Introduction
Hepatocellular carcinoma (HCC) is the sixth most
prevalent malignant cancer type, and results in ~800,000 fatalities
each year, making it the fourth leading cause of cancer-associated
mortality worldwide (1). In China,
HCC is commonly treated using surgical resection combined with
either chemo- or radiotherapy, and immunotherapy is used less
frequently (2). Despite the current
therapeutic strategies achieving promising results in certain
cases, it is reported that the 5-year overall and disease-free
survival rates are just 51 and 27%, respectively (3). High rates of tumor recurrence and
metastasis are significant barriers to further improving HCC
treatment, thus, early detection is crucial for successful
treatment. Numerous HCC biomarkers (such as α-fetoprotein, cancer
antigen 19-9 and glypican 3) have been identified and clinically
applied to improve the diagnosis of HCC. However, due to their low
sensitivity and specificity, the diagnostic accuracy of these
biomarkers is insufficient (4–6).
Therefore, it is necessary to further elucidate the underlying
mechanisms of HCC recurrence and metastasis, and to identify novel
biomarkers for the early diagnosis of HCC.
The C-type lectins comprise a large superfamily of
proteins, including selectins, mannose receptors (MRs) and liver
and lymph node sinusoidal endothelial cell C-type lectin (CLEC4G)
(7). The C-type lectins possess
several biological functions, such as mediation of the inflammatory
and immune responses (8), and
pathogen recognition using shared homologous
carbohydrate-recognition domains. Increasing evidence has suggested
that the dysregulation of C-type lectins is closely associated with
the progression of cancer. For example, serum E-selectin expression
was significantly higher in patients with colorectal cancer
compared with healthy subjects or patients with benign colorectal
diseases (9). Similarly, CLEC18B
expression was significantly altered in glioblastoma multiforme
tissue, and was identified as an independent predictor of patient
survival (10). Additionally, it has
been demonstrated that selectin influences the progression of
numerous cancer types, including colon carcinoma (11,12) and
gastric cancer (13). Coupland et
al (14) discovered that
P-selectin promoted the lung metastasis of breast cancer and
melanoma in vivo. Moreover, the interaction between
E-selectin and its ligands enhanced the adhesion of prostate cancer
cells to endothelial cells (15).
C-Type lectin domain family 4 member M, (CLEC4M;
also known as CD209L) belongs to the C-type lectin family and is
expressed in the endothelial cells of the lymph nodes and liver. It
exhibits ~80% amino acid homology with dendritic cell-specific
intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN)
(16). Unlike CLEC4M, DC-SIGN is
expressed on the surface of dendritic cells and a subset of
macrophages (17). It has been
demonstrated that cell surface expression of CLEC4M promotes HIV
(18), and hepatitis C (19) and hepatitis B (20) infection by increasing contact between
the virus and host cell. Several studies have demonstrated the
involvement of CLEC4M in colon and gastric cancer progression. For
example, CLEC4M is associated with poor prognosis in patients with
colon cancer, and it is involved in the adhesion, migration,
invasion and liver metastasis of colon cancer cells (21). Furthermore, CLEC4M exerts these
biological functions through the signal transducer and activator of
transcription 5/long non-coding RNA Heterogeneous Nuclear
Ribonucleoprotein K Pseudogene 2/C-X-C chemokine receptor type 4
axis (13). Although CLEC4M has been
examined in several malignant tumor types, to our knowledge, the
clinical significance of CLEC4M has never been investigated in HCC
so far.
Materials and methods
Patient information and specimen
collection
Fresh frozen tissues and pathological sections were
collected (in a double-blind manner) from 88 patients (82 males and
6 females) with HCC, who had completed a follow-up survey at the
Mengchao Hepatobiliary Hospital of Fujian Medical University
(Fujian, China) between December 2013 and July 2016. The ages of
enrolled patients range from 34 to 71 years (median age, 54 years).
