Introduction
Lung cancer is a type of malignant tumor with
unregulated and rapid proliferation that resulted in >1.6
million deaths worldwide in 2015 (1). Despite advances in clinical therapeutic
options, the 5-year survival rate of patients with lung cancer
remains <15% (2), which is
markedly lower than that of patients with breast, colon or prostate
cancer (3). Furthermore, advances in
treatments for lung cancer, including surgical excision, medical
treatment or radiotherapeutic intervention, have little effect on
the long-term survival rate (4).
Therefore, the identification of the underlying mechanisms of lung
cancer tumorigenesis and progression may aid clinical diagnosis and
treatment. Lung cancer tumorigenesis and development are closely
associated with dysregulation of microRNAs (miRNAs/miRs) (5,6).
miRNAs are small non-coding RNAs consisting of 19–25
nucleotides (7), which modulate gene
expression during cellular processes (8). An increasing number of studies suggest
that miRNAs act as either tumor suppressors (9) or oncogenes (10) in the progression of various types of
cancer, including lung cancer (11,12). A
previous study revealed that c-Myc-activated long non-coding RNA
H19 downregulated miR-107 and promoted the cell cycle progression
of non-small-cell lung carcinoma (NSCLC) cell lines (13). Another study revealed that the
expression of miR-107 was markedly reduced in pathological tissues
obtained from patients with lung cancer (14). Furthermore, Takahashi et al
(15) reported that the expression
levels of miR-107 were decreased in lung tumor tissues compared
with healthy tissues and that miR-107 induced cell cycle arrest in
NSCLC cell lines in vitro. However, the underlying mechanism
by which miR-107 functions in lung cancer progression and
development remains largely unknown.
In the current study, the mechanism of miR-107 and
its target gene TP53 regulated inhibitor of apoptosis 1 (TRIAP1) in
lung cancer was investigated. The results obtained revealed that
miR-107 decreased cell proliferation and induced cell apoptosis of
lung cancer cell lines in vitro, providing a novel
theoretical basis for the treatment of lung cancer.
Materials and methods
Specimens
A total of 45 pairs of lung cancer tumor tissues and
non-tumor adjacent lung tissues were obtained from Jingmen No. 2
People's Hospital (Jingmen, China) between July 2014 and April
2016. Among the patients that the tissues were obtained from, there
were 31 males and 14 females, and the average age was 62.3±6.8
years. All patients had been diagnosed with lung cancer and had not
undergone any other therapy. The present study was approved by the
Ethics Committee of Jingmen No. 2 People's Hospital and written
consent was acquired from each patient.
Cell culture
The A549 human NSCLC cell line (cat no. SCSP-503;
Type Culture Collection of the Chinese Academy of Sciences),
BESA-2B cell line (cat no. CL-0496; Procell Life Science &
Technology Co., Ltd.) and 293 cell line (cat no. GNHu43; Type
Culture Collection of the Chinese Academy of Sciences) were
cultured in RPMI-1640 (cat no. 11875093; Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (cat no. 10099-141;
Invitrogen; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin. The cells were maintained in a humid
atmosphere with 5% CO2 at 37°C.
Transfection efficiency
In order to determine the transfection efficiency,
cells were divided into three groups as follows: i) Control group
(untreated group); ii) microRNA-negative control (NC) mimics group
(to eliminate non-sequence-specific effects); and iii) miR-107
mimics group (transfected with miR-107 mimics). The miR-107 mimics
and NC oligonucleotides were purchased from Shanghai GenePharma
Co., Ltd. The cells were seeded into 6-well plates at a density of
5×105 cells/well, and 50 nM miR-107 mimics or NC mimics
were transfected into the cells using Lipofectamine®
2000 (cat no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Following transfection,
cells were cultured for another 24 h before RT-qPCR assays were
conducted. The primer sequences were as follows: miR-107 mimics
forward, 5′-ATACCGCTCGAGTGCCATGTGTCCACTGAAT-3′; miR-107 mimics
reverse, 5′-ATACCGCTCGAGTTCCATGCCTCAACTCCT-3′; miR-NC forward,
5′-ATAGCACAGCCTGGATAGCAACGTAC-3′; miR-NC reverse,
5′-CACCTTCTACAATGAGCTGCGTGTG-3′.
Dual-luciferase reporter assay
TRIAP1 was considered to be a predictive target gene
of miR-107 by TargetScan online tool (www.targetscan.org/vert_72). Subsequently, the 293
cell line was transfected with the wild-type TRIAP1 3′untranslated
region [UTR; (TRIAP1-3′UTR)] or mutant TRIAP1-3′UTR with either NC
mimics or miR-107 mimics using Lipofectamine® RNAiMax
Transfection reagent (cat no. 13778075; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Following
transfection, the cells were incubated for 4 h in 5% CO2
at 37°C. Subsequently, the firefly and Renilla luciferase
activities were detected using a dual-luciferase reporter kit
(Beyotime Institute of Biotechnology). Firefly luciferase activity
was normalized using Renilla luciferase activity.
Vector construction
In order to obtain pcDNA3.1-TRIAP1, a
TRIAP1-expression vector was constructed. In brief, TRIAP1 cDNA was
amplified by PCR (as will be described) from the cDNA of BEAS-2B
cells (cat no. CL-0496; Procell Life Science & Technology Co.,
Ltd.). The TRIAP1 cDNA (50 nM) was subsequently inserted into
pcDNA3.1 (cat no. VT8071; Yobio) to construct the pcDNA3.1-TRIAP1
expression vector. Subsequently, 50 nM pcDNA3.1 and pcDNA3.1-TRIAP1
plasmids were transfected into A549 cells using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.),
following the manufacturer's protocol, and incubated at 37°C for 24
h at 5% CO2 for 2 h. The transfected cells were
subsequently incubated at room temperature in 5% CO2 at
37°C for another 48 h, in order to determine cell proliferation and
apoptosis. The sequences were described as follows: TRIAP1 forward,
5′-TATCTTGCAGGAACTGTGTGCTA-3′, and TRIAP1 reverse
5′-AATTTAGGTTCTTCCTCCACAGC-3′.
Analysis of cell proliferation
In order to investigate the effect of miR-107 and
TRIAP1 on the proliferation of A549 cells, cells were divided into
four different groups as follows: i) control group; ii) microRNA-NC
mimics group; iii) miR-107 mimics group; iv) miR-107 mimics +
pcDNA3.1-TRIAP1 group. Briefly, the transfected cells were seeded
onto a 96-well plate at a density of 1×104 cells with
100 µl/well of fresh RPMI-1640 medium. The cells were incubated in
a humid atmosphere with 5% CO2 at 37°C for 48 h.
Following 12, 24 or 48 h, 10 µl Cell Counting Kit-8 (CCK-8)
solution (cat no. HY-P0093; MedChemExpress) was added to each well
and the cells were incubated for a further hour at room
temperature. The absorbance in each well was measured at a
wavelength of 490 nm in order to quantify the proliferation.
Analysis of cell apoptosis
In order to investigate the effect of miR-107 and
TRIAP1 on the apoptosis of A549 cells, cells were divided into four
different groups: i) Control group; ii) microRNA-NC mimics group;
iii) miR-107 mimics group; iv) miR-107 mimics + pcDNA3.1-TRIAP1
group. Briefly, the transfected cells in suspension were collected
at a density of 1×106 cells/ml. The cells were washed
with HEPES buffer solution (cat no. ACC0013A; Seebio Biotech, Inc.)
for 5 min at room temperature and centrifuged for 5 min at 5,000 ×
g on ice. Subsequently, 5 µl Annexin V-fluorescein isothiocyanate
(FITC) and 10 µl propidium iodide (BD Biosciences) were added to
the cells of the four different groups and the cells were incubated
for 10 min in the dark at room temperature. The apoptosis rate was
then measured using a flow cytometer (BD Biosciences) and analyzed
using FlowJo software (version 10; BD Biosciences).
