Introduction
Despite progress in both the treatment and
prevention of numerous cancer types, gastric cancer is still a
prevalent malignancy in both males and females (1). In 2018, there were 1,033,701 new
diagnoses of gastric cancer alone (5.7% of all cancer diagnoses),
which resulted in 782,685 mortalities (8.2% of all
cancer-associated mortalities) (2).
Lymph node metastasis and distant metastasis are common in patients
with gastric cancer and effective therapeutic approaches targeting
metastatic gastric cancer are ineffective (3,4),
resulting in a poor prognosis (3,4).
Helicobacter pylori infections and certain dietary
structures (including alcohol intake and vitamin C deficiency) are
major risk factors for gastric cancer (5,6).
However, the pathogenesis of this is poorly characterized.
Genetic alterations significantly contribute to the
development and pathogenesis of gastric cancer (7). Non-coding RNAs (ncRNAs), such as long
ncRNAs (lncRNAs) (>200 nt) and microRNAs (miR/miRNAs) regulate
gene expression to participate in the development of diverse types
of cancer, including gastric cancer (8–10). A
recent study showed that lncRNAs can interact with miRNAs to
regulate cancer cell behaviors (11). lncRNA-zinc finger protein (ZNF)281 is
a recently identified cancer-associated lncRNA in glioma (12). Our preliminary bioinformatics
analysis indicated that lncRNA-ZNF281 can bind the loop region of
miR-124 precursor, while miR-124 suppresses gastric cancer
(13). The present study was
performed to explore the interaction between lncRNA-ZNF281 and
miR-124 in gastric cancer and to examine the differential
expression of ZNF281 in gastric cancer using reverse
transcription-quantitative (RT-q)PCR. Moreover, the interaction
between ZNF281 and miR-124 was analyzed via overexpression
experiments. Transwell assays were conducted to analyze the roles
of ZNF281 and miR-124 in regulating cell invasion and migration. It
was revealed that lncRNA-ZNF281 may promote cancer cell migration
and invasion in gastric cancer cells via the downregulation of
miR-124.
Materials and methods
Gastric patients
A total of 72 patients with gastric cancer (44 males
and 28 females; median age, 50.1±6.3 years; range, 33–67 years)
were selected from 188 cases of gastric cancer admitted to the
Second People's Hospital of Liaocheng (Linqing, China) between
March 2015 and April 2019. Of the 72 patients, there were 38 cases
of adenocarcinoma and 34 cases of carcinoma. The current study was
approved by the Ethics Committee of the aforementioned hospital.
The inclusion criteria were as follows: i) Patients with newly
diagnosed gastric cancer; and ii) a diagnosis made using a
histopathological test (the gold standard). The exclusion criteria
were as follows: i) Patients with recurrent gastric cancer; ii)
prior initiation of therapy; and iii) other diagnosed clinical
disorders. According to the clinical data, the patients were
classified according to the American Joint Committee on Cancer
staging system (14). The results
revealed that there were a total of 12, 18, 22 and 20 patients at
clinical stages I, II, III and IV, respectively. The principle of
the experimental design was explained to all 72 patients, and all
provided written informed consent.
Gastric tissue specimens and cell
lines
Prior to therapy initiation, a gastric biopsy was
performed under the guidance of MRI to collect both tumor and
non-tumor gastric tissues from all included patients. Non-tumor
samples were collected ≤3 cm away from the cancerous tissues, and
all samples were validated via histopathological examination.
Additionally, two cell lines, AGS (gastric adenocarcinoma) and
SNU-1 (gastric carcinoma) from the American Type Culture Collection
(ATCC), were used in the present study. These cells were cultured
in a mixture of 10% FBS (ATCC) and 90% F-12K Medium (ATCC), at 95%
humidity, 37°C and 5% CO2.
Transient cell transfections
The PcDNA3.1 vector was used to construct an
lncRNA-ZNF281 expression vector (Sangon Biotech Co., Ltd.).
Negative control (NC; non-targeting cotntrol) miRNA
(5′-GUAGUCGAUGCUACGAUCGUAU-3′) and miR-124 mimic
(5′-CGUGUUCACAGCGGACCUUGAU-3′) were also purchased from Sangon
Biotech Co., Ltd. AGS and SNU-1 cells were harvested at 80%
confluence, followed by transfection with 10 nM vector (empty
vector as NC group) and 50 nM miRNA (NC miRNA as NC group) into
1×106 cells. All transfections were mediated by
Lipofectamine® 2000 reagent (Sangon Biotech Co., Ltd.).