The expression level of CLEC4M was detected using reverse
transcription-quantitative PCR (RT-qPCR) and immunohistochemical
staining. Additionally, 18 distant non-tumor hemangioma tissues
were collected, and the expression level of CLEC4M was determined
and used as a healthy hepatic tissue control. All of the enrolled
patients exhibited preoperative liver function of Child-Pugh score
A or B (without distant metastasis), and a follow-up was conducted
every 6 months via telephone or outpatient service, until July
2017. Western blotting was conducted on a total of 9 fresh frozen
HCC tissue samples (from the cohort of 88 patients as
aforementioned), in order to examine the differences in the
expression level of CLEC4M between patients in the
recurrence/metastasis group (R/M group) and the no R/M group (N R/M
group), according to their prognosis after surgery. The use of
human tissue samples in the present retrospective study was
approved by the ethics committee of Mengchao Hepatobiliary Hospital
of Fujian Medical University (Fujian, China) and written informed
consent was preoperatively obtained from each participant.
Immunohistochemical detection of
CLEC4M
Immunohistochemical analysis was performed to
determine the expression level of CLEC4M in 88 pathological
sections as previously described (22) with slight modification. Briefly, the
4-µm sections were deparaffinized in xylene and rehydrated in
graded ethanol. Afterwards, the slides were blocked with 1%
H2O2 for 30 min at room temperature.
Following rehydration, the slides were incubated with EDTA (0.1 M,
pH 9.0) antigen repairing buffer in a high-pressure cooker and
heated until steam was generated. The slides were further incubated
for 3 min, and then cooled off at room temperature. Thereafter, the
slides were probed with a rabbit anti-CLEC4M antibody (1:800; cat.
no. ab169783; Abcam) or mouse anti-CD31 antibody (1:200; cat. no.
3528S; CST Biological Reagents Co., Ltd.) overnight at 4°C. Then, a
commercial immunohistochemical staining kit (cat. no. KIT-5020;
Fuzhou Maixin Biotech Co., Ltd.), alexa 488-conjugated anti-rabbit
secondary antibody against CLEC4M (1:1,000; cat. no. A-11008;
Thermo Fisher Scientific, Inc.) and alexa 546-conjugated anti-mouse
secondary antibody against CD31 (1:1,000; cat. no. A-11003; Thermo
Fisher Scientific, Inc.) were used to incubate the slides
respectively, as the manufacturer's instructions. The
immunohistochemical score was based on the international German
immune response scoring system (German immunoreactive score, IRS)
(23) according to the intensity of
staining; two pathologists, who were blinded to the clinical
characteristics of the specimens, independently performed the
scoring. The final score for each section was based on the mean of
the scores obtained by the two pathologists, and were graded as
follows: 0, negative; 1+, weak positive; 2+, moderate positive; or
3+, strong positive.
RNA extraction and RT-qPCR
The expression level of CLEC4M mRNA in 88 fresh
frozen tumorous tissues, and their corresponding adjacent non-tumor
tissues, was calculated. Total RNA was extracted from tissues using
the TransZol up plus kit (Beijing Transgen Biotech Co., Ltd.)
according to the manufacturer's instructions. Following
quantification, 1 µg total RNA from each sample was
reverse-transcribed into cDNA using the Transcriptor First Strand
cDNA Synthesis Kit (Roche Diagnostics; cat. no. 04897030001)
according to the manufacturer's protocol. Subsequently, qPCR
detection was carried out under the following conditions: 40 cycles
of 95°C for 10 sec, 60°C for 60 sec and a final extension for 30
sec at 72°C. All PCR reactions were repeated ≥3 times.
β-actin was selected as the endogenous control and the
primer pairs are detailed in Table
I. The relative gene expression of CLEC4M was calculated using
the 2−ΔΔCq method (24).
 | Table I.Primer sequences used in RT-qPCR. |
Table I.