RNA extraction and reverse
transcription-quantitative (RT-q) PCR analysis
miRNA was isolated from the lung tumor and adjacent
non-tumor tissues and A549 cells using the miRNeasy Mini kit
(Qiagen, Inc.) according to the manufacturer's protocol. Following
isolation, the One-Step PrimeScript miRNA cDNA Synthesis kit
(Takara Biotechnology Co., Ltd.) was used to synthesize cDNA
according to the manufacturer's instructions. The total RNA was
extracted using TRIzol reagent (cat. no. 15596018; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Subsequently, 2 µl RNA was reverse transcribed into cDNA.
qPCR analysis was performed using the TaqMan MicroRNA RT kit (cat
no. 4366596; Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Briefly, 2 µl cDNA,
10 µl SYBRGreen RT-qPCR Master mix (cat. no. AB4106A; Applied
Biosystems; Thermo Fisher Scientific, Inc.), 1 µl primers and
nuclease-free water were combined during the PCR reaction. The
primer sequences were as follows: miR-107 forward,
5′-AGCAGCATTGTACAGGGCTATCA-3′; and reverse,
5′-GCGAGCACAGAATTAATACGAC-3′; U6 forward, 5′-AGAGCCTGTGGTGTCCG-3′;
and reverse, 5′-CATCTTCAAAGCACTTCCCT-3′; TRIAP1 forward,
5′-AGGATTTCGCAAGTCCAGAA-3′; and reverse, 5′-GCTGATTCCACCCAAGTAT-3′;
and GAPDH forward, 5′-AACGGATTTGGTCGTATTG-3′; and reverse,
5′-GGAAGATGGTGATGGGATT-3′. The PCR reaction conditions were as
follows: i) Denaturing for 3 min at 95°C; ii) denaturing for 30 sec
at 94°C, annealing for 30 sec at 56°C and extension for 30 sec at
72°C (35 cycles); and iii) extension for 10 min at 72°C. The
relative expression of miR-107 was normalized to that of U6 small
nuclear RNA (Thermo Fisher Scientific, Inc.) and the expression
levels of TRIAP1 mRNA were normalized to those of GAPDH according
to the 2−ΔΔCq method (16). U6 or GAPDH was used as the endogenous
control.
Western blot analysis
Radioimmunoprecipitation assay buffer (cat. no.
R0010; Beijing Solarbio Science & Technology Co., Ltd.) was
used to extract the proteins from tissues and cells. Following
centrifugation at 6,000 × g for 5 min at 4°C, a protease inhibitor
cocktail (cat. no. 78425; Thermo Fisher Scientific, Inc.) was added
to the protein lysates. The Bradford method was used to quantify
the concentration of proteins in the supernatant of the lysates.
The whole protein lysates (20 µg) were separated by SDS-PAGE (8%
gel; cat. no. LC26755; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Subsequently, 10 µg separated
protein was transferred to a polyvinylidene fluoride membrane
(Shanghai Ofluorine Chemical Technology Co., Ltd.) and the membrane
was blocked with 50 ml 5% nonfat milk for 50 min at 37°C. The
membrane was then incubated with the following primary antibodies
at 37°C for 50 min: Rabbit anti-proliferating cell nuclear antigen
(PCNA; 1:1,000; cat. no. ab18197; Abcam); rabbit anti-cyclin D1
(1:100; cat. no. ab16663; Abcam); rabbit anti-TRIAP1 (1:1,000; cat.
no. ABIN2970840; 4A Biotechnology Co., Ltd.), rabbit anti-BCL2
apoptosis regulator (BCL2; 1:1,000; cat. no. ab32124; Abcam),
rabbit anti-BCL2 associated X apoptosis regulator (BAX; 1:1,000;
cat. no. ab32503; Abcam), rabbit anti-tumor protein p53 (TP53;
1:1,000; cat. no. ab131442; Abcam), rabbit anti-caspase 3 (1:500;
cat. no. ab13847; Abcam) and rabbit anti-GAPDH (1:2,500; cat. no.
ab9485; Abcam). All antibodies were diluted in blocking buffer
(concentration, 10×; cat. no. ab126587; Abcam). After washing, the
membrane was then incubated for 1 h at 37°C with goat anti-rabbit
horseradish peroxidase (HRP) IgG H&L secondary antibody
(1:1,000; cat. no. ab7090; Abcam). Subsequently, 200 µl
chemiluminescent HRP substrate (cat. no. ab5801; Abcam) was added
to the surface of the membranes. The signals were captured and
exposed onto X-ray films. ImageJ software (version 1.49; National
Institutes of Health) was used to quantify the relative expression
levels of the proteins. GAPDH was used as the reference protein.
The procedures above were conducted in triplicate.
Statistical analysis
SPSS (version 22.0; IBM SPSS) was used to analyze
the results and the data are presented as the mean ± standard
deviation. The Student's t-test was used to compare two groups.
One-way analysis of variance followed by Newman-Keuls post hoc test
was used to distinguish differences among three and more groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
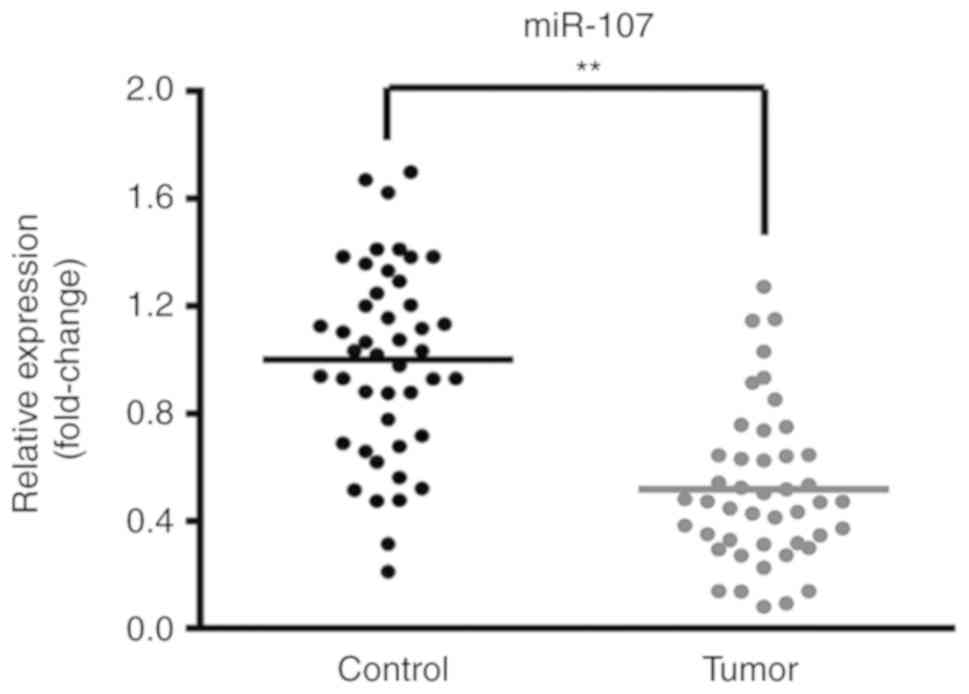
miR-107 is downregulated in lung
cancer tissues
In order to investigate the putative effects of
miR-107 in lung cancer, the expression levels of miR-107 in lung
cancer tumor tissues and adjacent non-tumor tissues from 45
patients were compared. As shown in Fig.
1, the expression levels of miR-107 were significantly reduced
in the lung cancer tumor tissues compared with the adjacent
non-tumor tissues (P<0.01).
