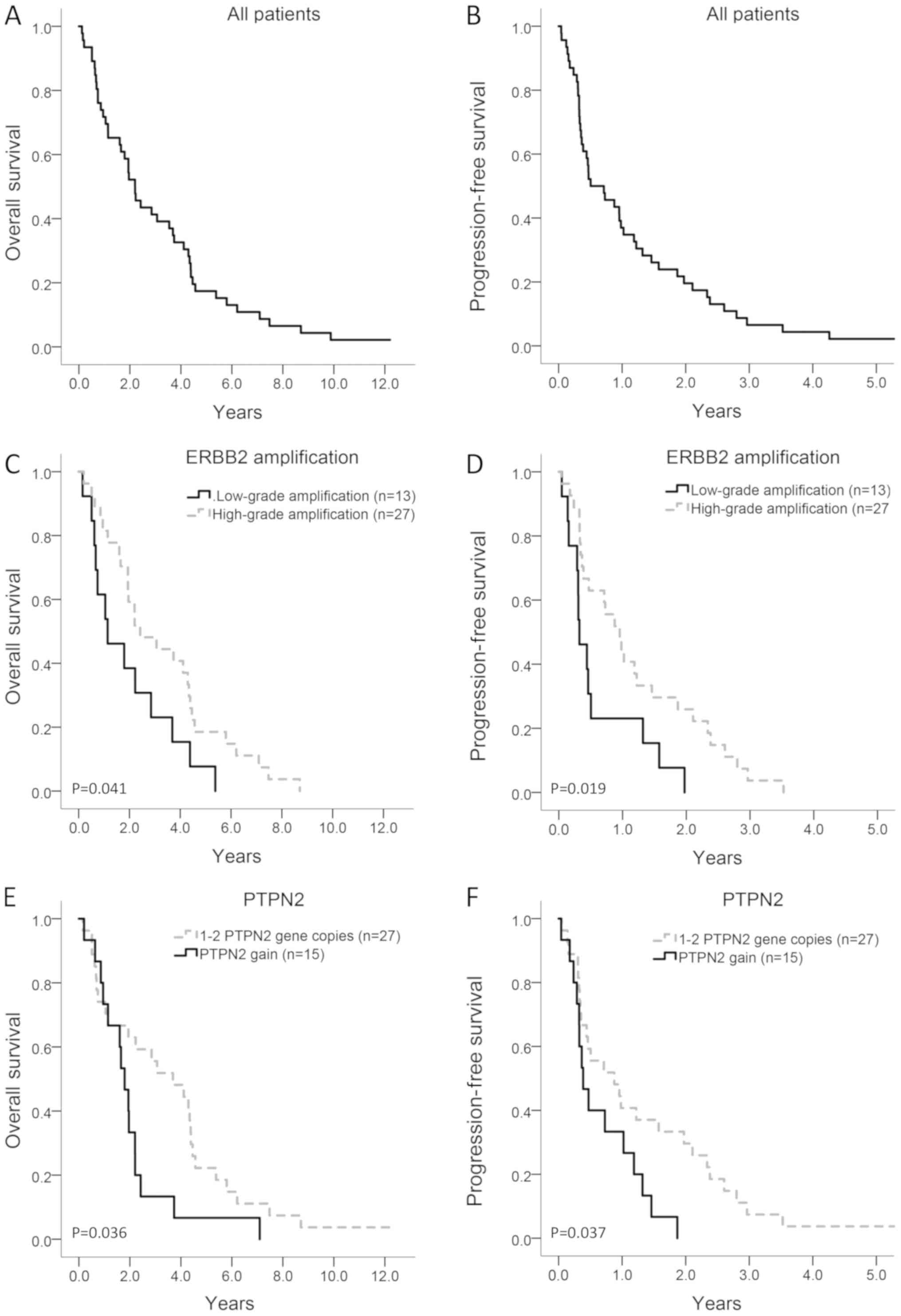
years with a median age of 57 years. Median OS and PFS were 2.23
years (95% CI: 1.63–2.84) and 0.51 years (95% CI: 0.43–0.98),
respectively (Fig. 2A and B). All but
one patient experienced progressive disease and breast
cancer-related death during the follow-up time. The patient who was
still alive at follow-up had then been monitored for twelve years
and had received a total of 159 cycles of trastuzumab.
 | Table III.Clinicopathological variables at
trastuzumab start and treatment regimen. |
Table III.