Primer sequences used in RT-qPCR.
| Gene name | Primer
sequence |
|---|
| CLEC4M-Forward |
5′-TGGGCCTCCTGGAAGAAGAT-3′ |
| CLEC4M-Reverse |
5′-GCGTCTTGCTCGGATTGTTC-3′ |
|
β-actin-Forward |
5′-GCCAACACAGTGCTGTCTGG-3′ |
|
β-actin-Reverse |
5′-GCTCAGGAGCAATGATCTTG-3′ |
Western blotting
Total protein was extracted from 9 HCC tissue
samples using RIPA lysis buffer (Beijing Solarbio Science &
Technology Co., Ltd.) supplemented with a protease inhibitor
cocktail (cat. no. 11697498001; Roche Diagnostics). The protein was
then quantified using a bicinchoninic acid protein assay kit
(Beijing Solarbio Science & Technology Co., Ltd.) according to
the manufacturer's instructions. Equal amounts (50 µg) of protein
were separated via SDS-PAGE on a 12% gel, and transferred to PVDF
membranes. Subsequently, the membranes containing the separated
proteins were blocked with 5% BSA for 2 h at room temperature.
Next, the membranes were probed with primary antibodies against
CLEC4M (1:1,000; cat. no. #ab169783; Abcam) overnight at 4°C. The
membranes were then incubated with horseradish
peroxidase-conjugated secondary antibodies (1:2,000 dilution; cat.
no. HS201-01; Beijing Transgen Biotech Co., Ltd.) for 1 h at room
temperature. Protein bands were visualized using an electrochemical
luminescence reagent (Thermo Fisher Scientific, Inc). β-actin
(anti-β-actin antibody; 1:1,000; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc.) was used as an endogenous control.
Densitometry of blot was analyzed by Image J software (version
1.45). Protein expression was subsequently quantified with
densitometry and normalized by β-actin.
Bioinformatics analysis
The expression of CLEC4M was also investigated in a
transcriptome dataset of our lab and The Cancer Genome Atlas (TCGA)
database (https://www.cancer.gov/tcga). The
transcriptome dataset included whole RNA-sequencing data of tumor
and non-tumor tissue samples collected from 65 HCC patients
receiving surgical resection in Mengchao Hepatobiliary Hospital of
Fujian Medical University. After mapping to reference genome
(GRCh37), annotation and quantification of gene expression were
conducted with STAR (version 2.5.3a) as previously described
(25). On the other hand, the mRNA
sequencing (mRNA-Seq) expression spectrum data (HiSeqV2), including
371 HCC patients was downloaded from the UCSC cancer browser
(https://genome-cancer.ucsc.edu) as
previously described (26). The
expression data (n=50) available for both primary HCC tissues and
paired non-tumor in TCGA dataset were extracted to analyze the
expression of CLEC4M.
Statistical analysis
Statistical analyses were conducted using SPSS
(V20.0, IBM Corp.) or Prism statistical software (GraphPad
Software, Inc.). The data were presented as the mean ± SD.
Comparisons between two groups were performed using the two-tailed
paired or unpaired Student's t-test. One-way ANOVA was used for
comparisons between groups, followed by Tukey's multiple
comparisons test as the post-hoc test. The χ2-test
and/or the Fisher's exact test were conducted to analyze the
association between CLEC4M expression level and various
clinicopathological features of patients with HCC. The Cox
proportional hazard model was used to analyze univariate and
multivariate survival time data. Kaplan-Meier analysis and the
Log-rank test were used to analyze the OS times of patients with
HCC. P<0.05 was considered to indicate a statistically
significant difference.
Results
CLEC4M expression is significantly
downregulated in HCC tissues
In a previous study, isobaric tags were used for the
relative and absolute quantitation of coupling, alongside 2-D
liquid chromatography-tandem mass spectrometry, to analyze the
proteomic differences between HCC and non-tumor tissues (data
unpublished). It was subsequently, determined that CLEC4M
expression was downregulated in HCC tumor tissues. To confirm these
proteomics results, RT-qPCR was conducted to detect the mRNA
expression level of CLEC4M in 88 pairs of tumor tissues and their
corresponding adjacent non-tumor tissues. As exhibited in Fig. 1A, the expression of CLEC4M was
significantly decreased in tumor tissues compared with paratumor
tissues (P<0.0001). Furthermore, the aforementioned results were
supported by analysis of the datasets retrieved from our
transcriptome database (P<0.0001; Fig. 1B) and TCGA dataset (P<0.0001;
Fig. 1C).