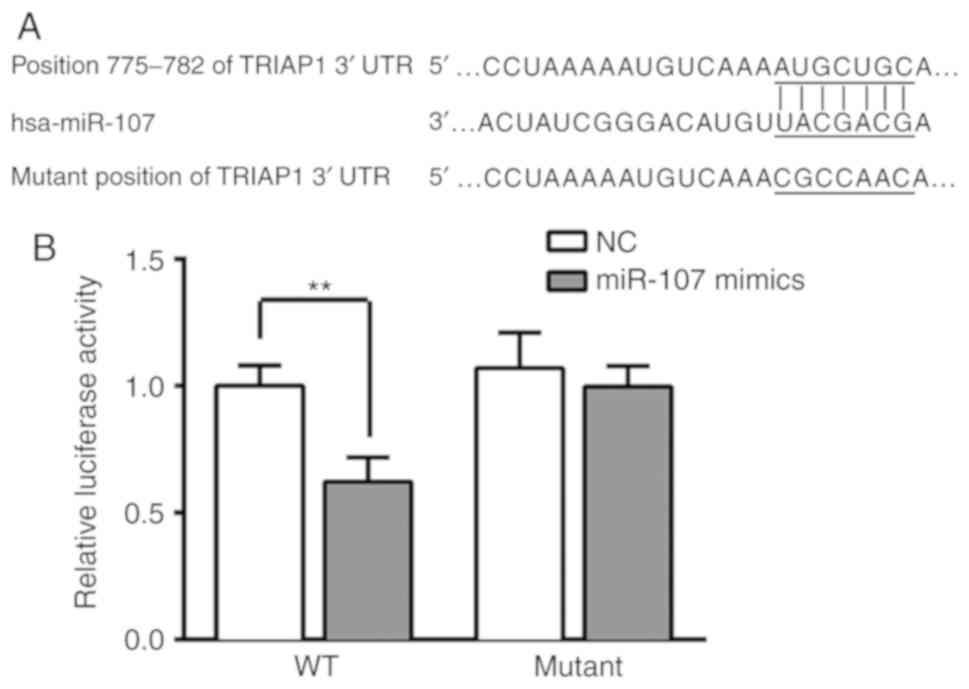
TRIAP1 is a direct target of miR-107
in lung cancer
A dual-luciferase reporter assay was conducted in
order to determine the association between miR-107 and TRIAP1. The
3′-UTR of the TRIAP1 gene was confirmed to contain binding
sequences for miR-107 (Fig. 2A),
suggesting that TRIAP1 may be a downstream target of miR-107.
Furthermore, the results demonstrated that transfection of miR-107
mimics markedly reduced the luciferase activity in the wild-type
TRIAP1-3′UTR plasmid-transfected cells; however, miR-107 had no
significant influence on the mutant TRIAP1-3′UTR
plasmid-transfected cells (P<0.01; Fig. 2B).
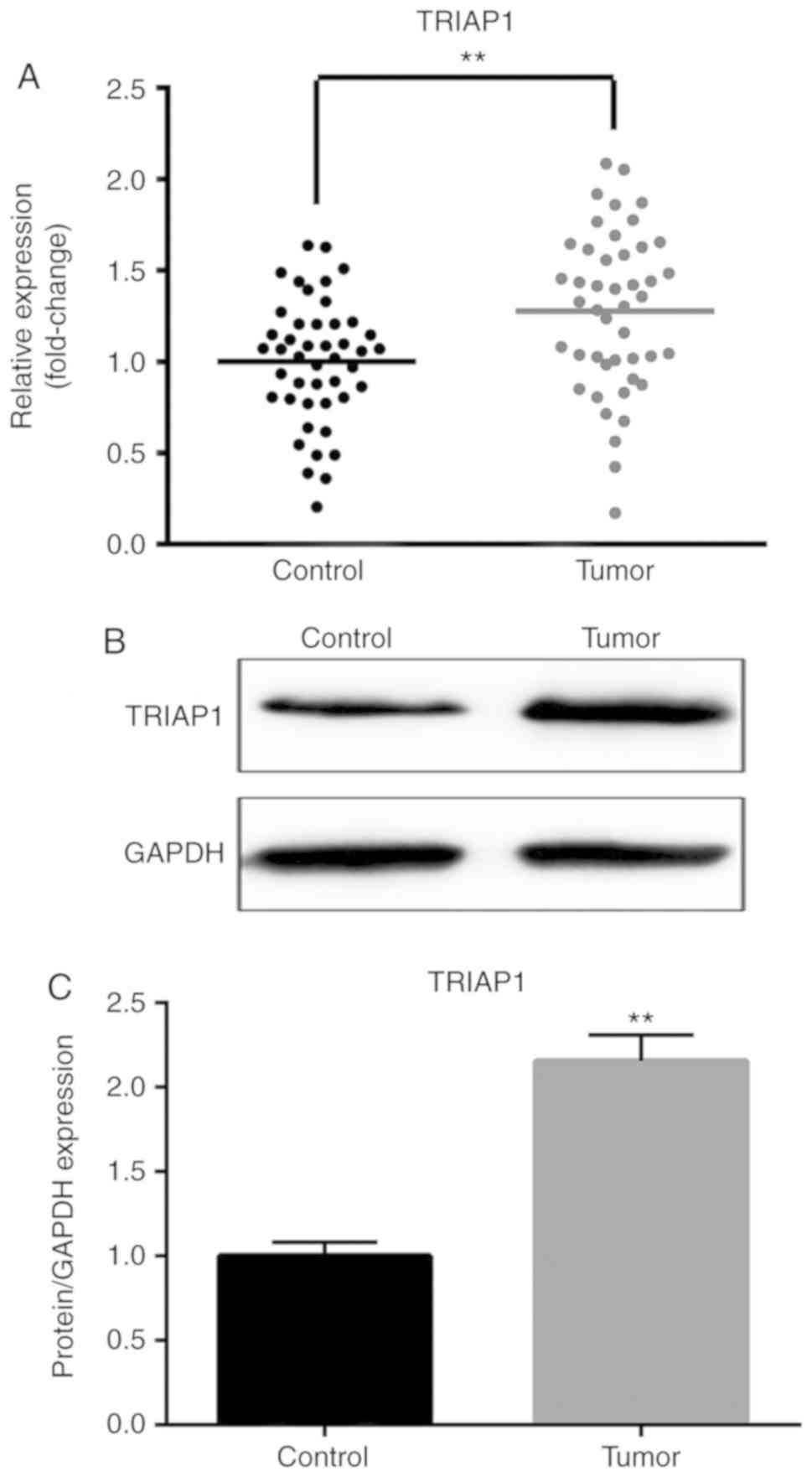
Expression levels of TRIAP1 in lung
cancer tumor tissues and adjacent non-tumor tissues
In order to further investigate the potential
function of TRIAP1 in lung cancer, the mRNA and protein expression
levels of TRIAP1 in lung cancer tumor tissues and adjacent
non-tumor tissues were quantified. As shown in Fig. 3, the mRNA and protein expression
levels of TRIAP1 were highly increased in the lung cancer tumor
tissues compared with the adjacent non-tumor tissues
(P<0.01).