Clinicopathological variables at
trastuzumab start and treatment regimen.
| Clinicopathological
variable | n (%) |
|---|
| Pre-Dominant
metastatic site |
|
|
Visceral | 38 (82.6) |
|
Non-viscerala | 8 (17.4) |
| Number of
metastatic sites |
|
| 1 | 20 (43.5) |
| 2 | 16 (34.8) |
| 3+ | 10 (21.7) |
| ER status |
|
|
Negative | 34 (73.9) |
|
Positive | 12 (26.1) |
| PR status |
|
|
Negative | 32 (69.6) |
|
Positive | 14 (30.4) |
| Trastuzumab
therapy |
|
| Single
therapy | 7 (15.2) |
|
Combination therapy | 39 (84.8) |
| Trastuzumab as
first-line |
|
| No | 25 (54.3) |
|
Yes | 21 (45.7) |
| Lapatinib |
|
| No | 36 (81.8) |
|
Yes | 8 (18.2) |
| Nottingham
histological grade |
|
| 1 | 2 (6.1) |
| 2 | 15 (45.4) |
| 3 | 16 (48.5) |
| Concomitant
treatment |
|
|
None | 7 (15.2) |
|
Vinorelbine | 32 (69.6) |
|
Paclitaxel | 3 (6.5) |
|
Docetaxel | 3 (6.5) |
|
Aromatase inhibitor | 1 (2.2) |
Concomitant treatment with trastuzumab and
chemotherapy was prescribed to 38 patients of whom 32 received
vinorelbine, three received docetaxel, and three paclitaxel. Seven
patients were treated with single trastuzumab and one patient with
trastuzumab and concomitant aromatase inhibitor (AI). Trastuzumab
treatment was continued beyond progression in 27 of the 46 patients
(58.7%). During the disease course, 28 of the 46 patients (60.8%)
developed metastases in the central nervous system (CNS).
Clinicopathological variables and
survival
Patients with more than one metastatic site at the
start of trastuzumab therapy, as compared with patients with a
single metastatic site, displayed significantly shorter OS and PFS
in univariable analysis (HR=1.50; 95% CI: 1.05–2.15, P=0.025 and
HR=1.45; 95%: 1.02–2.06, P=0.037, respectively). Patients receiving
lapatinib after failure on trastuzumab therapy had a significantly
improved OS (HR=0.39; 95% CI: 0.18–0.85, P=0.019) vs. those not
receiving lapatinib. Combination therapy (trastuzumab and
chemotherapy/AI) was significantly associated with improved PFS
(HR=0.40; 95% CI: 0.17–0.92, P=0.031) but not with OS (HR=0.50; 95%
CI: 0.22–1.20, P=0.11). First-line vs. second or later line of
trastuzumab showed a trend towards improved PFS (HR=0.57; 95% CI:
0.31–1.05, P=0.071). Lapatinib treatment after trastuzumab vs.
those receiving other or no therapy after trastuzumab was the only
clinicopathological variable keeping a significant association with
OS in the multiple Cox regression analysis. Likewise, concomitant
treatment was the only clinicopathological variable that remained
significant regarding PFS. The results of the multivariable
analyses are summarised in Table
IV.
 | Table IV.Multiple Cox regression analyses of
clinicopathological parameters, and ERBB2 and PTPN2
gene copy number. |
Table IV.
Multiple Cox regression analyses of
clinicopathological parameters, and ERBB2 and PTPN2
gene copy number.
|
|
| Overall
survival | Progression-free
survival |
|---|
|
|
|
|
|
|---|
| Variables | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| First-line
trastuzumab | 39 | 1.03
(0.48–2.20) | 0.95 | 0.62
(0.28–1.37) | 0.23 |
| Number of
metastasis at trastuzumab start | 39 | 1.33
(0.81–2.18) | 0.26 | 1.10
(0.67–1.80) | 0.70 |
| Combination
therapy | 39 | 0.55
(0.20–1.39) | 0.20 | 0.31
(0.12–0.82) | 0.018a |
| Visceral
metastasis | 39 | 0.56
(0.23–1.36) | 0.20 | 0.71
(0.26–1.96) | 0.51 |
| High S6K1
expression in cytoplasm | 39 | 0.85
(0.39–1.83) | 0.67 | 1.04
(0.53–2.04) | 0.91 |
| ERBB2
high-grade | 39 | 0.37
(0.18–0.79) | 0.010a | 0.35
(0.16–0.73) | 0.006a |
| PTPN2
gain | 39 | 3.40
(1.53–7.57) | 0.008a | 2.72
(1.30–5.71) | 0.008a |
| Lapatinib after
trastuzumab | 39 | 0.29
(0.12–0.73) | 0.008a | − | − |
Receptor tyrosine kinases, PTPN2, and
survival
High-grade ERBB2 amplification, defined as ≥6
gene copies, was found in 27 of the 40 (67.5%) tumours available
for ddPCR analysis and was significantly associated with improved
OS and PFS in univariable analysis (HR=0.49; 95% CI: 0.25–0.99,
P=0.045 and HR=0.44; 95% CI: 0.22–0.89, P=0.022, respectively;
Table V; Fig. 2C and D). Of the patients with
high-grade ERBB2 amplification, 78% (21/27) developed CNS
metastases compared with 31% (4/13) of the patients with low-grade
amplification (Fisher's exact test, P=0.006).
 | Table V.Gene copy numbers in relation to
overall survival and progression-free survival. |
Table V.
