Introduction
The hematopoietic microenvironment comprises a
mixture of bone marrow mesenchymal stromal cells (BM-MSCs),
fibroblasts, vascular endothelial cells, etc. BM-MSCs support the
proliferation and differentiation of hematopoietic stem cells, and
themselves undergo differentiation into various functional cells,
including adipocytes, osteocytes, and cells from the chondrogenic
lineage (1,2). Studies on hematologic malignancies have
demonstrated genetic abnormalities in neoplastic cells; however,
the role of the hematopoietic microenvironment in the pathogenesis
of human myeloid neoplasia is still unclear, therefore we attempted
to assess BM-MSCs. Considering the anatomical architecture, BM-MSCs
are components of the hematopoietic niche, and hematopoietic cells
lie in close proximity to the bone endosteal surface, which is in
direct contact with endothelial cells (1) and hence linked to drug sensitivity
(3). BM-MSCs also act as
immunosuppressive agents by inhibiting the proliferation of immune
cells (4,5). Therefore, BM-MSCs may have an important
effect on adjacent hematopoietic cells and immune control system in
hematologic neoplasia.
Recent studies have demonstrated that neoplastic
cells shed extracellular vesicles (EVs), including exosomes, and
endow BM-MSCs to facilitate progression of neoplastic cells
(6,7). Exosomes are EVs, about 50–150 nm in
size, and contain genetic elements such as messenger RNA, microRNA
(miR), and DNA (8–10). EVs act as cargos involved in
cell-to-cell communication and are associated with the
transformation of adjacent fibroblasts into cancer-associated
fibroblasts (CAFs), which are involved in cancer progression
(11,12). Tumor cell-derived EVs, including
exosomes, also act as angiogenesis agents in the cancer
microenvironment (10). Thus,
certain types of tumors have the ability to reconstruct the
surrounding environment to facilitate the progression of neoplastic
cells. On the contrary, BM-MSCs affect functions of neoplastic
cells associated with therapy resistance and disease progression
(4). Therefore, cell-to-cell
communication may exist between tumor cells and the surrounding
environment, including fibroblasts, microvessels, and other
adjacent tissues.
miRs are very short non-coding RNAs around 20 bp in
length and are detected in various cell types and body fluids,
including serum, urine, and saliva (9,10). miRs
are considered as the candidates of liquid biopsy. In the body
fluids, miRs are usually encapsulated in EVs in the stable forms
and are transported to other cells, wherein they control cellular
functions. As BM-MSCs are major players in the hematopoietic tissue
and their genetic alterations in myelodysplastic syndrome (MDS) is
plausible, we attempted to search EV-miRs in the BM-MSCs obtained
from patients with MDS and acute myeloid leukemia with
myelodysplasia-related changes (AML/MRC). We found that the
expression of EV-miR-101 in BM-MSCs was downregulated and related
to MDS progression.
Materials and methods
Patients
Twenty-nine consecutive patients with myeloid
malignancies (22 with MDS and 7 with AML/MRC) were enrolled in this
study (Table I). The diagnoses were
established as per the World Health Organization criteria, and
included three patients with MDS with single lineage dysplasia
(SLD), seven with multilineage dysplasia (MLD), two with
unclassifiable MDS, one with 5q-syndrome, five with excess blast
(EB)-1, and four with EB-2. Risk analysis was carried out based on
the revised International Prognostic Scoring System (IPSS-R)
(13) and risk categorization was
performed as per the NCCN guideline (14). Four patients with MDS with
intermediate risk were tentatively included in the low-risk MDS
group. Written informed consents were obtained from all patients
before the collection of specimens according to the Declaration of
Helsinki. The present study was validated by the internal review
boards of Tokyo Medical University (no. 2648, approved 22 April,
2014). As controls, human BM-MSCs from healthy donors (normal
BM-MSCs) were purchased from Lonza Inc. Control serum was obtained
from age-matched healthy individuals.
 | Table I.Hematologic features of
myelodysplastic syndrome patients of whom bone marrow mesenchymal
stroma cells were obtained. |
Table I.
Hematologic features of
myelodysplastic syndrome patients of whom bone marrow mesenchymal
stroma cells were obtained.
| UPN | Age (Year) | M/F | Dx (WHO) | Date of material
obtained | Transition to acute
leukemia | Outcome | IPSS | IPSS-R | WBC | Neutrophils | Hb | Plt | PB-blasts (%) | BM-blasts (%) | Karyotype |
|---|
| 33 | 51 | M | MDS-SLD | 2014/10/10 | No | Alive | Int-1 | Int | 5200 | 4030 | 7.6 | 135 | 0 | 0.8 | complex |