The control (C) in all transfections was untransfected cells. Cells
were harvested at 24 h post-transfection to perform the following
experiments.
RNA interaction prediction
IntaRNA 2.0 (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp)
was used to predict the interaction between lncRNA-ZNF281 and
miR-124 precursor. For the analysis, the lncRNA-ZNF281 sequence was
used as the long sequence, and the miR-124 precursor sequence was
used as the short sequence.
RT-qPCR
All tissue samples were stored in liquid nitrogen.
AGS and SNU-1 cells were harvested at 24 h post-transfection and
cells were counted. Total RNAs in 0.03-g tissue samples and
1×105 cells were extracted using RNAzol reagent
(Sigma-Aldrich; Merck KGaA). To harvest miRNAs, 85% ethanol was
used to precipitate and wash RNA samples. Total RNA was digested
using DNase I for 90 min at 37°C, before being reverse transcribed
into cDNA using TruScript Reverse Transcriptase kit (Norgen Biotek
Corp.). To measure the expression levels of lncRNA-ZNF281, all qPCR
assays were performed using Luna® Universal One-Step
RT-qPCR kit (SYBR; New England BioLabs, Inc.) with 18S rRNA as an
endogenous control. Primer sequences were as follows: lncRNA-ZNF281
forward, 5′-GAGGACACATAGTGGAGAAAAG-3′ and reverse,
5′-TGAGACAACACAGCCAGATTAC-3′; 18S rRNA forward,
5′-CTACCACATCCAAGGAAGC-3′ and reverse, 5′-TTTTCGTCACTACCTCCCC-3′.
To measure the expression levels of mature miR-124, the addition of
poly(A), miRNA reverse transcription and qPCR assays were also
performed using the All-in-One™ miRNA RT-qPCR Reagent kit
(GeneCopoeia, Inc.). The miR-124 forward sequence was:
5′-CGUGUUCACAGCGGACCUU-3′. Reverse primer and U6 primers were
included in the All-in-One™ miRNA RT-qPCR Reagent kit (cat. no.
QP015; GeneCopoeia, Inc.). with U6 used as an endogenous control.
All Cq values were processed using 2−ΔΔCq method
(15) and each PCR was repeated in
triplicate. All PCR reactions conditions were: 95°C for 1 min,
followed by 40 cycles of 95°C for 10 sec and 57°C for 40 sec.
Transwell assays
AGS and SNU-1 cells were harvested at 24 h
post-transfection. Transwell assays were performed to analyze the
effects of transfection with various molecules on the invasion and
migration of these cell lines. Briefly, single-cell suspensions
were prepared by mixing 3×104 cells with 1 ml serum-free
F-12K Medium. Membranes were coated with Matrigel at 37°C for 6 h
before being used in the invasion assay, and uncoated membranes
were used for the migration assay. The upper Transwell chamber was
loaded with 0.1 ml single-cell suspension, while the lower chamber
was filled with a mixture of 80% F-12K Medium and 20% FBS. Cells
were incubated for 12 h under the aforementioned conditions. After
that, membranes were stained using 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) at room temperature for 20 min, and
cells were counted under a light microscope (magnification,
40×).
Statistical analysis
All experiments were performed in 3 independent
biological replicates. The mean was then calculated and was used in
all subsequent statistical analyses. GraphPad Prism 6 (Graphpad
Software, Inc.) software was used for all data analysis.
Associations were analyzed using Linear regression. Differences
between 2 groups were analyzed using a Student's t-test and
differences between ≥3 groups were compared using ANOVA (one-way)
followed by Tukey's post hoc test. The 72 gastric cancer patients
were divided into high- and low-lncRNA-ZNF281 or -miR-124
expression level groups (n=36), with the median expression level of
the respective molecules in tumor tissues used as cutoff values
(4.72 and 2.12 for lncRNA-ZNF281 and miR-124, respectively). The
χ2 test was performed to analyze the association between
the expression levels of lncRNA-ZNF281 and miR-124 with patient
clinical data. P<0.05 was considered to indicate a statistically
significant difference.