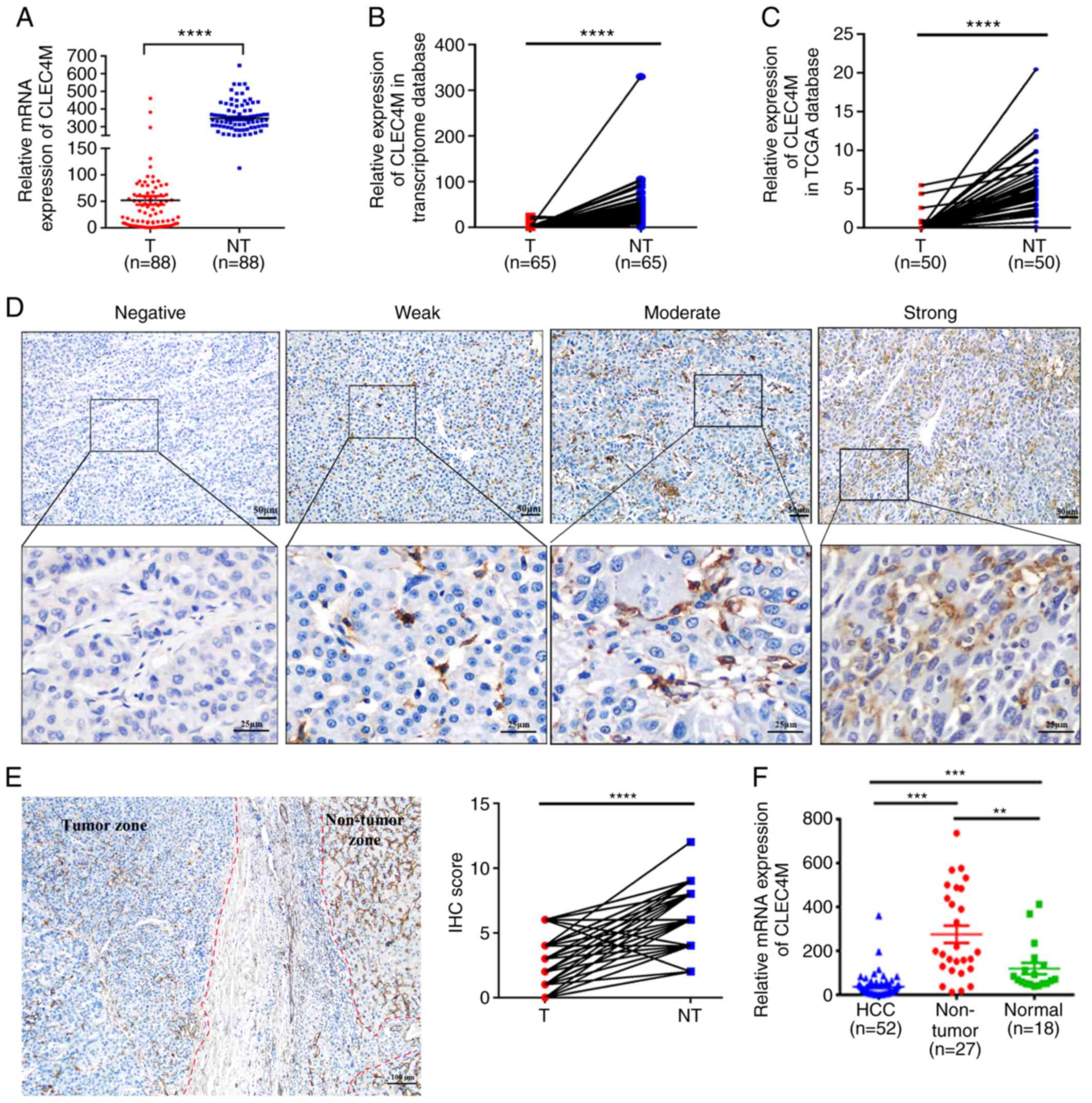 | Figure 1.Determination of CLEC4M expression
level in HCC tumor vs. adjacent non-tumor tissues. (A) RT-qPCR
results of CLEC4M mRNA expression in 88 pairs of HCC tumor samples
and their corresponding adjacent non-tumor tissues; paired
Student's t-test. (B) transcriptomic analysis of the relative
expression of CLEC4M in HCC tissues vs. adjacent non-tumor tissues.