| 54 | 58 | F | MDS-EB1 | 2014/12/16 | No | Alive | Int-1 | Int | 2400 | 1080 | 9.3 | 74 | 0 | 5.6 | 46,XX |
| 59 | 55 | M | MDS-MLD | 2015/1/27 | No | Alive | Low | Very low | 4700 | 2961 | 13.7 | 56 | 0 | 0 | 46,XY |
| 82 | 78 | M | MDS-MLD | 2015/4/30 | No | Unknown | Int-1 | Very low | 1900 | 1267 | 10.6 | 57 | 0 | 2 | 45,X,-Y |
| 95 | 68 | M | MDS-SLD | 2015/8/20 | No | Unknown | Low | Low | 10200 | 7905 | 12.1 | 18 | 0 | 1.2 | 46,XY |
| 93 | 82 | M | MDS-SLD | 2015/8/11 | No | Dead | Low | Very low | 2200 | 1606 | 10.4 | 105 | 0 | 0 | 46,XY,del
(20)(q11) |
| 101 | 77 | M | MDS-U based on
defining cytogenetic abnormality | 2015/9/17 | No | Alive | Low | Very low | 4600 | 2852 | 10 | 30 | 0 | 0 | 45,X,-Y |
| 102 | 47 | F | MDS-MLD | 2015/9/30 | No | Alive | Int-1 | Low | 2800 | 1274 | 8.3 | 15 | 0 | 0 | 46,XX |
| 103 | 22 | M | MDS-MLD | 2015/10/9 | No | Unknown | Low | Int | 2600 | 845 | 6.3 | 21 | 0 | 0 | 46,XY |
| 107 | 82 | F | MDS-MLD | 2015/10/22 | No | Alive | Low | Low | 6100 | 4148 | 11.2 | 187 | 0.5 | 0.4 | 47,XX,+8 |
| 64_2 | 46 | F | MDS-MLD | 2016/3/8 | No | Alive | Low | Very low | 4500 | 2835 | 13.1 | 299 | 0 | 2 | 46,XX |
| 141 | 84 | M | MDS with isolated
del(5q) | 2016/5/19 | No | Alive | Int-1 | Int | 3500 | 1295 | 9.5 | 344 | 0 | 0.5 | 46,XY,del(5q) |
| 115 | 50 | M | MDS-MLD | 2016/10/11 | No | Unknown | Int-1 | Low | 6600 | 5214 | 4.3 | 42 | 0.5 | 2 | 46,XY,
del(11)(q14) |
| 8 | 72 | M | MDS-EB1 | 2014/8/8 | No | Unknown | Int-1 | High | 1700 | 408 | 9 | 78 | 0 | 5.2 | 46,XY |
| 12 | 38 | M | MDS-EB2 | 2014/8/26 | No | Unknown | Int-2 | High | 3000 | 1410 | 10.3 | 95 | 0.5 | 10 | 45,XY,-7 |
| 58 | 71 | M | MDS-EB1 | 2015/1/22 | No | Unknown | Int-2 | Very high | 2900 | 2233 | 8.3 | 31 | 0 | 9.2 | complex |
| 22 | 66 | M | MDS-EB1 | 2014/11/27 | Yes | Unknown | Int-1 | High | 2000 | 910 | 7.6 | 16 | 0 | 7.6 | 46,XY |
| 44 | 72 | M | MDS-EB2 | 2015/1/22 | Yes | Dead | High | Very high | 16300 | 2853 | 9.1 | 97 | 5 | 16.8 | 46,XY,-7,+m/46,
XY,idem,del(12) (p13) |
| 38 | 56 | M | MDS-EB2 | 2014/10/14 | No | Dead | Int-2 | High | 5500 | 4895 | 8.3 | 164 | 0 | 12.4 | 46,XY |
| 120 | 68 | F | MDS-EB2 | 2016/3/10 | Yes | Dead | High | Very high | 1100 | 374 | 7.8 | 19 | 0 | 17.2 | complex |
| 108 | 87 | M | MDS-U with > 1%
PB blasts | 2015/10/26 | No | Dead | in-2 | High | 11400 | 10864 | 8.1 | 65 | 1.5 | 4.8 | 46,XY,del(20)
(q11)/46,XY, idem,-7,+8 |
| 175 | 68 | M | MDS-EB1 | 2016/2/3 | Yes | Dead | Int-2 | High | 2000 | 1110 | 8.8 | 30 | 2 | 4.4 | complex |
| 4 | 77 | M | AML with MRC | 2014/4/2 |
| Dead |
|
| 4500 | 393 | 8.9 | 23 | 17 | 49.5 | complex |
| 30 | 72 | M | AML with MRC | 2014/10/8 |
| Unknown |
|
| 800 | 248 | 7.7 | 50 | 2 | 21 | 45,X,-Y |
| 13 | 61 | M | AML (M6A) with
MRC | 2014/9/10 |
| Dead |
|
| 1300 | 338 | 7.6 | 17 | 12 | 12.8 | complex |
| 18 | 67 | F | AML with MRC | 2014/9/16 |
| Dead |
|
| 2000 | 520 | 7.9 | 7 | 2.5 | 66 | 45,XX,-7 |
| 127 | 64 | M | AML with MRC | 2016/4/11 |
| Alive |
|
| 1300 | 240 | 9.3 | 78 | 0 | 23.2 | 46,XY |
| 133 | 67 | F | AML with MRC | 2016/4/26 |
| Dead |
|
| 1400 | 400 | 7.3 | 25 | 8 | 34 | 45,XX,-18 |
| 98 | 42 | F | AML with MRC | 2015/9/9 |
| Dead |
|
| 2000 | 270 | 11.3 | 175 | 1.5 | 21 | 46,XX,inv(16)
(p13;q22)/47, XX,idem+22 |
Culture of BM-MSCs
BM-MSCs from patients with MDS and AML/MRC were
isolated using the conventional plastic adhesion method with a
minor modification (15). Briefly,
0.5 to 1 ml of freshy obtained heparinized BM aspirates were
cultured in equivalent volumes of Roswell Park Memorial Institute
(RPMI)-1640 medium (Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (FBS; HyClone) and 1%
penicillin-streptomycin (P/S; Thermo Fisher Scientific, Inc.) and
Dulbecco's modified Eagle's medium (DMEM) (Thermo Fisher
Scientific) containing 10% FBS (GE Healthcare), 1% of P/S, and 1%
non-essential amino acids (NEAAs; Thermo Fisher Scientific, Inc.).
Cells were cultured for 3 to 5 days, and the medium was changed to
DMEM (with 10% of FBS, 1% of P/S and 1% of NEAAs) for
non-hematopoietic expansion after the removal of non-adherent
cells. Cultured BM-MSC population was identified as
CD34−/CD45−/CD73+/CD90+/CD105+
with flow cytometry with <5% CD34+ and
CD45+ (15).
Isolation and miR profiling of
BM-MSCs
BM-MSCs (4×104 cells/cm2) were
cultured in 5 ml of DMEM and the culture supernatants were
harvested after 48 h of incubation. EV fraction was purified with
Exoquick-TC regent (System Biosciences) according to the
manufacturer's instructions. EVs were quantitated with a
nanoparticle tracking analysis (NanoSight LM10; Malven) (16–18).
Isolation of BM-MSC-derived EV-miRs or cellular-miR and serum
EV-miR was performed using the miRNeasy kit (Qiagen). Two-hundred
microliters of EV were diluted with 700 µl of QIAzol (Qiagen).