Gene copy numbers in relation to
overall survival and progression-free survival.
|
|
|
| Overall
survival | Progression-free
survival |
|---|
|
|
|
|
|
|
|---|
| Gene (protein) | Copy number
high | High/n (%) | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| CCND1 (Cyclin
D1) | ≥3 | 9/42 (21.4) | 1.6 (0.7–3.4) | 0.23 | 1.3 (0.6–2.8) | 0.46 |
| ERBB2
(HER2) | ≥6 | 27/40 (67.5) | 0.5 (0.3–0.9) | 0.045a | 0.4 (0.2–0.9) | 0.022a |
| MET
(Met) | ≥2 | 31/42 (73.8) | 1.2 (0.6–2.4) | 0.65 | 0.9 (0.5–1.9) | 0.84 |
| PTPN2
(PTPN2) | ≥3 | 15/42 (35.7) | 2.0 (1.0–4.0) | 0.040a | 2.1 (1.0–4.1) | 0.041a |
| RPS6KB1
(S6K1) | ≥3 | 8/42 (19.0) | 1.7 (0.8–3.8) | 0.19 | 2.0 (0.9–4.5) | 0.10 |
PTPN2 gain, defined as three or more gene
copies, was found in 15 of 42 (35.7%) patients available for ddPCR
analysis and was significantly associated with shorter OS and PFS
in univariable analysis (HR=2.0; 95% CI: 1.0–4.0, P=0.040 and
HR=2.1; 95% CI: 1.0–4.1, P=0.041, respectively; Table V, Fig. 2E
and F). Analysis of Met, pMet, HER3 and HER4 did not reveal any
significant prognostic values (Tables
V and VI).
 | Table VI.Protein expression levels in relation
to overall survival and progression-free survival. |
Table VI.
Protein expression levels in relation
to overall survival and progression-free survival.
|
|
|
| Overall
survival | Progression-free
survival |
|---|
|
|
|
|
|
|
|---|
| Protein | Localisation | High/n (%) | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| p4EBP1-S65 | Cytoplasmic | 34/40 (85.0) | 0.9 (0.4–2.1) | 0.75 | 0.8 (0.3–1.9) | 0.61 |
|
| Nuclear | 15/40 (62.5) | 1.2 (0.6–2.3) | 0.55 | 1.0 (0.5–1.9) | 0.98 |
| pAkt-S473 | Cytoplasmic | 36/42 (85.7) | 1.7 (0.7–4.1) | 0.23 | 1.8 (0.7–4.3) | 0.20 |
|
| Nuclear | 33/42 (78.6) | 1.2 (0.5–2.5) | 0.69 | 1.2 (0.6–2.6) | 0.60 |
| pAkt-T308 | Cytoplasmic | 15/41 (36.6) | 1.1 (0.6–2.2) | 0.75 | 1.0 (0.5–2.0) | 0.96 |
|
| Nuclear | 36/40 (90.0) | 1.9 (0.7–5.4) | 0.24 | 2.0 (0.7–5.9) | 0.18 |
| Cyclin D1 | Nuclear | 26/43 (60.5) | 1.1 (0.6–2.1) | 0.69 | 1.2 (0.7–2.3) | 0.49 |
| HER3 | Membranous | 33/44 (75.0) | 1.4 (0.7–2.9) | 0.32 | 1.3 (0.6–2.6) | 0.49 |
|
| Cytoplasmic | 15/44 (34.1) | 1.2 (0.6–2.3) | 0.57 | 1.0 (0.5–19) | 0.93 |
| HER4 | Membranous | 6/43 (14.0) | 1.4 (0.6–3.3) | 0.46 | 1.0 (0.4–2.3) | 0.93 |
|
| Cytoplasmic | 28/43 (65.1) | 1.0 (0.5–1.9) | 1.00 | 1.2 (0.7–2.4) | 0.51 |
| pMet-Y1349 | Membranous | 19/43 (44.2) | 1.1 (0.6–2.0) | 0.78 | 1.1 (0.6–2.0) | 0.84 |
|
| Cytoplasmic | 29/43 (67.4) | 0.7 (0.4–1.4) | 0.33 | 0.8 (0.4–1.5) | 0.48 |
| Met | Membranous | 7/40 (17.5) | 1.2 (0.5–2.8) | 0.66 | 1.3 (0.5–1.0) | 0.60 |
|
| Cytoplasmic | 22/40 (55.0) | 0.6 (0.3–1.2) | 0.17 | 0.5 (0.3–1.0) | 0.06 |
| PTEN | Cytoplasmic | 21/43 (48.5) | 1.1 (0.6–2.1) | 0.68 | 1.0 (0.5–1.8) | 1.0 |
| PTPN2 | Cytoplasmic | 31/42 (73.8) | 1.6 (0.8–3.2) | 0.19 | 1.6 (0.8–3.3) | 0.19 |
| pS6K1-T389 | Cytoplasmic | 16/43 (37.2) | 1.1 (0.6–2.1) | 0.71 | 1.2 (0.7–2.3) | 0.50 |
|
| Nuclear | 12/43 (27.9) | 0.6 (0.3–1.3) | 0.19 | 0.7 (0.3–1.4) | 0.30 |
| S6K1 | Cytoplasmic | 22/43 (51.2) | 0.5 (0.3–1.0) | 0.04a | 0.7 (0.4–1.3) | 0.24 |
|
| Nuclear | 13/43 (30.2) | 0.8 (0.4–1.6) | 0.57 | 1.1 (0.6–2.1) | 0.79 |
In the multiple Cox analysis, high-grade
ERBB2 amplification was significantly associated with
improved prognosis both in terms of OS and PFS. In contrast,
PTPN2 gain was significantly associated with shorter OS and
PFS (Table IV).