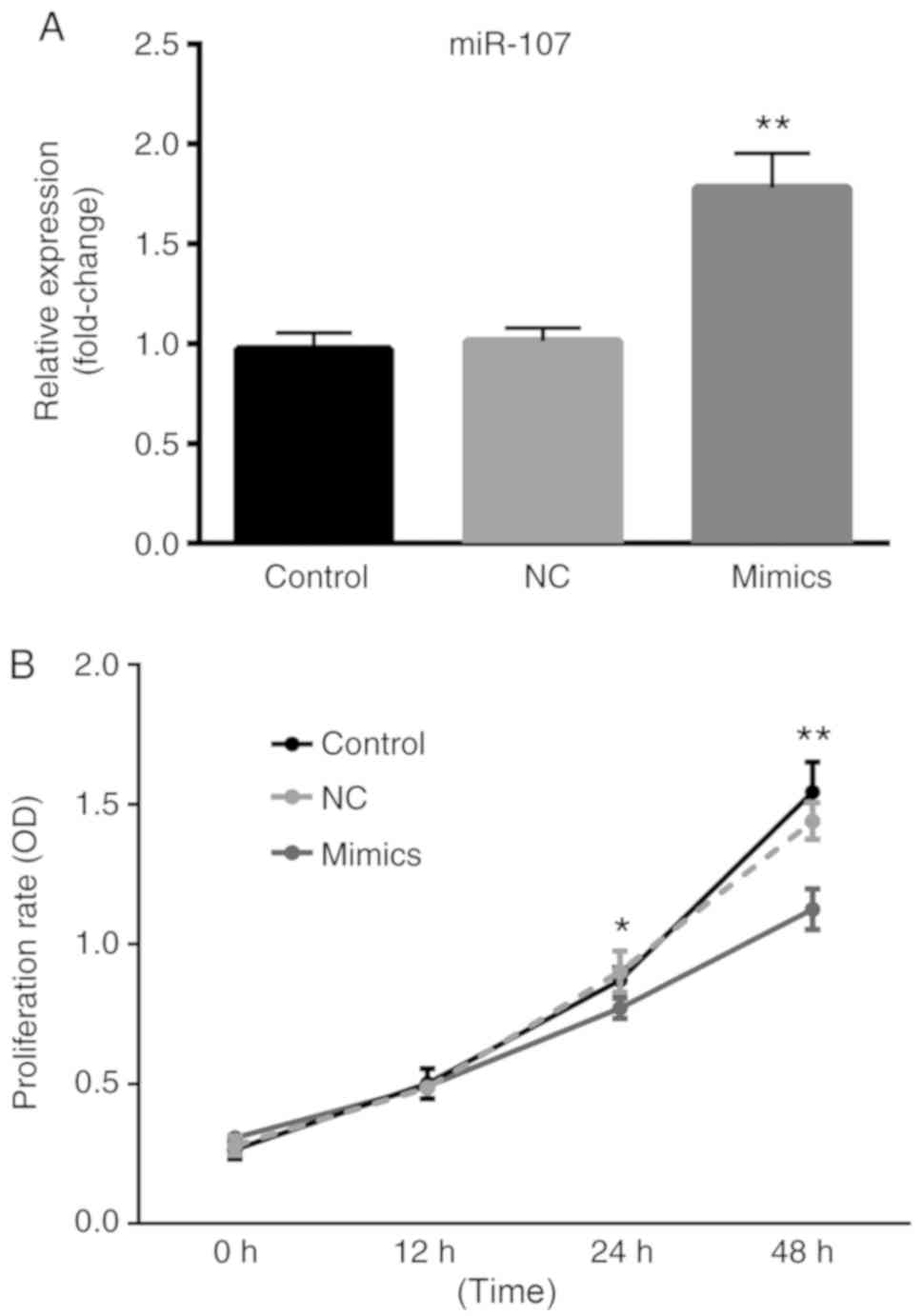
miR-107 inhibits lung cancer cell
proliferation
A549 cells were transfected with miR-107 mimics or
NCs and the transfection efficiency was demonstrated using RT-qPCR
analysis. As shown in Fig. 4A, there
was no significant difference between the control group and NC
group; however, compared with the NC group, the expression levels
of miR-107 in the miR-107 mimics group were significantly increased
(P<0.01). As demonstrated in Fig.
4B there was no significant difference in the proliferation of
cells between the control group and the NC group throughout the
whole experiment. As time increased, there was a significantly
reduced proportion of proliferative cells in the miR-107 mimics
group, in comparison with the control group (P<0.01). However,
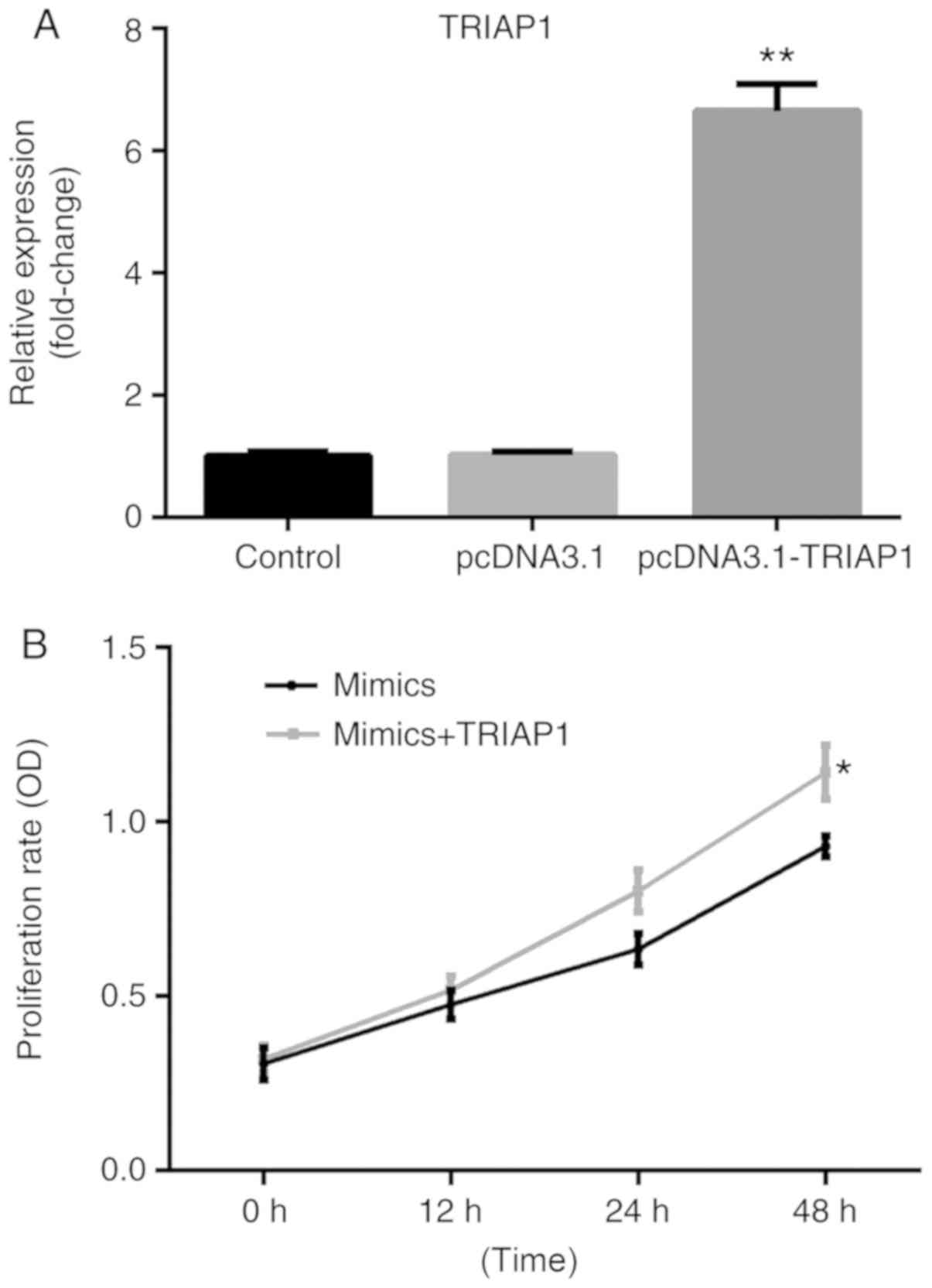
following transfection with pcDNA3.1 or pcDNA3.1-TRIAP1 (P<0.01;
Fig. 5A), the miR-107 mimics-reduced
A549 cell proliferation rate was reversed by co-transfection with
pcDNA3.1-TRIAP1 (P<0.05; Fig.