N=65, paired Student's t-test. (C) Analysis of CLEC4M mRNA
expression in The Cancer Genome Atlas database; n=50; paired
t-test. (D) CLEC4M expression levels in HCC tumor tissues. CLEC4M
expression was semi-quantitatively divided into four groups:
negative staining, weak-positive staining, moderate-positive
staining and strong-positive staining (magnification, ×40 and
×200); (E) representative immunohistochemical section of HCC border
indicating the decreased CLEC4M expression in the tumor tissue
(magnification, ×200; (F) RT-qPCR results of CLEC4M mRNA expression
in HCC (n=52), non-tumor tissues (n=27) and normal hepatic tissues
(n=18); One-way ANOVA combined with Tukey's multiple comparisons
test (post-hoc) was applied to perform the comparisons between
groups. **P<0.01, ***P<0.001 and ****P<0.0001. T, tumor
tissue; NT, non-tumor tissue. HCC, hepatocellular carcinoma;
CLEC4M, C-type lectin domain family 4 member M; RFS, recurrence
free survival; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; IHC, immunohistochemical. |
To investigate the clinical significance of CLEC4M,
its expression level in paraffin-embedded HCC samples (n=88) was
compared with normal hepatic tissue samples (n=18) using RT-qPCR.
The representative images of HCC tissues with negative, weakly
positive, moderate-positive and strong-positive CLEC4M staining are
exhibited in Fig. 1D. The
pathological results indicated that the expression level of CLEC4M
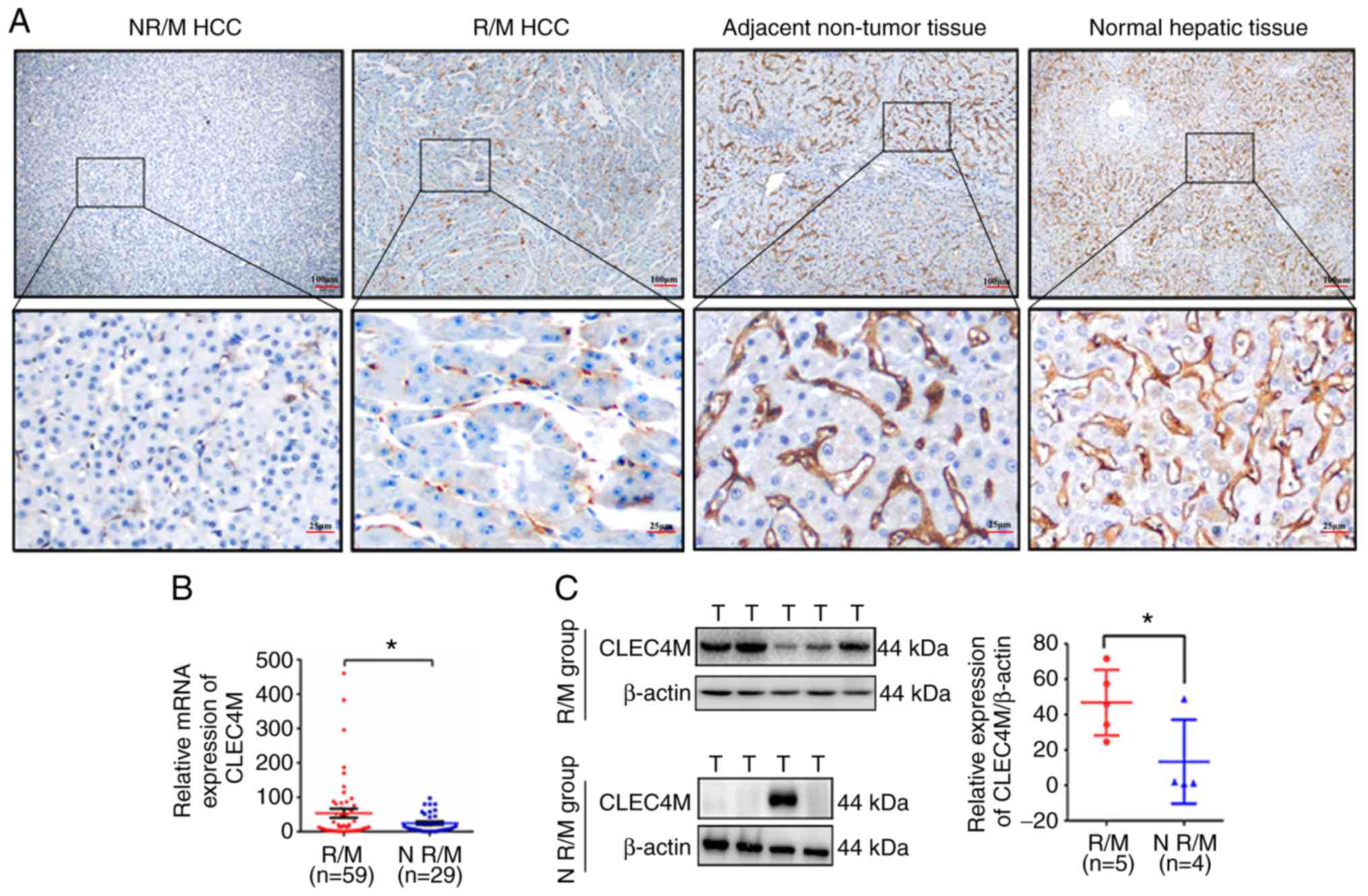
in both paratumor tissue and distant hepatic tissue is
significantly higher than that in HCC tissue (P<0.0001; Figs. 1E and 2A). Moreover, RT-qPCR conducted on another
group of 52 HCC tissue samples, 27 paratumor tissue samples and 18