After 5 min incubation, 1 nM ath-miR-159 (Hokkaido System Science)
was added [18]; the mixture was vortexed for 30 sec and incubated
on ice for 10 min. Phenol extraction and cartridge filtration were
subsequently performed according to the manufacture's
instruction.
Screening of candidate miRNA
miRNA profiling was carried out using a TaqMan
low-density miRNA array (TLDA; Thermo Fisher Scientific, Inc.), as
previously reported (16,17). PCR was done on a Applied Biosystem
7900 HT thermocycler (Carlsberg) according to the manufacturer's
recommended program (the reaction was first incubated at 95°C for 2
min, followed by 50 cycles of 95°C for 15 sec and 60°C for 1 min)
using SDS2.2 software and Data Assist (Thermo Fisher Sciences,
Inc.). The expression of miRNAs was calculated based on cycle
threshold (Ct) values using the 2−ΔΔCq method (19) normalized to those of ath-miR-159,
which was spiked in each sample. Data analysis was carried out
using GeneSpring software (Agilent Technologies). The
Benjamin-Hochberg algorithm was used for the estimation of false
discovery rates, as previously reported (16,17).
Validation of candidate miRNA
expression by qRT-PCR
To compare selected miRNA expression in various
fractions (EV-miRNA, cellular miRNA, and serum EV-miRNA),
quantitative real-time PCR was performed by TaqMan®
MicorRNA Assays (ThermoFisher Scientific, Inc.) using an ABI Prism
7900 sequence detection system (Applied Biosystem) according to the
manufacturer's instruction.
The microRNA specific stem-loop primers
(has-miR-101, cat. no. 000438; ath-miR-159, cat. no. 000338) were
purchased from ThermoFisher Scientific, Inc. The reaction was first
incubated at 95°C for 2 min, followed by 50 cycles of 95°C for 15
sec and 60°C for 1 min, as we have reported previously (18).
Statistical analysis and evaluation of
miR-101 targets
Data were expressed as means ± standard deviation
(SD). Mann-Whitney U and chi-square tests were used to determine
statistical significance for comparisons between the control and
test groups. Multiple groups were compared with one-way analysis of
variance (ANOVA). Statistical analysis was carried out using R and
GraphPad Prism software (v. 5c for Macintosh; GraphPad Software
Inc.).
miRs with a ΔCt value >1.0 or <-1.0, and
P-values <0.05 were considered to exhibit differential
expression. Following identification of differentially expressed
miRs, the predicted target genes for these altered miRs were
subjected to experimental validation using miR-target interaction
database MiRTarBase (http://mirtarbase.mbc.nctu.edu.tw/). In addition,
functional annotation of target genes was carried out with Database
for Annotation, Visualization, and Integrated Discovery (DAVID
Bioinformatics tools v6.7) (http://david.abcc.ncifcrf.gov).
Results
Downregulation of BM-MSC-derived EV
miR-101 in patients with high-risk MDS
To identify the association between EV-miRs from