5B).
miR-107 promotes lung cancer cell
apoptosis
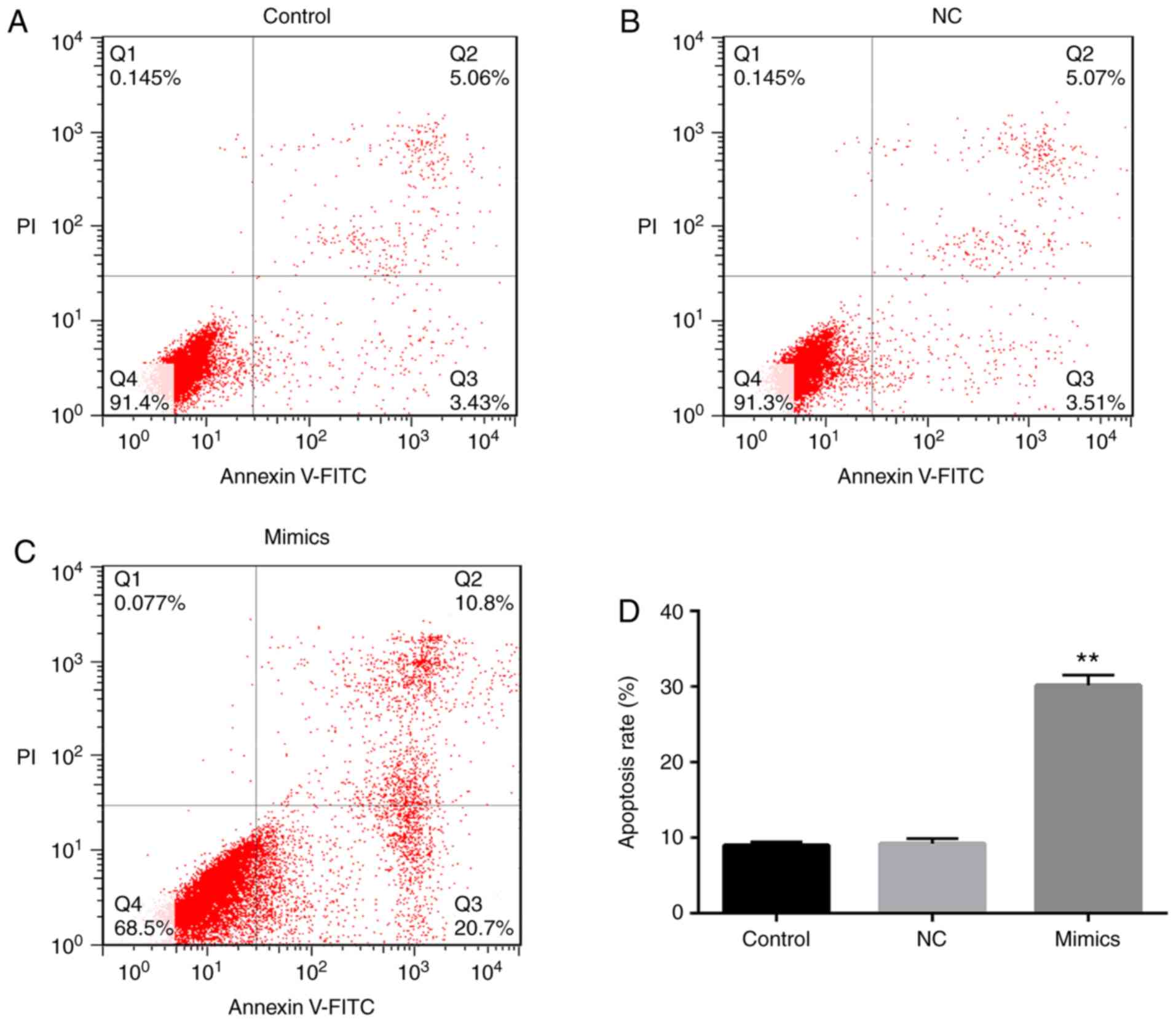
An Annexin V-FITC apoptosis kit was used to identify
the apoptotic rate of A549 cells in order to investigate the effect
of miR-107 on lung cancer cell apoptosis. The apoptotic rate of the
cells in the miR-107 mimics group was almost four times higher than
that of the cells in the control group or the NC group (P<0.01;
Fig. 6A-D). There was no distinct
difference in the apoptotic rate of the cells in the control group
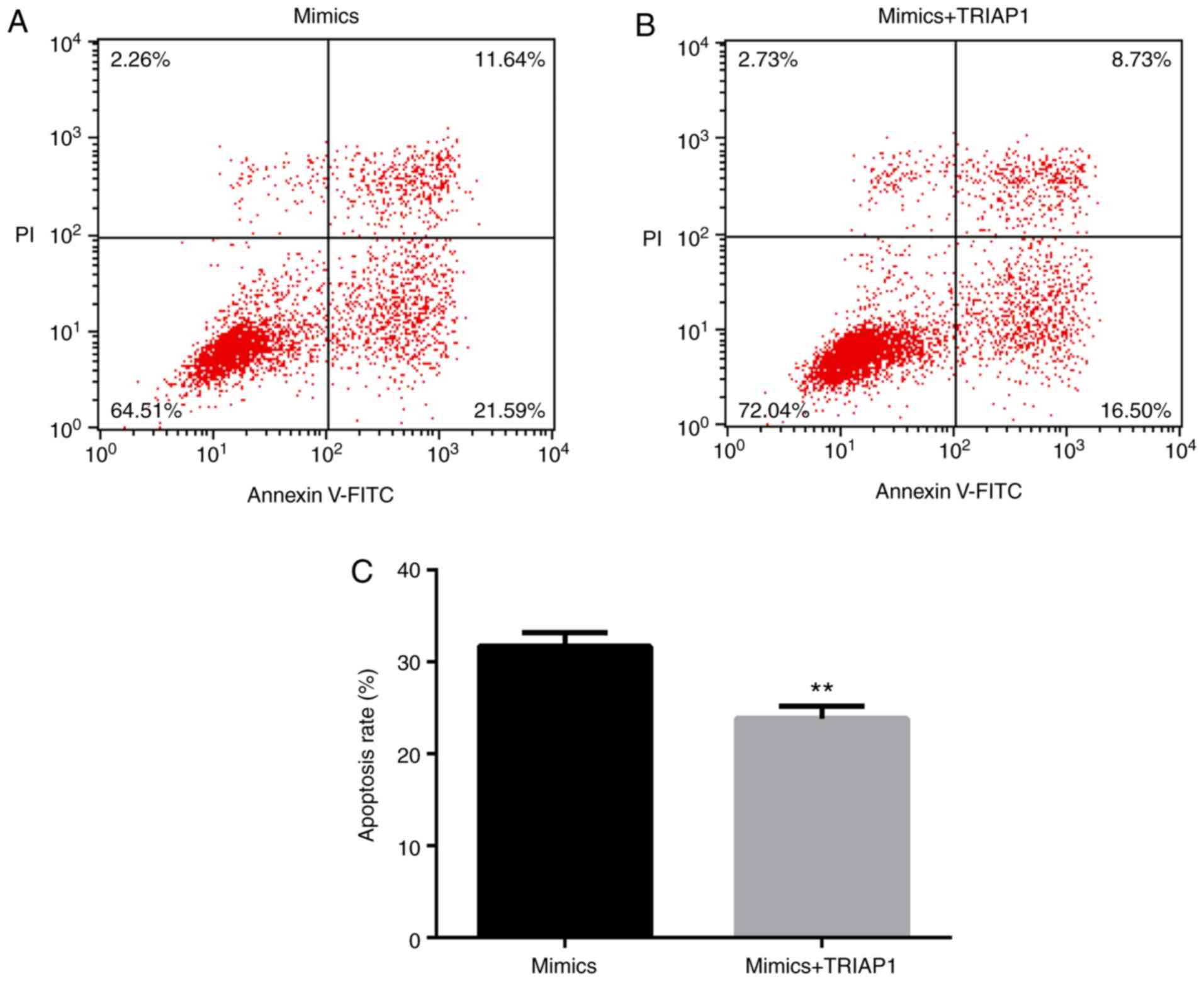
and the NC group. However, miR-107 mimics-induced A549 cell
apoptosis was reduced by co-transfection with pcDNA3.1-TRIAP1
(P<0.01; Fig. 7A-C).