normal hepatic samples supported the hypothesis that the expression
level of CLEC4M in HCC tissues is significantly lower compared with
paratumor and normal hepatic tissues (Fig. 1F).
The association between CLEC4M
expression level and various clinicopathological characteristics of
HCC
The correlation between CLEC4M protein expression
and various clinicopathological characteristics was investigated.
The data of multiple demographic parameters (including sex, age,
pathological diagnosis and certain tumor characteristics), as well
as follow-up data, were collected and recorded. The median
follow-up period was 34 months (range, 5–47 months). Subsequent
association analysis determined that high expression levels of
CLEC4M in HCC tissues significantly correlated with larger tumor
size (P=0.018), none encapsulation (P=0.0006), the presence of
microvascular invasion (P=0.008), and increased primary
differentiation (P=0.019). However, there was no significant
correlation between high CLEC4M expression levels and advanced
Barcelona clinic liver cancer stage, and α-fetoprotein (AFP)
expression, when the samples were divided into two groups,
including: negative and weak (n=59); and moderate and strong
(n=29), as summarized in Table II.
The results of the present study indicate that abnormal expression
levels of CLEC4M may be associated with the recurrence and
metastasis of HCC. Hence, the RT-qPCR data were further divided
into two groups (the N R/M group and the R/M group), depending on
whether the HCC had recurred or metastasized. The results suggested
that CLEC4M staining in the R/M group was dramatically stronger
compared with that of the N R/M group, but weaker than that in
non-tumor tissue and normal tissue (Fig.