BM-MSCs and risk in patients with MDS, we used TLDA to screen the
miR expression profile between low-risk group (n=13: Very low, low,
and intermediate risk; 9 males/4 females, mean age 61.5 years
(range 22 to 84 years old)) and high-risk group (n=9: High and very
high risk; 8 males/1 females, mean age 66.4 years (range 38 to 87
years old)) separated by the IPSS-R. Seven patients with AML-MRC
included 4 males and 3 females, mean age 64.3 years with ranging 42
to 77 years old). We extracted nine miRs (has-miR-375, has-miR-101,
has-miR-424, has-miR-548c-3p, has-miR-15b, has-miR-485-3p,
has-miR-579, has-miR-195, and has-miR-369-3p) that were
differentially expressed between low-risk and high-risk groups
using R software-T test (Figs. 1 and
S1) (GSE133276). We then compared
the expression patterns of BM-MSC-derived EV-miRs detected with
TLDA between normal controls (Lonza) and low-risk group or
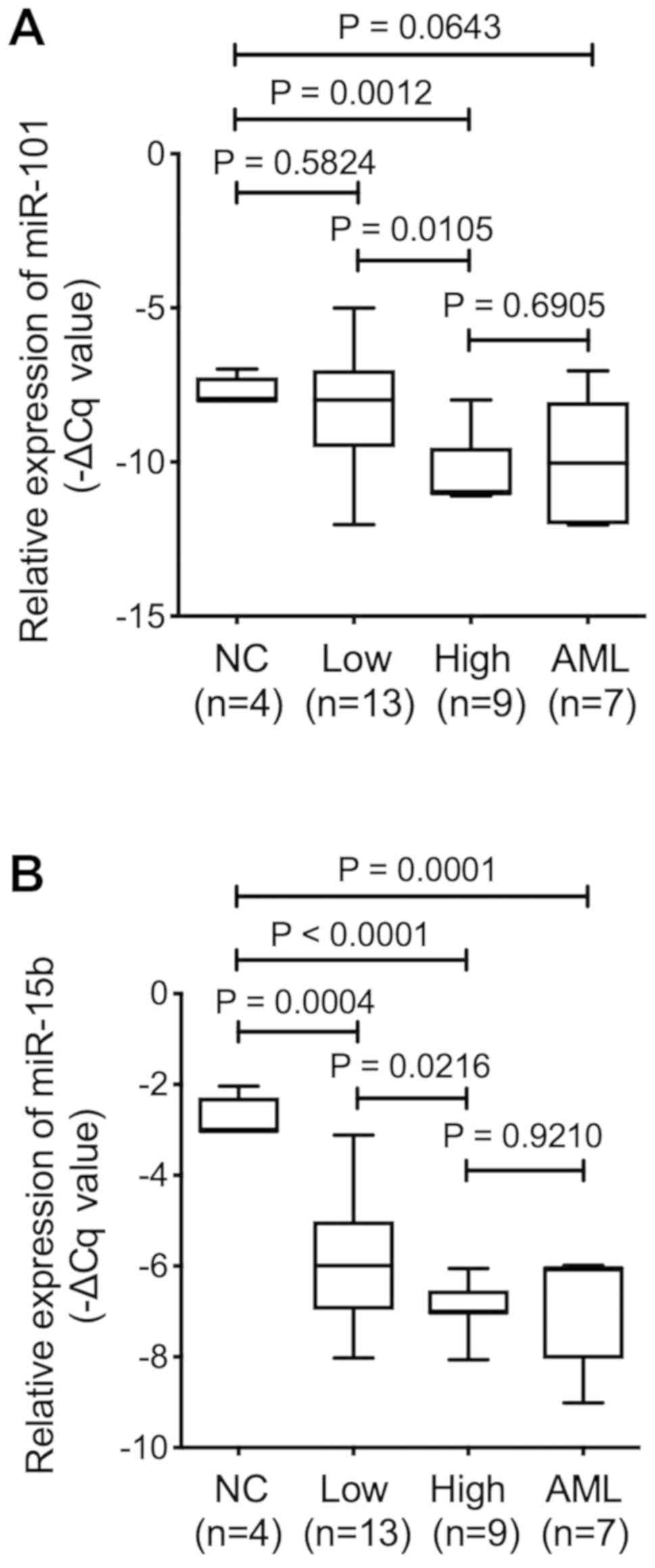
high-risk group. Of the nine EV-miRs, EV-miR-101 showed no
significant difference between normal control and low-risk groups
(P=0.5824), while its expression was downregulated in the high-risk
group (P=0.0012). We separated 13 MDS patients as low-risk group,
including 5 with very low MDS by IPSS-R with marrow blasts <2%,
therefore, we considered the character of BM-MSC may overlap of
those from normal control. On the other hand, EV-miR-15b expression
was downregulated in both the low-risk (P=0.0004) and high-risk
(P=0.0001) groups as compared with that in the control BM-MSCs,
suggesting that EV-miR-15b could serve as a biomarker for MDS
rather than that for disease progression (Fig. 1).
To confirm the observation of low expression level
of BM-MSC-derived EV-miR-101 in high-risk group and patients with
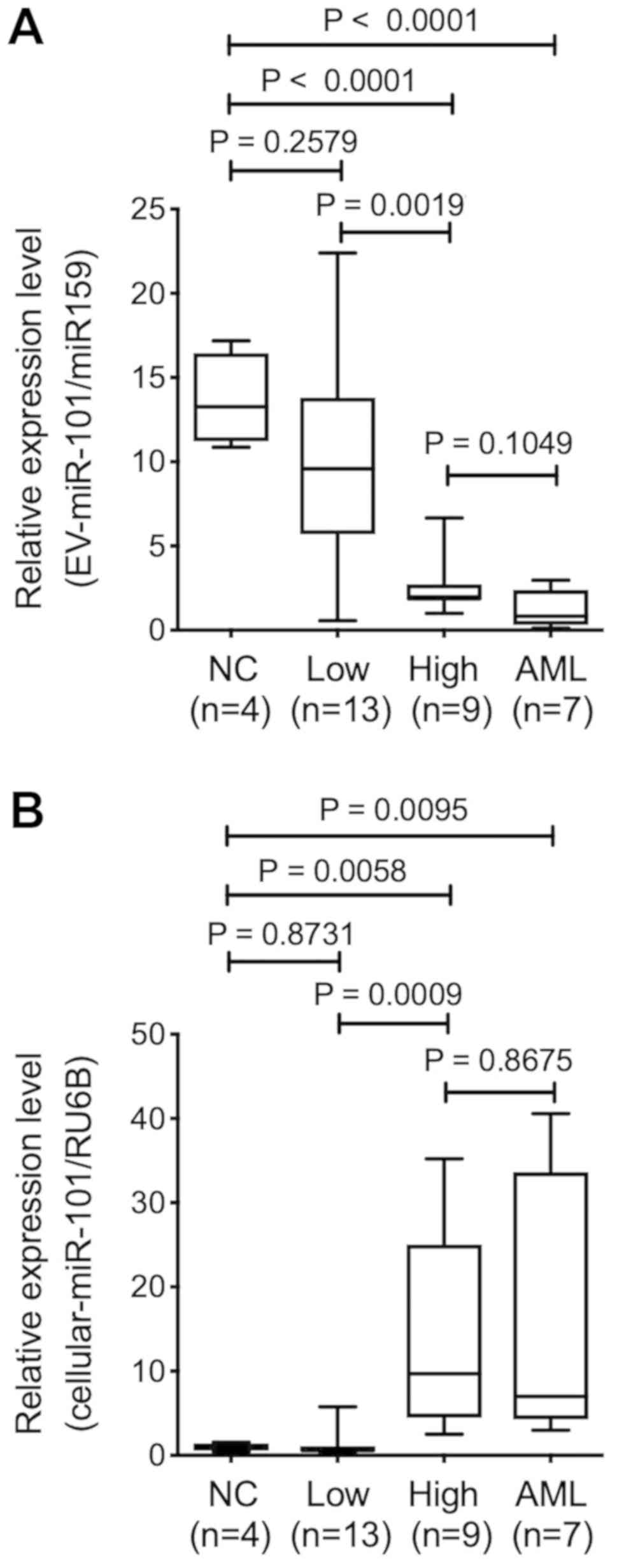
AML/MRC, we quantified the expression of EV-miR-101 with qRT-PCR.
As a result, we found no significant difference in EV-miR-101
expression level in BM-MSCs from normal control and low-risk groups
(P=0.2579). In contrast, EV-miR-101 expression significantly
decreased in the high-risk group (P<0.0001) and patients with
AML/MRC (P<0.0001) (Fig. 2A),
indicating that BM-MSC-derived EV-miR-101 expression level was
associated with disease severity in patients with MDS.
Expression pattern of miR-101 in
BM-MSCs and serum EV-miR-101
We assessed miR-101 level in BM-MSCs using qRT-PCR,
as the pattern of BM-MSC-derived EV-miR-101 was related to MDS
disease progression. The relative expression level of miR-101 in
control BM-MSCs (Lonza) was lower than that reported in the BM-MSCs
from the high-risk group (P=0.0058) and patients with AML/MRC
(P=0.0095). No significant difference was reported between normal
control and low-risk groups (P=0.8713). Therefore, the expression
profile of cellular miR-101 seemed to be a mirror image of the
EV-miR-101 obtained from BM-MSCs (Fig.