Results
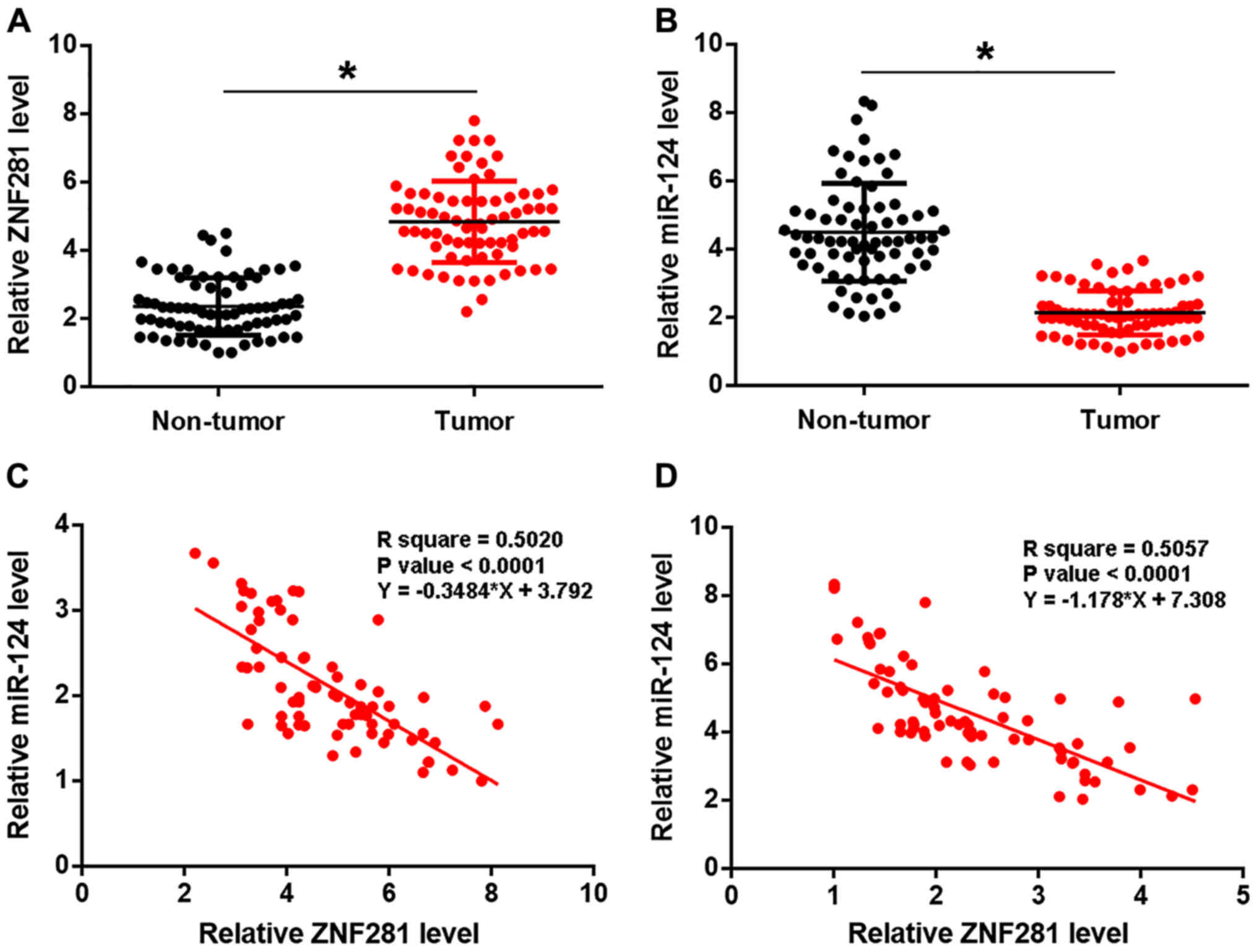
lncRNA-ZNF281 expression is inversely
correlated with miRNA expression in gastric tumor tissues
Expression levels of lncRNA-ZNF281 and miR-124 in
two types of tissues were measured using qPCR and compared using
the paired Student's t-test. lncRNA-ZNF281 expression was
upregulated (Fig. 1A), while miR-124
was downregulated (Fig. 1B) in tumor
tissues, compared with adjacent non-tumor tissues (P<0.05).