2A). Furthermore, high expression of CLEC4M in tumor tissue was
positively associated with the recurrence and metastasis of HCC
(3*2 χ2 test, P=0.004, Table III), which was further confirmed by
RT-qPCR analysis (P=0.035; Fig. 2B)
and western blotting (P=0.049; Fig.
2C).
 | Table II.Association between CLEC4M expression
and certain clinicopathological features. |
Table II.
Association between CLEC4M expression
and certain clinicopathological features.
|
|
| Association with
CLEC4M expression |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | Patients, n
(%) | Negative and weak
(%) | Moderate and strong
(%) | P-value |
|---|
| Sex |
|
|
| 0.984 |
|
Female | 6 (6.82) | 4 (66.7) | 2 (33.3) |
|
|
Male | 82 (93.18) | 55 (67.07) | 27 (32.93) |
|
| Age, years |
|
|
| 0.788 |
|
≤50 | 29 (32.95) | 20 (68.97) | 9 (31.03) |
|
|
>50 | 59 (67.05) | 39 (66.10) | 20 (33.90) |
|
| HBsAg |
|
|
| 0.668 |
|
Negative | 11 (14.29) | 8 (72.72) | 3 (27.28) |
|
|
Positive | 77 (85.71) | 51 (66.23) | 26 (33.77) |
|
| AFP, ng/ml |
|
|
| 0.408 |
|
≤20 | 40 (45.45) | 25 (62.50) | 15 (37.50) |
|
|
>20 | 48 (54.55) | 34 (70.83) | 14 (29.17) |
|
| Liver
cirrhosis |
|
|
| 0.466 |
| No | 32 (36.36) | 23 (78.88) | 9 (21.12) |
|
|
Yes | 56 (63.64) | 36 (64.29) | 20 (35.71) |
|
| Tumor number |
|
|
| 0.412 |
|
Single | 75 (85.23) | 49 (62.82) | 26 (37.18) |
|
|
Multiple | 13 (14.77) | 10 (76.92) | 3 (23.08) |
|
| Tumor size, cm |
|
|
| 0.018a |
| ≤5 | 52 (59.09) | 40 (81.13) | 12 (18.87) |
|
|
>5 | 36 (40.91) | 19 (52.78) | 17 (47.22) |
|
| Tumor
encapsulation |
|
|
|
<0.001c |
|
Complete | 58 (65.91) | 47 (81.03) | 11 (18.97) |
|
|
None | 30 (34.09) | 12 (40.00) | 18 (60.00) |
|
| Microvascular
invasion |
|
|
| 0.008b |
|
Absence | 42 (47.73) | 34 (80.95) | 8 (19.05) |
|
|
Present | 46 (52.27) | 25 (54.35) | 21 (45.65) |
|
| TNM stage |
|
|
| 0.019a |
|
I+II | 46 (52.27) | 36 (78.26) | 10 (21.74) |
|
|
III+IV | 42 (47.73) | 23 (54.76) | 19 (45.24) |
|
| BCLC stage |
|
|
| 0.548 |
|
0+A | 70 (79.54) | 48 (68.57) | 22 (31.43) |
|
|
B+C | 18 (20.46) | 11 (61.11) | 7 (38.89) |
|
| HBV-DNA |
|
|
| 0.703 |
|
Negative | 42 (47.73) | 29 (69.05) | 13 (30.95) |
|
|
Positive | 46 (52.27) | 30 (65.22) | 16 (34.78) |
|
 | Table III.Immunohistochemical analysis of
CLEC4M expression in 88 hepatocellular carcinoma tissues. |
Table III.