2B).
We next assessed EV-miR-101 level obtained from the
serum using qRT-PCR. Serum was obtained from identical patients
with MDS assessed by TLDA, and normal control serum was obtained
from age-matched healthy individuals (n=4). The expression level of
serum EV-miR-101 was significantly downregulated in low-risk group
(n=10) as compared with that in healthy control (P=0.0070), while
no significant difference was observed in high-risk group (n=7)
(P=0.3232) or patients with AML/MRC (n=6) (P=0.2731) as compared
with healthy subjects (Fig. 2C).
Serum EV-miR-101 expression pattern correlated with neither
BM-MSC-derived EV-miR-101 nor BM-MSC-derived cellular-miR-101 level
(data not shown).
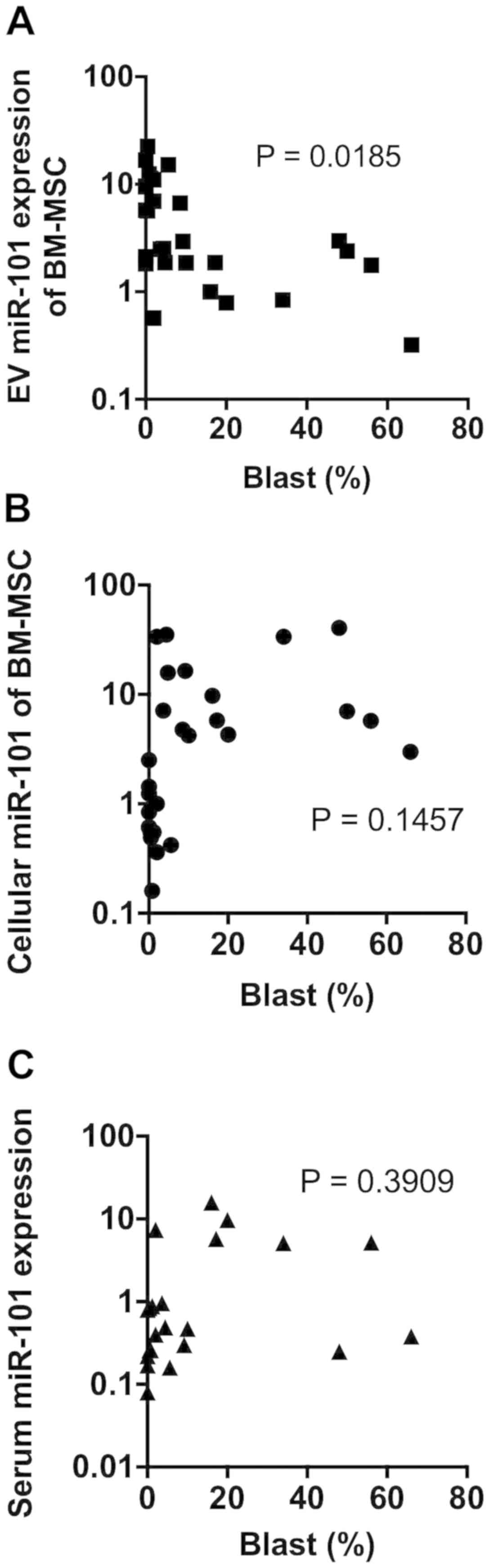
MiR-101 expression level and BM blast
percentage
We assessed the association between miR-101
expression levels and percentage of blasts in the BM. The
BM-MSC-derived EV-miR-101 level negatively correlated with the
occupancy of blasts in the BM (P=0.0185) (Fig. 3A). No significant correlations were
observed between BM blast percentage and BM-MSC-derived cellular
miR-101 level (P=0.1457) (Fig. 3B)
or serum EV-miR-101 level (P=0.3909) (Fig. 3C).
Target for miR-101
MiRTarBase was used to identify the predicted target
genes of miR-101 to determine its biological significance. More
than 50 target genes were extracted by MiRTarBase, and the target
genes that showed a strong evidence are summarized in Supplementary
Table I. The epigenetic regulator
genes, including enhancer of zester homolog 2 (EZH2), part
of polycomb repressive complex subunit (SUZ12), ten-eleven
translocation oncogene family member 2 (TET2), and
proto-oncogenes (c-FOS and c-MYC), were
experimentally validated with a luciferase reporter assay in the
literature. Functional annotation analysis with DAVID revealed the
most affected biological pathways of these genes downregulated in
the high-risk group, including cell-cycle related pathway and
epigenetic regulation pathway (Table
II).
 | Table II.The gene ontology terms enriched in
targets of miR-101. The terms with P-value <0.05 and false
discovery rate <0.05 were listed. |
Table II.
The gene ontology terms enriched in
targets of miR-101. The terms with P-value <0.05 and false
discovery rate <0.05 were listed.