Subsequently, the association between lncRNA-ZNF281 and miR-124
expression was analyzed using linear regression and revealed a
significant and inverse association in both tumor (Fig. 1C) and adjacent non-cancerous
(Fig. 1D) tissues. The χ2
test revealed that lncRNA-ZNF281 and miR-124 expression levels were
not significantly associated with patient age, sex or gastric
cancer subtype (adenocarcinoma or carcinoma), but were
significantly correlated with clinical stage (P<0.05; Tables I and II).
 | Table I.Association between the expression
level of lncRNA-ZNF281 and patient clinicopathological data. |
Table I.
Association between the expression
level of lncRNA-ZNF281 and patient clinicopathological data.
| Characteristic | Cases, n | High expression,
n | Low expression,
n | χ2
value | P-value |
|---|
| Age, years |
|
|
| 0.48 | 0.50 |
|
>50 | 37 | 20 | 17 |
|
|
|
<50 | 35 | 16 | 19 |
|
|
| Sex |
|
|
| 0.94 | 0.33 |
| Male | 44 | 24 | 20 |
|
|
|
Female | 28 | 12 | 16 |
|
|
| AJCC stage |
|
|
| 14.67 | <0.01 |
| I | 12 | 3 | 9 |
|
|
| II | 18 | 4 | 14 |
|
|
| III | 22 | 15 | 7 |
|
|
| IV | 20 | 14 | 6 |
|
|
| Subtypes |
|
|
| 0.89 | 0.35 |
|
Adenocarcinoma | 38 | 17 | 21 |
|
|
|
Carcinoma | 34 | 19 | 15 |
|
|
 | Table II.Association between the expression
level of miR-124 with patient clinicopathological data. |
Table II.
Association between the expression
level of miR-124 with patient clinicopathological data.
| Characteristic | Cases, n | High expression,
n | Low expression,
n | χ2
value | P-value |
|---|
| Age, years |
|
|
| 0.06 | 0.81 |
|
>50 | 37 | 18 | 19 |
|
|
|
<50 | 35 | 18 | 17 |
|
|
| Sex |
|
|
| 0.23 | 0.63 |
| Male | 44 | 21 | 23 |