PI3K/Akt pathway and survival
High cytoplasmic expression level of the S6K1
protein was significantly associated with improved OS in
univariable analysis (Table VI).
However, gene copy number variation of the S6K1-encoding gene
RPS6KB1 did not show a significant prognostic value and high
S6K1 expression in the cytoplasm was not prognostic in the multiple
Cox analysis (Tables V and VI). Protein expression levels of PTEN,
pAkt, p4EBP1, and Cyclin D1 had no significant prognostic value
(Table VI).
Discussion
The present study was performed on a cohort of the
first 46 consecutive patients with recurring HER2-positive breast
cancer who received palliative treatment with trastuzumab at
Linköping University Hospital. The results reveal that the
prognosis remains poor for patients with HER2-positive MBC even
after the introduction of trastuzumab therapy. While the overall
survival, in general, was comparable with the outcome data of the
early trastuzumab studies (30), a
small number of long-term responders were identified in the present
cohort.
Analyses of the tumour material with ddPCR revealed
that high-grade ERBB2 amplification and gain of PTPN2
appear as prognostic biomarkers in patients with HER2-positive MBC.
High-grade ERBB2 amplification was significantly associated
with prolonged OS and PFS in both uni- and multivariable analyses.
Gene copy number analysis with ddPCR has been confirmed to have a
high correlation with FISH/ISH when determining the level of
ERBB2 amplification (31–33). The
cut-off determined for high-grade ERBB2 amplification in
this study was derived from the distribution of data and could be
argued to be low when compared with the current cut-off to
determine HER2-positivity with ISH/FISH (>4 gene copies). Both
similar and higher cut-offs have been used to define high-grade
ERBB2 amplification and further research is needed to
determine the optimal cut-off for FISH and ddPCR (34–36).
The positive prognostic value of high-grade
ERBB2 amplification is in concordance with previous studies
on MBC, although, to our knowledge, no previous study has utilised
ddPCR quantification of ERBB2 amplification to discriminate
between patients with good vs. poor prognosis in this setting
(34,35,37,38). A
recent meta-analysis, focused on ERBB2 amplification status
in patients with loco-regional HER2-positive breast cancer treated
with adjuvant trastuzumab, co-analysed three cohort studies with
1360 patients in total. In contrast to our results, Xu et al
(35) concluded that ERRB2
amplification level was not prognostic in the adjuvant setting.
This implies that the prognostic value of ERBB2
amplification status appears to vary depending on the disease
stage. Further studies should focus on the prognostic impact of
ERBB2 amplification status in metastatic rather than
loco-regional HER2-positive breast cancer.
The high prevalence of CNS metastases (61%) in our
cohort may be explained by the long follow-up period and the nature
of HER2-positive disease; similar results have been found in other
long-term follow-up studies (3).
Interestingly, high-grade ERBB2 amplification was associated
with increased prevalence of CNS-metastases (78% vs. 31%), but not
the time to the first presentation of CNS-metastasis (data not
shown). If confirmed in other studies, the high prevalence of
CNS-metastasis found in this population could warrant routine
radiological evaluation of the brain in suitable intervals for all
HER2-positive MBC patients, especially in patients with high-grade
ERBB2 amplification.