| Term | P-value | Benjamini |
|---|
| Positive regulation
of transcription from RNA polymerase II promoter |
9.90×1010 |
1.30×106 |
| Negative regulation
of transcription from RNA polymerase II promoter |
4.10×107 |
2.60×104 |
| Positive regulation
of gene expression |
2.00×106 |
8.70×104 |
| Negative regulation
of cell proliferation |
5.00×106 |
1.60×103 |
| Positive regulation
of transcription, DNA-templated |
5.70×106 |
1.50×103 |
| Response to
estradiol |
1.20×105 |
2.50×103 |
| Positive regulation
of neuroblast proliferation |
3.60×105 |
6.60×103 |
| Cochlea
morphogenesis | 4.90
×105 | 7.80
×103 |
| Branching
morphogenesis of an epithelial tube | 5.60
×105 | 8.00
×103 |
| Positive regulation
of mesenchymal cell proliferation |
8.20×105 |
1.00×102 |
| Protein
phosphorylation |
1.10×104 |
1.30×102 |
| Positive regulation
of epithelial to mesenchymal transition |
1.70×104 |
1.80×102 |
| Epithelial to
mesenchymal transition |
1.90×104 |
1.80×102 |
| Response to
lipopolysaccharide |
2.00×104 |
1.80×102 |
| Coronary artery
morphogenesis |
2.20×104 |
1.80×102 |
| Cellular response
to hypoxia |
2.70×104 |
2.20×102 |
| Heart
development |
3.30×104 |
2.40×102 |
| Thymus
development |
3.70×104 |
2.60×102 |
| Positive regulation
of apoptotic process |
4.30×104 |
2.90×102 |
| Response to
drug |
4.60×104 |
2.90×102 |
| Positive regulation
of cellular component movement |
4.70×104 |
2.80×102 |
| Negative regulation
of epithelial cell differentiation |
5.70×104 |
3.30×102 |
| Negative regulation
of gene expression, epigenetic |
5.80×104 |
3.20×102 |
| Regulation of
apoptotic process |
6.50×104 |
3.40×102 |
| Positive regulation
of protein phosphorylation |
7.90×104 |
4.00×102 |
| Positive regulation
of epithelial cell proliferation |
1.00×103 |
4.80×102 |
Discussion
The present study demonstrates that EV-miR-101
expression level in BM-MSCs was different depending on MDS
severity. High-risk group and patients with AML/MRC showed
significantly lower expression of BM-MSC-derived EV-miR-101 than
the low-risk group or normal controls. In contrast, the expression
of miR-101 derived from BM-MSCs was upregulated in patients from
the high-risk group, indicative of the dissociation between miR-101
expression level and EV from BM-MSCs in patients with MDS. Although
no relationship was observed between BM blast percentage and
BM-MSC-derived cellular-miR-101 or serum EV-miR-101 expression
level, EV-miR-101 level from BM-MSCs exhibited reverse correlation
with BM blast percentage. These findings suggest the possibility
that some specific miRs from BM-MSCs may be interrupted from
translocation into EVs (or intra-cellular accumulation) and this
phenomenon may be linked with the increase in the number of blasts
in patients with MDS. The dissociation between cellular miR and
EV-miR (or exosomal miR) has been observed in neoplastic cells
(9), including leukemia cells
(18). However, the current findings
deny the possible utility of serum miR measurement as liquid biopsy
for various diseases. The importance of BM-MSC-derived EV-miR may
indicate the nature of EV-miR, which may participate in the
communication between adjacent cells, especially between BM-MSCs
and hematologic malignant cells.
Hematologic neoplasia-derived exosomes affect the
microenvironment in the BM. For instance, chronic lymphocytic
leukemia (CLL) cell-derived exosomes induce the transition of BM
stroma cells into CAFs (20). These
present enriched miR-150 and miR-146a in CLL-derived exosomes and
an elevated miR-150 and miR-146a in BM-MSCs after co-culture with
primary CLL cells. CLL exosomes induced the proliferation and
migration of BM-MSCs with angiogenesis (20). We also reported that EV-miR-135b from
multiple myeloma cells transfers and induces angiogenesis (18). In solid tumors, the phenomenon of
transfiguration of the surrounding fibroblasts into CAF has been
reported, and this change is known to affect cancer, leading to
tumor progression and drug resistance (21). In hematologic neoplasia, the similar
situation (CAF induction of BM-MSCs) could be plausible; however,
such phenomenon is only limited in lymphoid neoplasia, i.e., CLL
and multiple myeloma (20,22).
Ozdogan et al demonstrated the low expression
level of DICER1 gene and miR dysregulation in the BM-MSCs
obtained from patients with MDS and AML along with the
downregulated expression of miRs (miR-30d-5p, miR-222-3p and
miR-30a-3p) and overexpression of miR-4426 in MDS-derived BM-MSCs
(23). Downregulation of
DICER1 expression in the BM-MSCs from patients with MDS
promoted cellular senescence and decreased the supportive
capability of hematopoietic stem cells along with the
downregulation of the expression of miR-17 family (miR-17-5p,
miR-20a/b, miR-106a/b, and miR-93) (24). Studies on BM-MSC-derived EV
(including exosomes) have focused on limited fields of hematologic
neoplasia. Wang et al demonstrated that mouse BM-MSC
(5T33)-derived exosomes increased the viability and proliferation
and decreased the apoptosis of mouse myeloma cells (5T33 MM model)
in response to bortezomib resistance (3). Moreover, these authors showed that
human myeloma BM-MSC-derived exosomes increased the viability of
human myeloma cells (RPMI-8226) in a dose-dependent manner
(3). Umezu et al also
reported that the BM-MSC-derived EV-miR-340 from normal young donor
(Lonza), rather than old donor, inhibited myeloma-induced
angiogenesis via hepatocyte growth factor/c-MET signaling (25). In this study, no particular
correlation was observed in the expression level between
BM-MSC-derived EV-miR-101 and serum EV-miR-101, suggesting that the
BM-MSC-derived EV-miR-101 may affect adjacent cells, including MDS
cells.
miR-101 is known to exert anti-neoplastic effects
and facilitate apoptosis (26). For
instance, miR-101 exhibited suppressive effects on bladder cancer
via c-FOS (27), colorectal cancer
via EZH2 (28), and breast cancer
via EYA1 (29). miR-101 is also
known to exert suppressive effects on vascular endothelial growth
factor-C (30,31). Recent reports found that miR-101
regulates the development and progression of myeloid- and
T-lymphoid lineage leukemia (32,33) and
suppresses the proliferation of neoplastic cells by targeting genes
related to epigenetic regulation (Tables II and SI). Thus, we speculate the possibility
that EV-miR-101 from BM-MSCs transfer to MDS cells and control
their proliferation via epigenetic regulation. In patients with
high-risk MDS, the decrease in the expression level of EV-miR-101
may result in the release of the anti-proliferative effect on MDS
cells. Another plausible possibility is that the more malignant
clone during MDS progression may induce BM-MSCs to suppress
EV-miR-101 expression. miR-101 antisense transfection assay showed
enhanced proliferation of human myeloid leukemia (HL-60) cells with
an increase in c-FOS expression (unpublished observation), thus
suggesting that the downregulation of EV-miR-101 expression in
patients with high-risk MDS may affect the proliferation of MDS
cells.