Effect of miR-107 on the expression
levels of cyclin D1 and PCNA
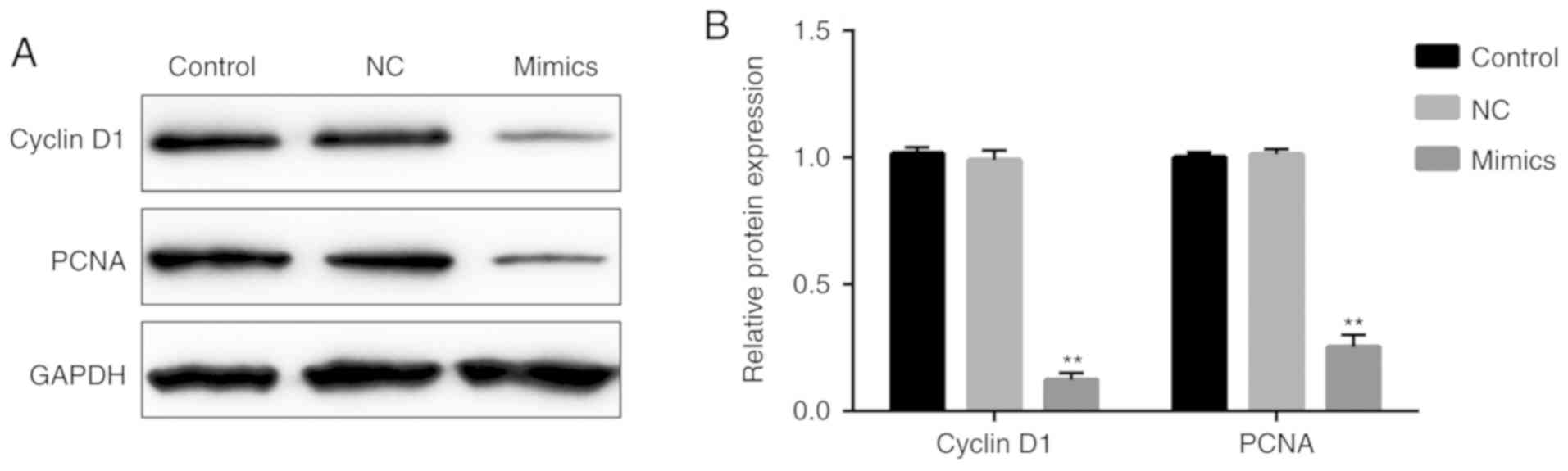
In order to further demonstrate the effect of
miR-107 on regulating A549 cell proliferation, the expression
levels of cyclin D1 and PCNA were measured. As demonstrated in
Fig. 8, the protein expression
levels of cyclin D1 and PCNA were significantly decreased in the
miR-107 mimics group, as compared with the NC group (P<0.01).
However, there were no significant differences between the NC and
control groups.
Effect of miR-107 on the protein
expression of BCL2, BAX, TP53 and caspase 3
In order to further investigate the association
between miR-107 and its target gene TRIAP1, a western blot assay
was performed to examine the protein expression levels of TRIAP1
and related apoptotic proteins among the different groups. As shown
in Fig. 9, the protein expression
levels of TRIAP1 and BCL2 were significantly reduced and the
expression levels of BAX, TP53 and caspase 3 were significantly
increased in the miR-107 mimics group, compared with those in the
NC group (P<0.01). The protein expression levels of BCL2, BAX,
TP53 and caspase 3 in the control group and the NC group were
similar.
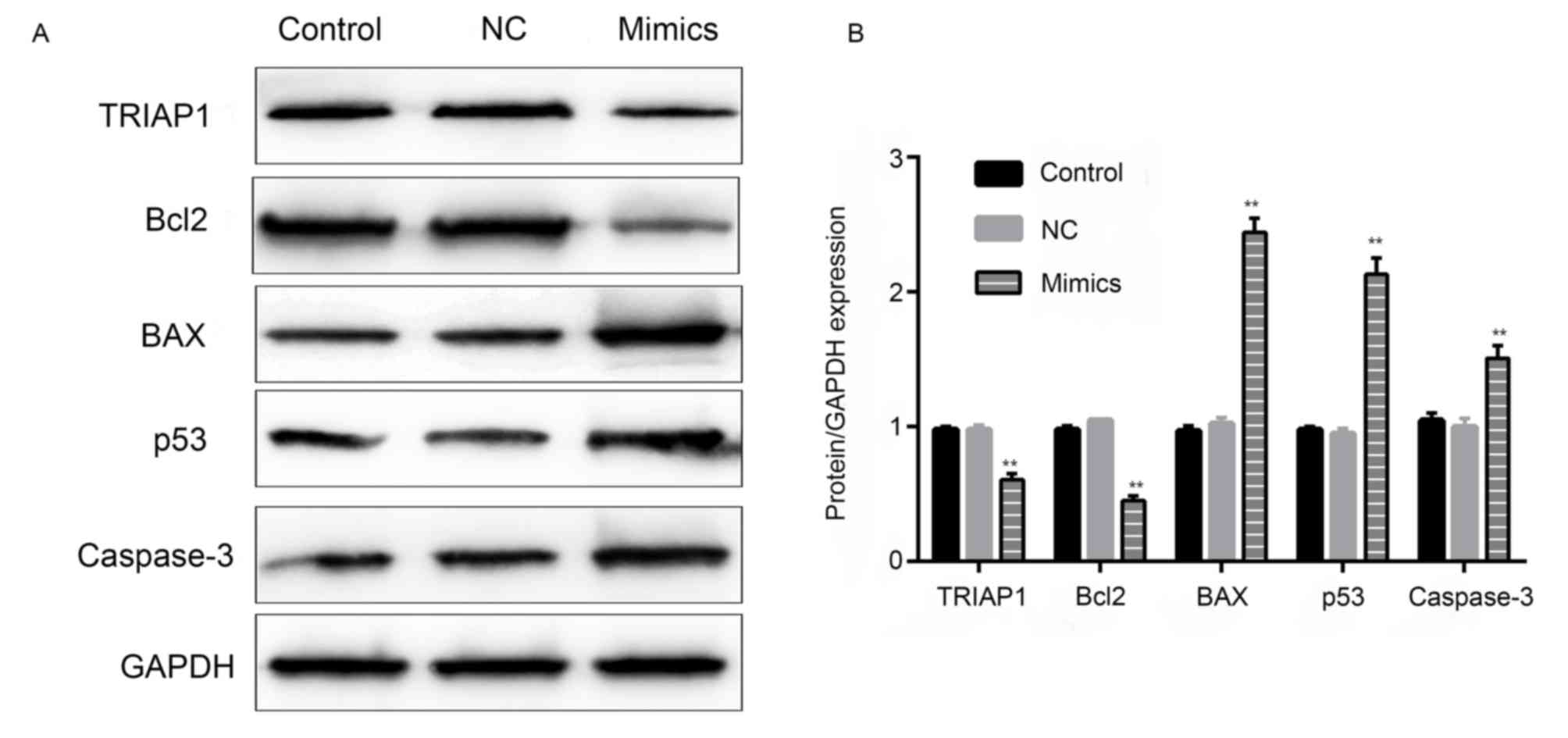 | Figure 9.Effect of miR-107 on the expression
of TRIAP1, BCL2, BAX, TP53 and caspase 3. (A) Protein expression
level of TRIAP1, BCL2, BAX, TP53 and caspase 3 in different groups.