|
|
|
Female | 28 | 15 | 13 |
|
|
| AJCC stages |
|
|
| 11.73 | <0.01 |
| I | 12 | 8 | 4 |
|
|
| II | 18 | 14 | 4 |
|
|
|
III | 22 | 8 | 14 |
|
|
| IV | 20 | 6 | 14 |
|
|
| Subtypes |
|
|
| 0.22 | 0.64 |
|
Adenocarcinoma | 38 | 20 | 18 |
|
|
|
Carcinoma | 34 | 16 | 18 |
|
|
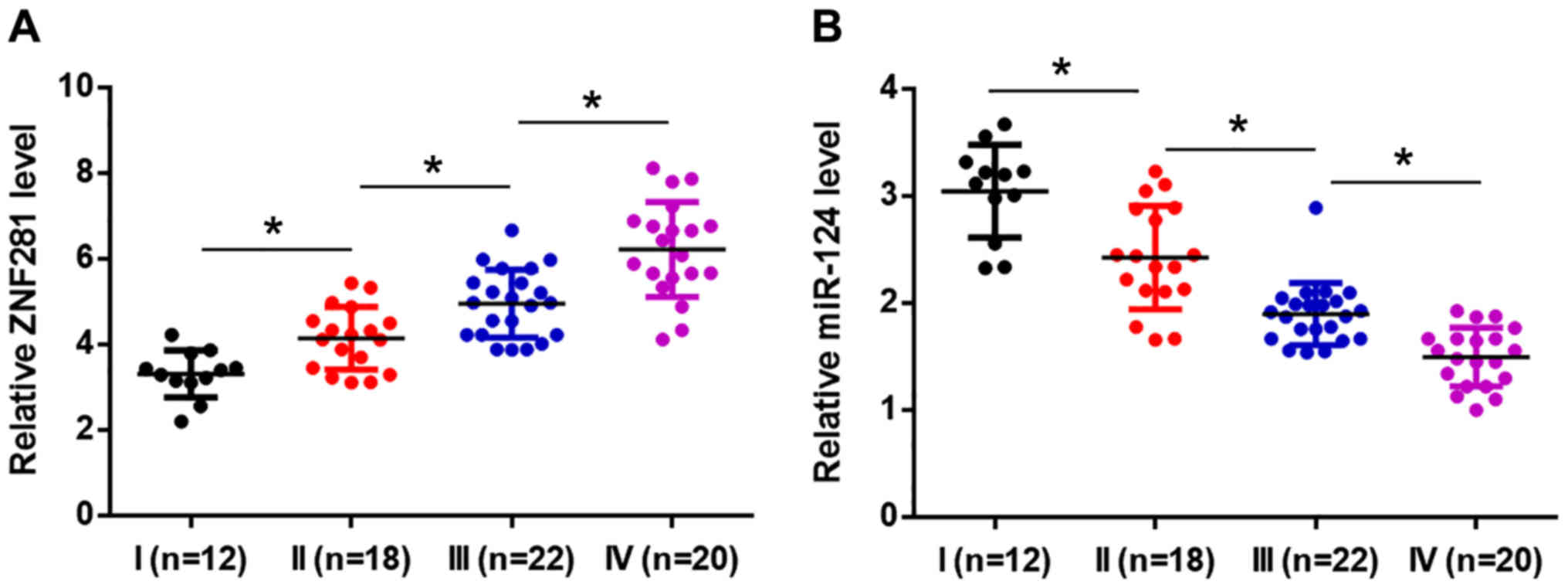
lncRNA-ZNF281 and miR-124 expression
levels are significantly associated with clinical stage
Based on the American Joint Committee on Cancer
staging system, the 72 patients presented with a total of 12, 18,
22 and 20 cases at clinical stages I, II, III and IV, respectively.
Expression levels of lncRNA-ZNF281 and miR-124 were compared
between the 4 clinical stages by performing a one-way ANOVA,
followed by Tukey's post hoc test. It was revealed that the
expression levels of the lncRNA were higher in patients at a more
advanced clinical stage (P<0.05; Fig.
2A). Conversely, expression levels of miR-124 decreased
significantly in patients at a more advanced clinical stage
(P<0.05; Fig. 2B).
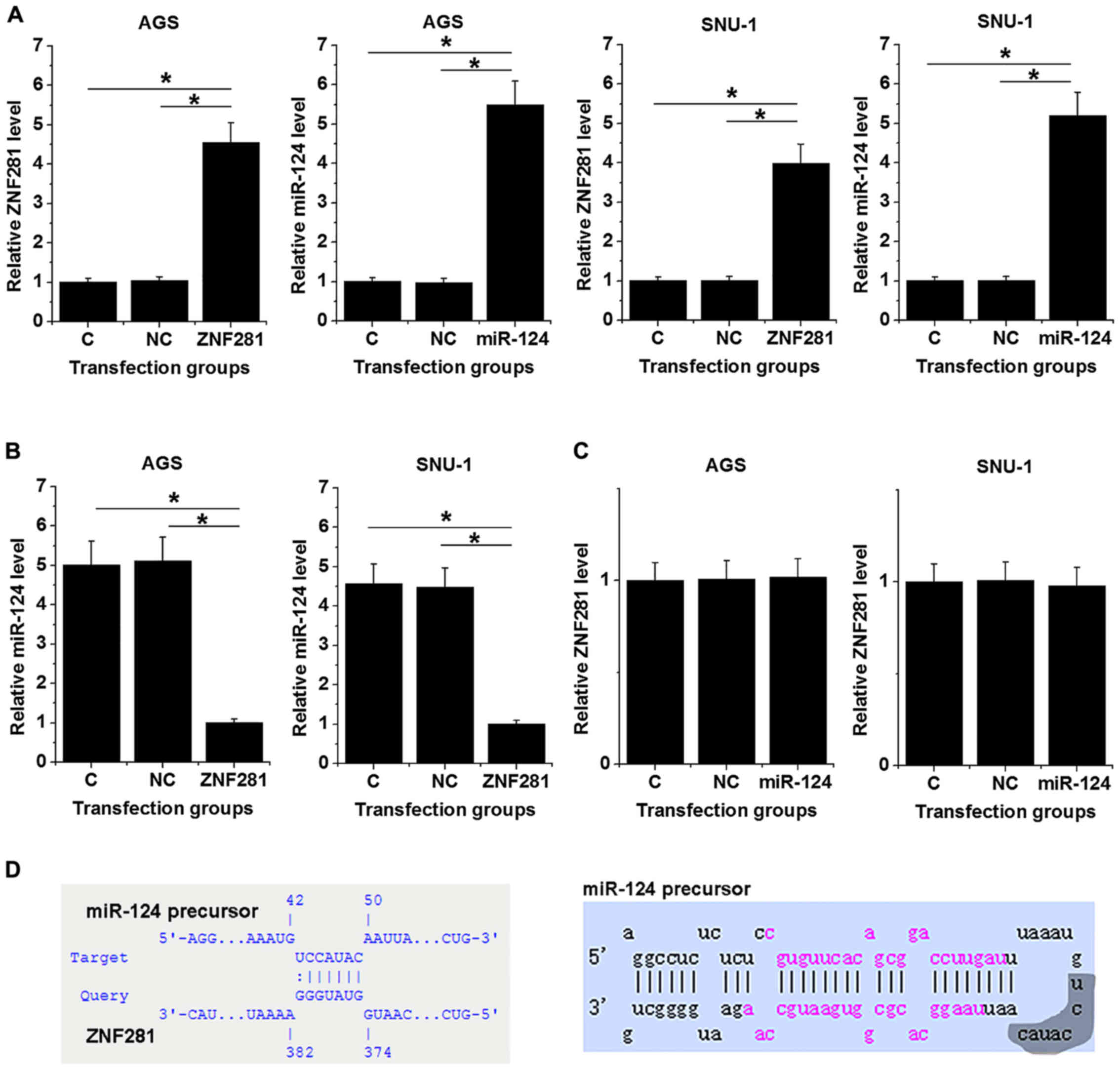
lncRNA-ZNF281 may act as a molecular
sponge of the miR-124 precursor molecule, downregulating miR-124
expression
To further investigate the interactions between
lncRNA-ZNF281 and miR-124, AGS and SNU-1 cells were transfected
with an lncRNA-ZNF281 expression vector or miR-124 mimic.
Expression levels of lncRNA-ZNF281 and miR-124 were measured at 24
h post-transfection. Compared with the NC (empty vector or NC miRNA
transfection) and C groups, expression levels of lncRNA-ZNF281 and
miR-124 were significantly increased in transfected cells
(P<0.05; Fig. 3A). Moreover,
lncRNA-ZNF281 overexpression mediated the downregulation of