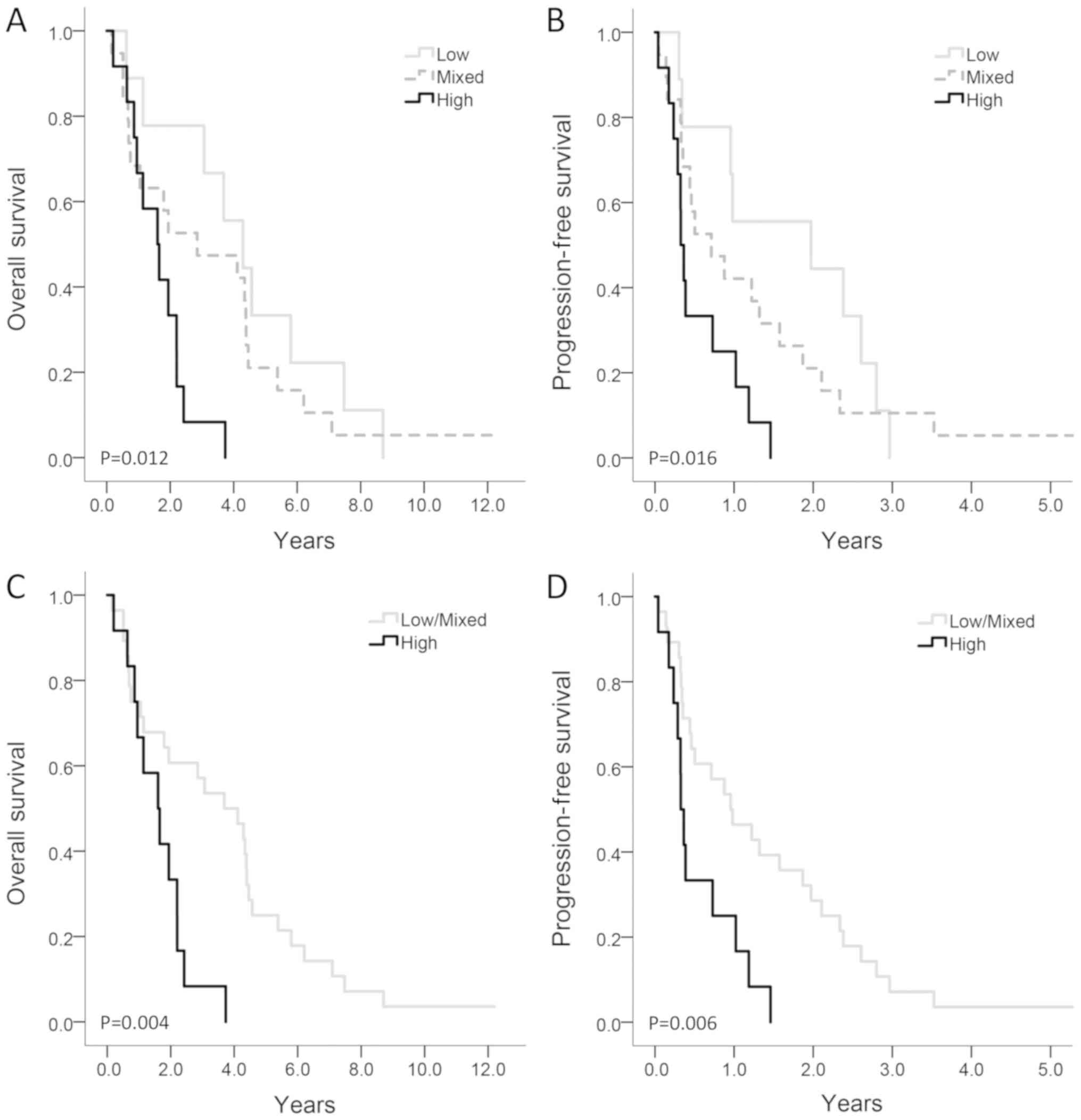
Copy gain of PTPN2 was significantly
associated with shorter OS and PFS in both uni- and multivariable
analyses. Likewise, high protein expression of PTPN2 was associated
with a shorter OS and PFS, though not statistically significant.
Combining protein expression with gene copy number increased the
prognostic value (Fig. 3) indicating
that further research of both protein and gene expression could be
valuable. In contrast with these results, PTPN2 has previously been
described as a tumour suppressor and loss of PTPN2 was shown
to correlate with shorter breast cancer survival in patients with
early breast cancer, including those with HER2-positive disease
(23). Unlike the patients in our
cohort, these patients did not receive trastuzumab. This suggests
that the effect seen in our study could be due to an interaction
between PTPN2 gain, HER2 positivity, and trastuzumab
treatment. A recent in vivo study of malignant melanoma in
mice suggested that loss of PTPN2 may sensitise tumours to
immunotherapy through the regulation of antigen presentation and
recruitment of cytotoxic CD8+ T-cells (39). Hypothetically, PTPN2 could have a
similar effect on the ADCC caused by trastuzumab, which would
explain the negative prognostic value of PTPN2 gain in this
study.
Previous studies report conflicting data regarding
the prognostic significance of the PI3K/Akt pathway. In
vitro and in vivo PTEN loss has been previously
identified as a mechanism of trastuzumab resistance (40,41). Here,
no prognostic value of PTEN was found, in line with a more recent
large study of patients treated in the adjuvant setting (42). Furthermore, no prognostic value of
other key proteins and genes within the PI3K/Akt pathway was
evident in this study, making them less likely to be suitable
biomarkers for patients with HER2-positive MBC receiving
trastuzumab. Although it would have been relevant to explore the
prognostic value of the components of the PI3K/Akt pathway in
relation to ER-status, this was beyond the scope of the present
study and the limited size of our cohort did not permit such
subgroup analyses.