(B) The quantified protein expression levels of TRIAP1, BCL2, BAX,
TP53 and caspase 3 (n=3). **P<0.01 vs. the miR-107
NC-transfected cells. miR, microRNA; TRIAP1, TP53-regulated
inhibitor of apoptosis 1; BCL2, BCL2 apoptosis regulator; BAX, BCL2
associated X, apoptosis regulator; TP53, tumor protein 53; NC,
negative control; GAPDH, glyceraldehyde-3-phosphate
hydrogenase. |
Discussion
A previous study revealed that miR-107 promoted the
proliferation of hepatocellular carcinoma cells by targeting axin 2
(17). Furthermore, miR-107 promoted
the proliferation and invasion of gastric adenocarcinoma cells
through large tumor suppressor kinase 2 (18). However, miR-107 inhibited cell
proliferation and metastasis in gastric cancer (19) and osteosarcoma (20). Therefore, miR-107 acts as a tumor
suppressor or oncogene in different types of cancer. The present
study demonstrated that miR-107 may function as a tumor suppressor
in lung cancer, which corresponds with previous studies (17–20).
In the present study, the results of the CCK-8 and
flow cytometry assays demonstrated that the overexpression of
miR-107 reduced proliferation and promoted apoptosis in lung cancer
cells in vitro. A previous study demonstrated that
miR-107-5p suppressed NSCLC by directly targeting the oncogene
epidermal growth factor receptor (21). Another study indicated that miR-107
inhibited tumor growth by targeting the brain derived neurotrophic
factor-mediated PI3K/AKT signaling pathway in human NSCLC (14). The results of the present study were
consistent with those of previous studies; however, a target gene
of miR-107, which may provide novel approaches for clinical
treatment, was also identified.
miRNAs regulate cell proliferation and apoptosis by
targeting specific genes during cellular processes (22). According to previous studies, TRIAP1
was predicted to be a candidate oncogene in various types of
cancer, including ovarian cancer (23), nasopharyngeal carcinoma (24) and lung cancer (25). In the present study, TRIAP1 was
revealed to be upregulated in lung cancer tumor tissues compared
with adjacent non-tumor tissue. TRIAP1 was subsequently predicted
to be a target gene of miR-107 using TargetScan and a
dual-luciferase reporter assay. The CCK-8 and flow cytometry assays
further demonstrated that the effects on A549 cell proliferation
and apoptosis induced by miR-107 mimics were reversed by
co-transfection with pcDNA3.1-TRIAP1. Collectively, miR-107 may
inhibit cell proliferation and promote cell apoptosis in lung
cancer by targeting TRIAP1. However, the association between
miR-107 and TRIAP1 requires further investigation.
In order to demonstrate the function of miR-107 on
cell proliferation of lung cancer, the expression levels of the
proliferation-associated factors, cyclin D1 and PCNA were measured
via a western blot assay. Previous studies reported that inhibitory
expression of cyclin D1 decreased lung cancer cell proliferation
(26,27). Furthermore, the decreased expression
of PCNA may inhibit lung cancer cell proliferation (28,29). In
the present study, the expression levels of cyclin D1 and PCNA were
significantly decreased in the miR-107 mimics group compared with
the miR-NC mimics group. Thus, it was hypothesized that the
overexpression of miR-107 may inhibit lung cancer cell
proliferation by reducing the expression levels of cyclin D1 and
PCNA.
In the current study, the expression levels of the
apoptosis-associated factors BAX, TP53, caspase 3 and BCL2 were
measured using a western blot assay. A previous study demonstrated
that overexpression of caspase 3 may inhibit the apoptosis of lung
cancer cells (30). Another study
reported that activation of BAX contributed to the apoptosis of
lung cancer cells (31). TP53
functions as a tumor suppressor and blocks cancer progression
(32). Overexpression of miR-107 may
increase the expression levels of caspase 3 (33), BAX (34) and TP53 (35); while the inhibition of TRIAP1 may
increase the expression levels of TP53 and caspase 3 (36,37). In
the present study, the protein expression levels of BAX, TP53 and
caspase 3 were significantly increased, and those of TRIAP1 were
decreased, in the miR-107 mimics group compared with the NC group.
Therefore, it was hypothesized that miR-107 may induce the
apoptosis of lung cancer cells by targeting TRIAP1 and increasing
the expression levels of BAX, TP53 and caspase 3, which are three
known tumor suppressors in lung cancer (32,33).
Furthermore, previous studies have demonstrated that BCL2 knockdown
decreased the viability and increased apoptosis of lung cancer
cells (38), while overexpression of
BCL2 decreased apoptosis of human lung cancer cells (39). Therefore, BCL2 may act as a tumor
promoter in lung cancer. In the present study, the expression
levels of TRIAP1 and BCL2 were decreased in the miR-107 mimics
group compared with the NC group. Therefore, the inhibition of
TRIAP1 induced by the overexpression of miR-107 may reduce the
expression of BCL2, a known tumor promoter in lung cancer (38,39),
which contributes to apoptosis in the process of lung cancer.
However, the underlying mechanisms involved in TRIAP1 and
apoptosis-associated proteins require further investigation.
In conclusion, miR-107 may inhibit A549 lung cancer
cell proliferation and promote apoptosis by targeting TRIAP1,
indicating that miR-107 may be a novel target in lung cancer
treatment. However, the current study had the following
limitations: i) The association and interaction of miR-107 and its
target TRIAP1 require further investigation; ii) the effects of
miR-107 and other targets on regulating lung cancer cell
proliferation and apoptosis requires further study; iii) the
histopathological patterns of lung cancer tissues were not
distinguished; and iv) further experiments on different lung cancer
cell lines and in in vivo models are required.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PC collected lung cancer tumor and non-tumor
adjacent tissues, analyzed and interpreted the patient data. JL,
GC, BP, LY and BZ analyzed all the experimental data. YY performed
all the cell experiments and was a major contributor in writing the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Jingmen No. 2 People's Hospital (Jingmen, China) and
written consent was acquired from each patient.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Didkowska J, Wojciechowska U, Mańczuk M
and Łobaszewski J: Lung cancer epidemiology: Contemporary and
future challenges worldwide. Ann Transl Med. 4:1502016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nadel E, Truini A, Nakata A, Lin J, Reddy
RM, Change AC, Ramnath N, Gotoh N, Beer DG and Chen G: A novel
serum-4-microRNA signature for lung cancer detection. Sci Rep.
23:124642015. View Article : Google Scholar
|
|
3
|
Tarver T: American cancer society cancer
facts & figures 2014. Jour Consum Heal Intern. 16:366–367.
2012. View Article : Google Scholar
|
|
4
|
Nanavaty P, Alvarez MS and Alberts WM:
Lung cancer screening: Advantages, controversies, and applications.
Cancer Control. 21:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fortunato O, Boeri M, Moro M, Verri C,
Mensah M, Conte D, Calecal L, Roz L, Pastorino U and Sozzi G:
Mir-660 is downregulated in lung cancer patients and its
replacement inhibits lung tumorigenesis by targeting MDM2-p53
interaction. Cell Death Dis. 11:e15642014. View Article : Google Scholar
|
|
6
|
Huang J, Sun C, Wang S, He Q and Li D:
MicroRNA miR-10b inhibition reduces cell proliferation and promotes
apoptosis in non-small cell lung cancer (NSCLC) cells. Mol Biosyst.
11:2051–2059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as a tumor suppressors and oncogenes.
Oncogene. 25:6199–6196. 2006. View Article : Google Scholar
|
|
8
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Imamura T, Komatsu S, Ichikawa D, Miyamae
M, Okajima W, Ohashi T, Kiuchi J, Nishibeppu K, Konishi H, Shiozaki
A, et al: Depleted tumor suppressor miR-107 in plasma relates to
tumor progression and is a novel therapeutic target in pancreatic
cancer. Sci Rep. 7:57082017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu P, Qi X, Bian C, Yang F, Lin X, Zhou
S, Xie C, Zhao X and Yi T: MicroRNA-18a inhibits ovarian cancer
growth via directly targeting TRIAP1 and IPMK. Oncol Lett.