miR-124, compared with both control groups (P<0.05; Fig. 3B), while miR-124 overexpression did
not significantly influence lncRNA-ZNF281 expression (Fig. 3C). Bioinformatics analysis (performed
using IntaRNA) revealed that lncRNA-ZNF281 may bind to a
complementary base sequence in the hairpin loop of miR-124
precursor (Fig. 3D).
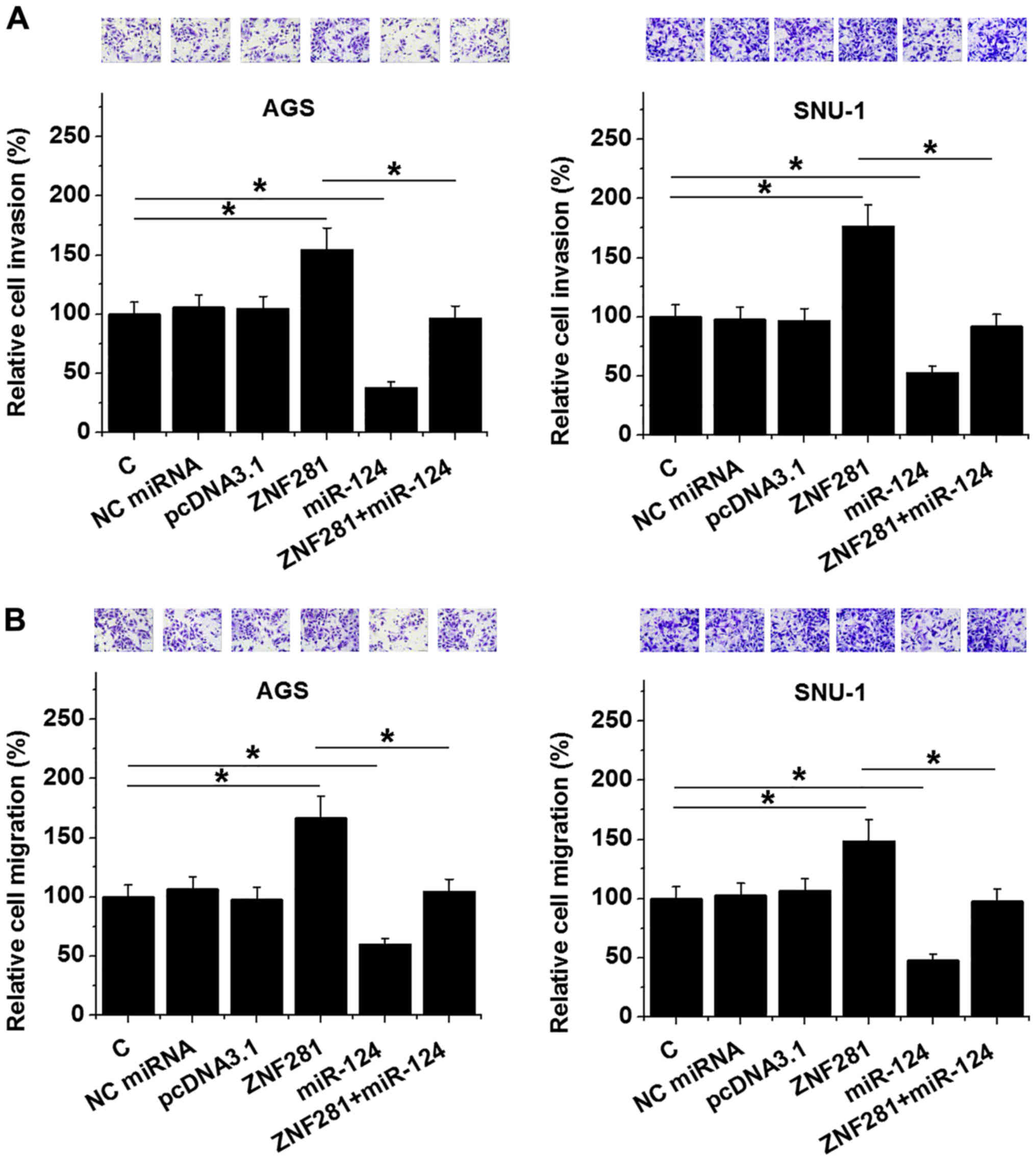
lncRNA-ZNF281 suppresses the invasion
and migration of gastric cancer cells, via miR-124
downregulation
Transwell invasion and migration assays were
performed to analyze the effects of lncRNA-ZNF281 and miR-124
overexpression on the invasion (Fig.
4A) and migration (Fig. 4B) of
both AGS and SNU-1 cells. Compared with the NC and C groups,
lncRNA-ZNF281 overexpression led to promotion of both migration and
invasion of gastric cancer cells, while miR-124 overexpression led
to their inhibition (P<0.05). In addition, miR-124
overexpression partially recovered the action of lncRNA-ZNF281
overexpression (P<0.05).
Discussion
The present study investigated the functional role
of lncRNA-ZNF281 in the progression of gastric cancer, and
determined that lncRNA-ZNF281 was downregulated in gastric cancer
tissues, promoting the invasion and migration of cells via
downregulation of miR-124 expression.
The majority of lncRNAs serve a similar role in
numerous cancer types; for instance, lncRNA HOX transcript
antisense RNA is upregulated in all cancer types, and regulates
chromatin dynamics to promote cancer progression (16). However, it has also been observed
that lncRNAs sometimes exhibit opposite functions in certain cancer
types. For example, lncRNA taurine upregulated gene 1 (TUG1) is