Development of MDS is known to be associated with
the accumulation of genetic abnormalities by aging, so-called
‘Clonal Hematopoiesis of Indeterminate Potential (CHIP)’, and
additional mutations in critical genes, including epigenetic
regulator genes, accelerate AML (34). In addition to genetic abnormalities
in MDS cells, BM-MSCs are also shown to have synonymous or
non-synonymous mutations (15,35,36) such
as ‘genetic injury.’ The unresolved issue is whether demethylating
agents available for patients with MDS could exert any effect on
BM-MSCs, especially BM-MSC-derived EV, as miR-101 targets many
epigenetic regulator genes. Poon et al demonstrated that a
hypomethylating agent (azacytidine) could also target BM-MSCs in
dysplastic MDS and contribute to the restoration of active
hematopoiesis, whereas patients with MDS that failed to respond to
hypomethylating treatment had disease progression (37). We could not reproduce the peculiarity
of BM-MSC-derived miR concerning the anatomical architecture of
MDS.
In conclusion, the current study demonstrates that
the BM-MSCs obtained from patients with MDS were heterogenous with
respect to the expression of EV-miR. Patients with high-risk MDS
and AML/MRC had lower expression of BM-MSC-derived EV-miR-101 than
those with low-risk MDS. This tendency was reversible as compared
with the miR-101 level from BM-MSCs; the serum EV-miR-101
expression showed no association. Although we failed to clarify the
exact mechanism underlying the downregulation of EV-miR-101
expression from BM-MSCs in patients with high-risk MDS, targeting
miR-101 may regulate neoplastic proliferation. Thus, the
downregulation of EV-miR-101 expression may affect adjacent
cell-to-cell communication and accelerate the malignant process in
MDS cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Platform
Project for Practical Research for Innovative Cancer Control from
the Japan Agency for Medical Research and Development (grant no.
15Ack0106073h0002), the Private University Strategic Research-Based
Support Project (grant no. S1311016) from the Ministry of
Education, Culture, Sports, Science and Technology and JSPS KAKENHI
(grant no. 19K16811).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the Gene Expression Omnibus
repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133276.
Authors' contributions
YS, MA, SY, SK, TS, HF, DA and KO treated the
patients; TU and CKK performed TLDA, PCR and data analysis; SI
established BM-MSCs; TU contributed essential reagents or tools; YS
and KO wrote the manuscript; KO and JHO designed the research and
supervised the project. All authors contributed to data analysis
and drafting of the manuscript.
Ethics approval and consent to
participate
Patient data was used in the present study according
to the ethical principles for medical research involving human
subjects of The Declaration of Helsinki (38). The study protocols were approved by
The Regional Ethics Review Board of Tokyo Medical University
(Tokyo, Japan), and all patients provided written informed consent
prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BM
|
bone marrow
|
|
BM-MSC
|
bone marrow mesenchymal stromal
cell
|
|
miR
|
microRNA
|
|
MDS
|
myelodysplastic syndrome
|
|
AML
|
acute myeloid leukemia
|
|
AML/MRC
|
acute myeloid leukemia with
myelodysplasia-related changes
|
|
TLDA
|
TaqMan-low density array
|
|
CAF
|
cancer-associated fibroblast
|
|
IPSS-R
|
revised International Prognostic
Scoring System
|
References
|
1
|
Morrison SJ and Scadden DT: The bone
marrow niche for haematopoietic stem cells. Nature. 505:327–334.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anthony BA and Link DC: Regulation of
hematopoietic stem cells by bone marrow stromal cells. Trends
Immunol. 35:32–37. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Hendrix A, Hernot S, Lemaire M, De
Bruyne E, Van Valckenborgh E, Lahoutte T, De Wever O, Vandenrkerken
K and Menu E: Bone marrow stroma cell-derived exosomes as
communications in drug resistance in multiple myeloma cells. Blood.
124:555–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bernardo ME and Fibbe WE: Mesenchymal
stromal cells: Sensors and switches of inflammation. Cell Stem
Cell. 13:392–402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nauta AJ and Fibbe WE: Immunomodulatory
properties of mesenchymal stromal cells. Blood. 110:3499–3506.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Whiteside TL: Exosome and mesenchymal stem
cell cross-talk in the tumor microenvironment. Semin Immunol.
35:69–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin LY, Du LM, Cao K, Huang Y, Yu PF,
Zhang LY, Li FY, Wang Y and Shi YF: Tumor cell-derived exosomes
endow mesenchymal stromal cells with tumour-promotion capabilities.
Oncogene. 35:6038–6042. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Makarova JA, Shkumikov MU, Wicklein D,
Lange T, Samatov TR, Turchinovich AA and Tonevitsky AG:
Intracellular and extracellular microRNA: An update on localization
and biological role. Prog Histochem Cytochem. 51:33–49. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ohyashiki JH, Umezu T and Ohyashiki K:
Extracellular vesicle-mediated cell-cell communication in
haematological neoplasms. Philos Trans R Soc Lond B Biol Sci.
373(pii): 201604842018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Azmi AS, Bao B and Sarkar FH: Exosomes in
cancer development, metastasis and drug resistance: A comprehensive
review. Cancer Metastasis Rev. 32:623–642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang X, Li Y, Zou L and Zhu Z: Role of
exosomes in crosstalk between cancer-associated fibroblasts and
cancer cells. Front Oncol. 9:3562019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Greenberg PL, Tuechler H, Schanz J, Sanz
G, Garcia-Manero G, Solé F, Bennett JM, Bowen D, Fenaux P, Dreyfus
F, et al: Revised international prognostic scoring system for
myelodysplastic syndromes. Blood. 120:2454–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Greenberg PL, Stone RM, Al-Kli A, Barta
SK, Bejar R, Bennett JM, Carraway H, De Castro CM, Deeg HJ, DeZern
AE, et al: Myelodysplastic syndrome, version 2. 2017, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw. 15:60–87.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Azuma K, Umezu T, Imanishi S, Asano M,
Yoshizawa S, Katagiri S, Ohyashiki K and Ohyashiki JH: Genetic
variations of bone marrow mesenchymal stromal cells derive from
acute leukemia and myelodysplastic syndrome by targeted deep
sequencing. Leuk Res. 62:23–28. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Umezu T, Ohyashiki K, Kuroda M and
Ohyashiki JH: Leukemia cell to endothelial cell communication via
exosomal miRNAs. Oncogene. 32:2747–2755. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshizawa S, Umezu T, Saitoh Y, Gotoh M,
Akahane D, Kobayashi C, Ohyashiki JH and Ohyashiki K: Exosomal
miRNA signatures for late-onset acute graft-versus host disease in
allogeneic hematopoietic stem cell transplantation. Int J Mol Sci.