13:4039–4046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang JZ, Bian L, Hou JG and Wang HY:
MiR-550a-3p promotes non-small cell lung cancer cell proliferation
and metastasis through down-regulating TIMP2. Eur Rev Med Pharmacol
Sci. 22:4156–4165. 2018.PubMed/NCBI
|
|
12
|
Han L, Chen W, Xia Y, Song Y, Zhao Z,
Cheng H and Jiang T: MiR-101 inhibits the proliferation and
metastasis of lung cancer by targeting zinc finger E-box binding
homeobox 1. Am J Transl Res. 10:1172–1183. 2018.PubMed/NCBI
|
|
13
|
Cui J, Mo J, Luo M, Yu Q, Zhou S, Li T,
Zhang Y and Luo W: c-Myc-activated long non-coding RNA h19
downregulates miR-107 and promotes cell cycle progression of
non-small cell lung cancer. Int J Clin Exp Pathol. 8:12400–12409.
2015.PubMed/NCBI
|
|
14
|
Xia H, Li Y and Lv X: MicroRNA-107
inhibits tumor growth and metastasis by targeting the BDNF-mediated
PI3K/AKT pathway in human non-small lung cancer. Int J Oncol.
49:1325–1333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takahashi Y, Forrest AR, Maeno E,
Hashimoto T, Daub CO and Yasuda J: MiR-107 and miR-185 can induce
cell cycle arrest in human non small cell lung cancer cell lines.
PLoS One. 4:e66772009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression (data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang JJ, Wang CY, Hua L, Yao KH, Chen JT
and Hu JH: MiR-107 promotes hepatocellular carcinoma cell
proliferation by targeting Axin2. Int J Clin Exp Pathol.
8:5168–5174. 2015.PubMed/NCBI
|
|
18
|
Zhang M, Wang X, Li W and Cui Y: MiR-107
and miR-25 simultaneously target LATS2 and regulate proliferation
and invasion of gastric adenocarcinoma (GAC) cells. Biochem Biophys
Res Commun. 460:806–812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheng F, Yang Z, Huang F, Yin L, Yan G and
Gong G: MicroRNA-107 inhibits gastric cancer cell proliferation and
metastasis by targeting PI3K/AKT pathway. Microb Pathog.
121:110–114. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu M, Guo D, Cao Z, Xiao L and Wang G:
Inhibitory effect of microRNA-107 on osteosarcoma malignancy
through regulation of wnt/β catenin signaling in vitro. Cancer
Invest. 36:175–184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang P, Liu X, Shao Y, Wang H, Liang C,
Han B and Ma Z: MicroRNA-107-5p suppresses non-small cell lung
cancer by directly targeting oncogene epidermal growth factor
receptor. Oncotarget. 8:57012–57023. 2017.PubMed/NCBI
|
|
22
|
Chen JF, Mandel EM, Thomson JM, Wu QL,
Callis T, Hammond SM, Conlon FL and Wang DZ: The role of microRNA-1
and microRNA-133 in skeletal muscle proliferation and
differentiation. Nat Genet. 38:228–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu P, Qi X, Bian C, Yang F, Lin X, Zhou
S, Xie C, Zhao X and Yi T: MicroRNA-18a inhibits ovarian cancer
growth via directly targeting TRIAP1 and IPMK. Oncol Lett.
13:4039–4046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He Q, Yang X, Ren X, Wen X, Zhang J, Wang
Y, Liu N and Ma J: Overexpression of mitochondria mediator gene
TRIAP1 by miR-320b loss is associated with progression in
nasopharyngeal carcinoma. PLoS Genet. 12:e10061832016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang B, Zuo ZJ, Lv F, Zhao L, Du MJ and
Gao YS: MiR-107 inhibits proliferation of lung cancer cells through
regulating TP53 regulated inhibitor of apoptosis 1 (TRIAP1). Open
Life Sci. 12:200–205. 2017.
|
|
26
|
Tian XP, Jin XH, Li M, Huang WJ, Xie D and
Zhang JX: The depletion of PinX1 involved in the tumorigenesis of
non-small cell lung cancer promotes cell proliferation via
p15/cyclin D1 pathway. Mol Cancer. 16:742017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Du B, Wang Z, Zhang X, Feng S, Wang G, He
J and Zhang B: MicroRNA-545 suppresses cell proliferation by
targeting cyclin D1 and CDK4 in lung cancer cells. PLoS One.
9:e880222014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Chen T, Huang H, Jiang Y, Yang L,
Lin Z, He H, Liu T, Wu B, Chen J, et al: miR-363-3p inhibits tumor
growth by targeting PCNA in lung adenocarcinoma. Oncotarget.
8:20133–20144. 2017.PubMed/NCBI
|
|
29
|
Wang X, Shi W, Shi H, Lu S, Wang K, Sun C,
He J, Jin W, Lv X, Zhou H and Shu Y: TRIM11 overexpression promotes
proliferation, migration and invasion of lung cancer cells. J Exp
Clin Cancer Res. 35:1002016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xue Y, Wu L, Liu Y, Ma Y, Zhang L, Ma X,
Yang Y and Chen J: ENTPD5 induces apoptosis in lung cancer cells
via regulating caspase 3 expression. PLoS One. 10:e01200462015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu JJ, Qiao KS, Sun P, Chen P and Li Q:
Study of EGCG induced apoptosis in lung cancer cells by inhibiting
PI3K/Akt signaling pathway. Eur Rev Med Pharmacol Sci.
22:4557–4563. 2018.PubMed/NCBI
|
|
32
|
Stegh AH: Targeting the p53 signaling
pathway in cancer therapy-the promises, challenges and perils.
Expert Opin Ther Targets. 16:67–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang ZC, Liu JX, Shao ZW, Pu FF, Wang BC,
Wu Q, Zhang YK, Zeng XL, Guo XD, Yang SH and He TC: In vitro effect
of microRNA-107 targeting Dkk-1 by regulation of Wnt/β-catenin
signaling pathway in osteosarcoma. Medicine (Baltimore).
96:e72452017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sirotkin AV, Laukova M, Ovcharenko D,
Brenaut P and Mlyncek M: Identification of microRNAs controlling
human ovarian cell proliferation and apoptosis. J Cell Physiol.
223:49–56. 2010.PubMed/NCBI
|
|
35
|
Yamakuchi M, Lotterman CD, Bao C, Hruban
RH, Karim B, Mendell JT, Huso D and Lowenstein CJ: P53-induced
microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad
Sci USA. 107:6334–6339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Andrysil Z, Kim J, Tan AC and Espinosa JM:
A genetic screen identifies TCF3/ESA and TRIAP1 as pathway-specific
regulators of the cellular response to p53 activation. Cell Rep.
3:1346–1354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu P, Qi X, Bian C, Yang F, Lin XJ, Zhou
S, Xie C, Zhao X and Yi T: MicroRNA-18a inhibits ovarian cancer
growth via directly targeting TRIAP1 and IPMK. Oncol Lett.
13:4039–4046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao J, Li X, Zou M, He J, Han Y, Wu D,
Yang H and Wu J: MiR-135a inhibition protects A549 cells from
LPS-induced apoptosis by targeting Bcl-2. Biochem Biophys Res
Commun. 452:951–957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun SY, Yue P, Zhou JY, Wang Y, Choi Kim
HR, Lotan R and Wu GS: Overexpression of BCL2 blocks TNF-related
apoptosis-inducing ligand (TRAIL)-induced apoptosis in human lung
cancer cells. Biochem Biophys Res Commun. 280:788–797. 2001.
View Article : Google Scholar : PubMed/NCBI
|