downregulated in glioma and acts as a tumor suppressor by promoting
glioma cell apoptosis (17). TUG1 is
also upregulated in osteosarcoma and promotes cancer cell
proliferation (18). Furthermore,
lncRNA-ZNF281 was downregulated in glioma and inhibited cancer cell
invasion (12). By contrast, the
present study revealed that lncRNA-ZNF281 was upregulated in
gastric cancer tissues and promoted the invasion and migration of
cancer cells. Therefore, lncRNA-ZNF281 may serve different, or even
opposite roles depending on cancer type.
It has been revealed that lncRNAs are able to act as
molecular sponges of miRNAs, attenuating their effects on
downstream genes (19); however, the
sponging of mature miRNAs by lncRNAs does not affect the expression
levels of miRNAs. Notably, in the present study, it was discovered
that lncRNA-ZNF281 downregulated miR-124 expression. It has
previously been reported that miR-124 can repress Snail2 to inhibit
cancer cell invasion in gastric cancer (13). Therefore, miR-124 may represent a
link between Snail2 and lncRNA-ZNF281. There was no indication of
significant interaction between mature miR-124 and lncRNA-ZNF281;
instead, lncRNA-ZNF281 was revealed to form a strong base pairing
with the loop region of the secondary structure of miR-124
precursor. It is known that the hairpin structure of miRNA
precursors is critical for miRNA maturation (20). Therefore, lncRNA-ZNF281 may prevent
the formation of the hairpin structure of miR-124 precursor and
suppress its maturation, thereby downregulating the expression
level of mature miR-124. However, more experiments are needed to
further validate this conclusion.