19(pii): E24932018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Umezu T, Tadokoro H, Azuma K, Yoshizawa S,
Ohyashiki K and Ohyashiki JH: Exosomal miR-135b shed from hypoxic
multiple myeloma cells enhances angiogenesis by targeting
factor-inhibiting HIF-1. Blood. 124:3748–3757. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Paggetti J, Haderk F, Seiffert M, Janji B,
Distler U, Ammerlaan W, Kim YJ, Adam J, Lichter P, Solary E, et al:
Exosomes released by chronic lymphocytic leukemia cells induce the
transition of stromal cells into cancer-associated fibroblasts.
Blood. 126:1106–1117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Orimo A and Weinberg RA: Stromal
fibroblasts in cancer: A novel tumor-promoting cell type. Cell
Cycle. 5:1597–1601. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De Veirman K, Rao L, De Bruyne E, Menu E,
Van Valckenborgh E, Van Riet I, Frassanito MA, Di Marzo L, Vacca A
and Vanderkerken K: Cancer associated fibroblasts and tumor growth:
Focus on multiple myeloma. Cancers (Basel). 6:1363–1381. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ozdogan H, Gur Dedeoglu B, Oztemur
Islakoglu Y, Aydos A, Kose S, Ataly A, Yegin ZA, Avcu F, Uckan
Cetinkaya D and Ihan O: DICER1 gene and miRNA dysregulation in
mesenchymal stem cells of patients with myelodysplastic syndrome
and acute myeloblastic leukemia. Leuk Res. 63:62–71. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao Y, Wu D, Fei C, Guo J, Gu S, Xu F,
Zhang Z, Wu L, Li X and Chang C: Down-regulation of Dicer1 promotes
cellular senescence and decreases the differentiation and stem
cell-supporting capacities of mesenchymal stromal cells in patient
with myelodysplastic syndrome. Haematologica. 100:194–204. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Umezu T, Imanishi S, Azuma K, Kobayashi C,
Yoshizawa S, Ohyashiki K and Ohyashiki JH: Replenishing exosomes
from older bone marrow stromal cells with miR-340 inhibits
myeloma-related angiogenesis. Blood Adv. 1:812–823. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Long Y, Wu Z, Yang X, Chen L, Han Z, Zhang
Y, Liu L, Liu W and Liu X: MicroRNA-101 inhibits the proliferation
and invasion of bladder cancer cells via targeting c-FOS. Mol Med
Rep. 14:2651–2656. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peng Z and Zhang Y: Methyl jasmonate
induces the apoptosis of human colorectal cancer cells via down
regulation of EZH2 expression by microRNA-101. Mol Med Rep.
15:957–962. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guan H, Dai Z, Ma Y, Wang Z, Liu X and
Whang X: MicroRNA-101 inhibits cell proliferation and induces
apoptosis by targeting EYA1 in breast cancer. Int J Mol Med.
37:1643–1651. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deng G, Teng Y, Huang F, Nie W, Zhu I,
Huang W and Xu H: MicroRNA-101 inhibits the migration and invasion
of intrahepatic cholangiocarcinoma cells via direct suppression of
vascular endothelial growth factor-C. Mol Med Rep. 12:7079–7085.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li G, Yang F, Gu S, Li Z and Xue M:
MicroRNA-101 induces apoptosis in cisplatin-resistant gastric
cancer cells by targeting VEGF-C. Mol Med Rep. 13:572–578. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gonzales-Aloy E, Connerty P, Salik B, Liu
B, Woo AW, Haber M, Norris MD, Wand J and Wand JY: MiR-101
suppresses the development of MLL-rearranged acute myeloid
leukemia. Haematologica. 104:e296–e300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qian L, Zhang W, Lei B, He A, Ye L, Li X
and Don X: MicroRNA-101 regulates T-cell acute lymphoblastic
leukemia progression and chemotherapeutic sensitivity by targeting
Notch1. Oncol Rep. 36:2511–2526. 2015. View Article : Google Scholar
|
|
34
|
Ghobrial IM, Detappe A, Anderson KC and
Steensma SP: The bone-marrow niche in MDS and MGUS: Implications
for AML and MM. Nat Rev Clin Oncol. 15:219–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Flores-Figueroa E, Arana-Trejo RM,
Gutierrez-Espindola G, Perez-Cabrera A and Mayani H: Mesenchymal
stem cells in myelodysplastic syndromes: Phenotypic and cytogenetic
characterization. Leuk Res. 29:215–224. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Blau O, Hofmann WK, Baldus CD, Thiel G,
Serbent GV, Schumann E, Thiel E and Blau IW: Chromosomal
aberrations in bone marrow mesenchymal stroma cells from patients
with myelodysplastic syndrome and acute myeloblastic leukemia. Exp
Hematol. 35:221–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Poon Z, Dighe N, Venkatesan SS, Cheung
AMS, Fan X, Bari S, Hota M, Ghosh S and Hwang WYK: Bone marrow MSCs
in MDS: Contribution towards dysfunctional hematopoiesis and
potential targets for disease response to hypomethylating therapy.
Leukemia. 33:1487–1500. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
WMA declaration of Helsinki: Ethical
principles for medical research involving human subjects. Retrieved
from. https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/World
Medical Association; 2013
|