The only clinicopathological variable where a
significant impact on OS was evident in multivariable analysis was
the presence of later line lapatinib treatment after failure of
trastuzumab. Although the number of patients receiving lapatinib
was low (n=8), these data support the idea that the continuation of
HER2-targeted therapy is valuable even after progression on
trastuzumab (43).
The weaknesses of this study include its
retrospective nature and the small population size, and any results
should be interpreted in this context. The main strengths of this
study are the extensive follow-up duration, with endpoint events
(OS and PFS) reported for all but one patient, and the wide
inclusion criteria mirroring the real world authentic clinical
setting.
In conclusion, our results suggest that high-grade
ERBB2 amplification is a potential positive prognostic
factor and that PTPN2 gene copy gain is a potential negative
prognostic factor in HER2-positive MBC patients receiving
trastuzumab treatment. Furthermore, the ERBB2 amplification
level may identify patients at high risk of developing
CNS-metastases in this subgroup of patients.
Acknowledgements
The authors would like to thank Mrs Birgitta
Holmlund (Department of Clinical and Experimental Medicine and
Department of Oncology, Linköping University) for help with
constructing the tissue microarrays.
Funding
The present study was supported by grants from the
Swedish Cancer Society (Grant no. 17-0479), Medical Research
Council of Southeast Sweden (Grant no. FORSS-757671), ALF Grants
Region Östergötland (Grant no. LIO-795201), and Stiftelsen
Onkologiska Klinikernas Forskningsfond i Linköping (Grant no.
2016-06-21).
Availability of data and materials
The datasets generated and/or analysed during the
present study are available from the corresponding author on
reasonable request.
Authors' contribution
SE, MS, AM, SW and OS conceptualised the design of
the study. SE and AM collected clinical data from patient records.
SE, OS, CV, GPT, VF and JG performed and analysed
immunohistochemistry results. CV and KG performed ddPCR analyses.
SE, CV, ALH, NE and OS interpreted the data. SE and CV were major
contributors of drafting the manuscript. All authors read and
critically revised the manuscript and later approved the final
version to be published.
Ethics approval and consent to
participate
The present study was approved by the Regional
Ethical Review Board in Linköping (M140-06 and updated in 2014,
2014/163-32). The local ethical review board concluded that
individual consent was not required.
Patient consent for publication
Not applicable. Due to the retrospective nature of
the study, the non-invasive design and the anonymisation of
personal data, the local ethics review board concluded that
patients would be unlikely to object to publication and that the
public interest outweighed the potential harm to the individuals'
integrity.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Slamon DJ, Godolphin W, Jones LA, Holt JA,
Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al:
Studies of the HER-2/neu proto-oncogene in human breast and ovarian
cancer. Science. 244:707–712. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dawood S, Broglio K, Buzdar AU, Hortobagyi
GN and Giordano SH: Prognosis of women with metastatic breast
cancer by HER2 status and trastuzumab treatment: An
institutional-based review. J Clin Oncol. 28:92–98. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Olson EM, Najita JS, Sohl J, Arnaout A,
Burstein HJ, Winer EP and Lin NU: Clinical outcomes and treatment
practice patterns of patients with HER2-positive metastatic breast
cancer in the post-trastuzumab era. Breast. 22:525–531. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Balduzzi S, Mantarro S, Guarneri V,
Tagliabue L, Pistotti V, Moja L and D'Amico R:
Trastuzumab-containing regimens for metastatic breast cancer.
Cochrane Database Syst Rev. CD0062422014.PubMed/NCBI
|
|
5
|
Yeo B, Kotsori K, Mohammed K, Walsh G and
Smith IE: Long-term outcome of HER2 positive metastatic breast
cancer patients treated with first-line trastuzumab. Breast.
24:751–757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yardley DA, Tripathy D, Brufsky AM, Rugo
HS, Kaufman PA, Mayer M, Magidson J, Yoo B, Quah C and Ulcickas
Yood M: Long-term survivor characteristics in HER2-positive
metastatic breast cancer from registHER. Br J Cancer.
110:2756–2764. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Murthy P, Kidwell KM, Schott AF, Merajver
SD, Griggs JJ, Smerage JD, Van Poznak CH, Wicha MS, Hayes DF and
Henry NL: Clinical predictors of long-term survival in
HER2-positive metastatic breast cancer. Breast Cancer Res Treat.
155:589–595. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arnould L, Gelly M, Penault-Llorca F,
Benoit L, Bonnetain F, Migeon C, Cabaret V, Fermeaux V, Bertheau P,
Garnier J, et al: Trastuzumab-based treatment of HER2-positive
breast cancer: An antibody-dependent cellular cytotoxicity
mechanism? Br J Cancer. 94:259–267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vu T and Claret FX: Trastuzumab: Updated
mechanisms of action and resistance in breast cancer. Front Oncol.