Notably, as a result of a lack of available
resources, the current study failed to produce an miR-124 mutant,
which may help to verify the interaction between lncRNA-ZNF281 and
miR-124. This should be incorporated into future studies. In
addition, preliminary cell proliferation assay results demonstrated
no effects of lncRNA-ZNF281 overexpression on the proliferation of
gastric cancer cells (data not shown). Therefore, lncRNA-ZNF281 may
only regulate certain behaviors of gastric cancer cells, and may
not influence factors associated with proliferation.
In conclusion, the present study revealed that
lncRNA-ZNF281 is upregulated in gastric cancer and able to
downregulate miR-124 expression, resulting in the suppression of
cancer cell invasion and migration.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SL, HG and PL designed and performed experiments.
YH, JW, XL and JZ collected and analyzed data. SL drafted the
manuscript, which was approved by all authors.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of the Second People's Hospital of Liaocheng (Linqing,
China) and written informed consent was obtained from each
patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interest.
References
|
1
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Riihimäki M, Hemminki A, Sundquist K,
Sundquist J and Hemminki K: Metastatic spread in patients with
gastric cancer. Oncotarget. 7:52307–52316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gotoda T, Yanagisawa A, Sasako M, Ono H,
Nakanishi Y, Shimoda T and Kato Y: Incidence of lymph node
metastasis from early gastric cancer: Estimation with a large
number of cases at two large centers. Gastric Cancer. 3:219–225.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Forman D and Burley VJ: Gastric cancer:
Global pattern of the disease and an overview of environmental risk
factors. Best Pract Res Clin Gastroenterol. 20:633–649. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mocellin S, Verdi D, Pooley KA and Nitti
D: Genetic variation and gastric cancer risk: A field synopsis and
meta-analysis. Gut. 64:1209–1219. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang J, Song YX and Wang ZN: Non-coding
RNAs in gastric cancer. Gene. 560:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li XT, Li JC, Feng M, Zhou YX and Du ZW:
Novel lncRNA-ZNF281 regulates cell growth, stemness and invasion of
glioma stem-like U251s cells. Neoplasma. 66:118–127. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li SL, Gao HL, Lv XK, Hei YR, Li PZ, Zhang
JX and Lu N: MicroRNA-124 inhibits cell invasion and
epithelial-mesenchymal transition by directly repressing Snail2 in
gastric cancer. Eur Rev Med Pharmacol Sci. 21:3389–3396.
2017.PubMed/NCBI
|
|
14
|
Ikoma N, Blum M, Estrella JS, Das P,
Hofstetter WL, Fournier KF, Mansfield P, Ajani JA and Badgwell BD:
Evaluation of the American joint committee on cancer 8th edition
staging system for gastric cancer patients after preoperative
therapy. Gastric Cancer. 21:74–83. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bhan A and Mandal SS: LncRNA HOTAIR: A
master regulator of chromatin dynamics and cancer. Biochim Biophys
Acta. 1856:151–164. 2015.PubMed/NCBI
|
|
17
|
Li J, Zhang M, An G and Ma Q: LncRNA TUG1
acts as a tumor suppressor in human glioma by promoting cell
apoptosis. Exp Biol Med (Maywood). 241:644–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yun-Bo F, Xiao-Po L, Xiao-Li L, Guo-Long
C, Pei Z and Fa-Ming T: LncRNA TUG1 is upregulated and promotes
cell proliferation in osteosarcoma. Open Med (Wars). 11:163–167.
2016.PubMed/NCBI
|
|
19
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Krol J, Sobczak K, Wilczynska U, Drath M,
Jasinska A, Kaczynska D and Krzyzosiak WJ: Structural features of
microRNA (miRNA) precursors and their relevance to miRNA biogenesis
and small interfering RNA/short hairpin RNA design. J Biol Chem.
279:42230–42239. 2004. View Article : Google Scholar : PubMed/NCBI
|