2:622012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park YH, Lee SJ, Cho EY, Choi YL, Lee JE,
Nam SJ, Yang JH, Shin JH, Ko EY, Han BK, et al: Clinical relevance
of TNM staging system according to breast cancer subtypes. Ann
Oncol. 22:1554–1560. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Holbro T, Beerli RR, Maurer F, Koziczak M,
Barbas CF III and Hynes NE: The ErbB2/ErbB3 heterodimer functions
as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor
cell proliferation. Proc Natl Acad Sci USA. 100:8933–8938. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kurokawa H and Arteaga CL: ErbB (HER)
receptors can abrogate antiestrogen action in human breast cancer
by multiple signaling mechanisms. Clin Cancer Res. 9:511S–515S.
2003.PubMed/NCBI
|
|
13
|
Liu P, Cheng H, Roberts TM and Zhao JJ:
Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev
Drug Discov. 8:627–44. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Musgrove EA, Caldon CE, Barraclough J,
Stone A and Sutherland RL: Cyclin D as a therapeutic target in
cancer. Nat Rev Cancer. 11:558–572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Caldon CE, Daly RJ, Sutherland RL and
Musgrove EA: Cell cycle control in breast cancer cells. J Cell
Biochem. 97:261–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Berns K, Horlings HM, Hennessy BT,
Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM,
Stemke-Hale K, Hauptmann M, et al: A functional genetic approach
identifies the PI3K pathway as a major determinant of trastuzumab
resistance in breast cancer. Cancer Cell. 12:395–402. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luque-Cabal M, Garcia-Teijido P,
Fernandez-Perez Y, Sanchez-Lorenzo L and Palacio-Vazquez I:
Mechanisms behind the resistance to trastuzumab in HER2-amplified
breast cancer and strategies to overcome it. Clin Med Insights
Oncol. 10 (Suppl 1):S21–S30. 2016.
|
|
18
|
Zhang S, Huang WC, Li P, Guo H, Poh SB,
Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z, et al: Combating
trastuzumab resistance by targeting SRC, a common node downstream
of multiple resistance pathways. Nat Med. 17:461–469. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ha Thi HT, Choi SW, Kim YM, Kim HY and
Hong S: Protein tyrosine phosphatase N2 is a positive regulator of
lipopolysaccharide signaling in Raw264.7 cell through derepression
of Src tyrosine kinase. PLoS One. 11:e01627242016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Finn RS: Targeting Src in breast cancer.
Ann Oncol. 19:1379–1386. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Soysal S, Obermann EC, Gao F, Oertli D,
Gillanders WE, Viehl CT and Muenst S: PTP1B expression is an
independent positive prognostic factor in human breast cancer.
Breast Cancer Res Treat. 137:637–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rivera Franco MM, Leon Rodriguez E,
Martinez Benitez B, Villanueva Rodriguez LG, de la Luz Sevilla
Gonzalez M and Armengol Alonso A: Association of PTP1B with
outcomes of breast cancer patients who underwent neoadjuvant
chemotherapy. Breast Cancer (Auckl). 10:177–184. 2016.PubMed/NCBI
|
|
23
|
Karlsson E, Veenstra C, Emin S, Dutta C,
Pérez-Tenorio G, Nordenskjöld B, Fornander T and Stål O: Loss of
protein tyrosine phosphatase, non-receptor type 2 is associated
with activation of AKT and tamoxifen resistance in breast cancer.
Breast Cancer Res Treat. 153:31–40. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shields BJ, Wiede F, Gurzov EN, Wee K,
Hauser C, Zhu HJ, Molloy TJ, O'Toole SA, Daly RJ, Sutherland RL, et
al: TCPTP regulates SFK and STAT3 signaling and is lost in
triple-negative breast cancers. Mol Cell Biol. 33:557–570. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McShane LM, Altman DG, Sauerbrei W, Taube
SE, Gion M and Clark GM; Statistics Subcommittee of the NCI-EORTC
Working Group on Cancer Diagnostics: Reporting recommendations for
tumor marker prognostic studies (REMARK). J Natl Cancer Inst.
97:1180–1144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Veenstra C, Pérez-Tenorio G, Stelling A,
Karlsson E, Mirwani SM, Nordenskoljd B, Fornander T and Stal O: Met
and its ligand HGF are associated with clinical outcome in breast
cancer. Oncotarget. 7:37145–37159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bostner J, Karlsson E, Pandiyan MJ,
Westman H, Skoog L, Fornander T, Nordenskjöld B and Stål O:
Activation of Akt, mTOR, and the estrogen receptor as a signature
to predict tamoxifen treatment benefit. Breast Cancer Res Treat.
137:397–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Karlsson E, Pérez-Tenorio G, Amin R,
Bostner J, Skoog L, Fornander T, Sgroi DC, Nordenskjold B, Hallbeck
AL and Stål O: The mTOR effectors 4EBP1 and S6K2 are frequently
coexpressed and associated with a poor prognosis and endocrine
resistance in breast cancer: A retrospective study including
patients from the randomised Stockholm tamoxifen trials. Breast
Cancer Res. 15:R962013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bostner J, Karlsson E, Eding CB,
Perez-Tenorio G, Franzen H, Konstantinell A, Fornander T,
Nordenskjöld B and Stål O: S6 kinase signaling: Tamoxifen response
and prognostic indication in two breast cancer cohorts. Endocr
Relat Cancer. 22:331–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Slamon DJ, Leyland-Jones B, Shak S, Fuchs
H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M,
et al: Use of chemotherapy plus a monoclonal antibody against HER2
for metastatic breast cancer that overexpresses HER2. N Engl J Med.
344:783–792. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Belgrader P, Tanner SC, Regan JF, Koehler
R, Hindson BJ and Brown AS: Droplet digital PCR measurement of HER2
copy number alteration in formalin-fixed paraffin-embedded breast
carcinoma tissue. Clin Chem. 59:991–994. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heredia NJ, Belgrader P, Wang S, Koehler
R, Regan J, Cosman AM, Saxonov S, Hindson B, Tanner SC, Brown AS
and Karlin-Neumann G: Droplet Digital™ PCR quantitation of HER2
expression in FFPE breast cancer samples. Methods. 59:S20–S23.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Otsuji K, Sasaki T, Tanaka A, Kunita A,
Ikemura M, Matsusaka K, Tada K, Fukayama M and Seto Y: Use of
droplet digital PCR for quantitative and automatic analysis of the
HER2 status in breast cancer patients. Breast Cancer Res Treat.
162:11–18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Giuliani R, Durbecq V, Di Leo A, Paesmans
M, Larsimont D, Leroy JY, Borms M, Vindevoghel A, Jerusalem G,
D'Hondt V, et al: Phosphorylated HER-2 tyrosine kinase and
Her-2/neu gene amplification as predictive factors of response to
trastuzumab in patients with HER-2 overexpressing metastatic breast
cancer (MBC). Eur J Cancer. 43:725–735. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu QQ, Pan B, Wang CJ, Zhou YD, Mao F, Lin
Y, Guan JH, Shen SJ, Zhang XH, Xu YL, et al: HER2 amplification
level is not a prognostic factor for HER2-positive breast cancer
with trastuzumab-based adjuvant treatment: A systematic review and
meta-analysis. Oncotarget. 7:63571–63582. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim JW, Kim JH, Im SA, Kim YJ, Han HS, Kim
JS, Lee KH, Kim TY, Han SW, Jeon YK, et al: HER2/CEP17 ratio and
HER2 immunohistochemistry predict clinical outcome after first-line
trastuzumab plus taxane chemotherapy in fluorescence in situ
hybridization-positive metastatic breast cancer. Cancer Chemother
Pharmacol. 72:109–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Carey LA, Berry DA, Cirrincione CT, Barry
WT, Pitcher BN, Harris LN, Ollila DW, Krop IE, Henry NL, Weckstein
DJ, et al: Molecular heterogeneity and response to neoadjuvant
human epidermal growth factor receptor 2 targeting in CALGB 40601,
a randomized phase III trial of paclitaxel plus trastuzumab with or
without lapatinib. J Clin Oncol. 34:542–549. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fumagalli D, Venet D, Ignatiadis M, Azim
HA Jr, Maetens M, Rothé F, Salgado R, Bradbury I, Pusztai L,
Harbeck N, et al: RNA sequencing to predict response to neoadjuvant
anti-HER2 therapy: A secondary analysis of the NeoALTTO randomized
clinical trial. JAMA Oncol. Sep 29–2016.(Epub ahead of print).
PubMed/NCBI
|
|
39
|
Manguso RT, Pope HW, Zimmer MD, Brown FD,
Yates KB, Miller BC, Collins NB, Bi K, LaFleur MW, Juneja VR, et
al: In vivo CRISPR screening identifies Ptpn2 as a cancer
immunotherapy target. Nature. 547:413–418. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nagata Y, Lan KH, Zhou X, Tan M, Esteva
FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, et al: PTEN
activation contributes to tumor inhibition by trastuzumab, and loss
of PTEN predicts trastuzumab resistance in patients. Cancer Cell.
6:117–127. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Park YH, Jung HA, Choi MK, Chang W, Choi
YL, Do IG, Ahn JS and Im YH: Role of HER3 expression and PTEN loss
in patients with HER2-overexpressing metastatic breast cancer (MBC)
who received taxane plus trastuzumab treatment. Br J Cancer.
110:384–391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Perez EA, Cortés J, Gonzalez-Angulo AM and
Bartlett JM: HER2 testing: Current status and future directions.
Cancer Treat Rev. 40:276–284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Geyer CE, Forster J, Lindquist D, Chan S,
Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A,
Kaufman B, et al: Lapatinib plus capecitabine for HER2-positive
advanced breast cancer. N Engl J Med. 355:2733–2743. 2006.
View Article : Google Scholar : PubMed/NCBI
|