Immunohistochemical analysis of
CLEC4M expression in 88 hepatocellular carcinoma tissues.
|
|
| CLEC4M
expression |
|---|
|
|
|
|
|---|
| Group | Patients, n
(%) | Moderate and strong
(%) | Negative and weak
(%) | P-value |
|---|
| R/M | 54 (61.36) | 24 (44.44) | 30 (55.56) | 0.004a |
| N R/M | 34 (38.64) | 5 (14.71) | 29 (85.29) |
|
High CLEC4M expression levels in HCC
tissues indicate poor prognosis in patients with HCC
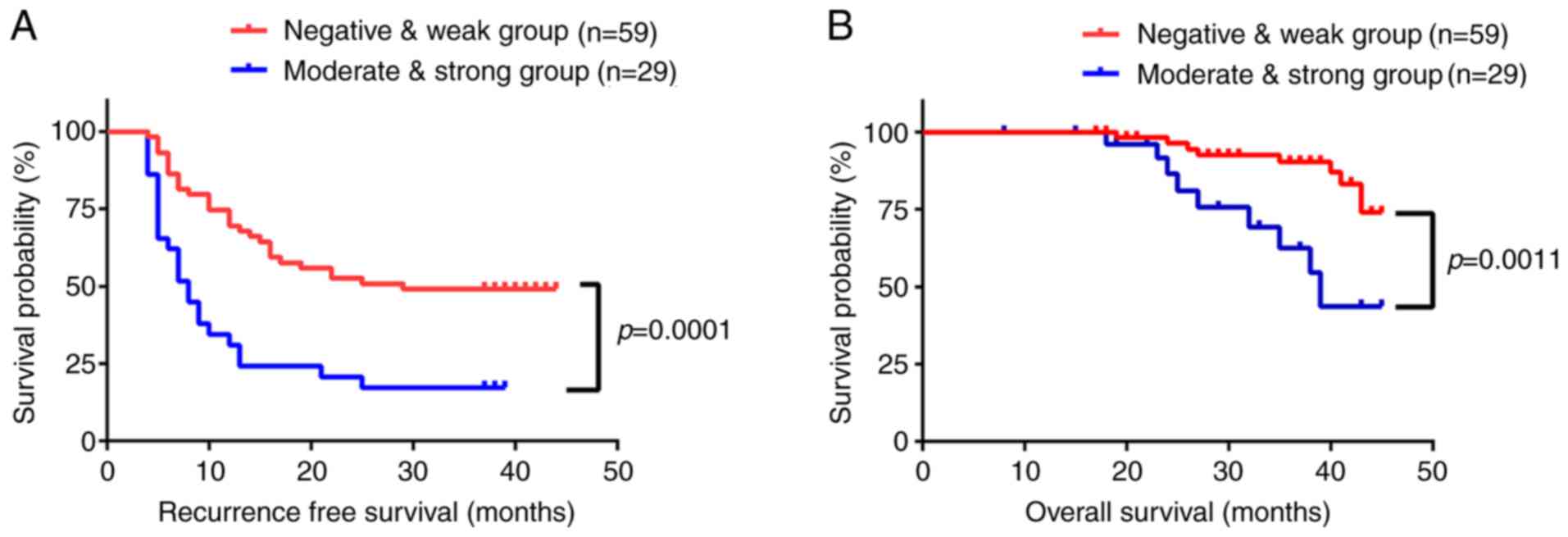
The influence of CLEC4M expression on recurrence
free survival (RFS) and overall survival (OS) time was then
investigated using Kaplan-Meier analysis. The RFS and OS times were
significantly lower in patients with a high expression level of
CLEC4M, compared with those with low expression levels (P=0.0001
and P=0.0011, respectively; Fig. 3A and
B). The mean RFS time was 14 months in the moderate- and
strong-positive group, and 26 months in negative- and weak-positive
group. Additionally, the mean OS time was 29 months in the
moderate- and strong-positive group, compared with 36 months in the
negative- and weak-positive group. Subsequently, Cox's multivariate
regression analysis was performed using recurrence/metastasis as
the dependent variable. Age, sex, tumor size, tumor number,
encapsulation, microvascular invasion, differentiation, AFP
expression and disease stage were listed as independent variables.
The results of the analysis revealed that high expression levels of
CLEC4M independently predicted RFS (P=0.032) and OS times (P=0.011;
Table IV) of patients with HCC. The
results of the present study suggest that CLEC4M is a promising
prognostic biomarker for HCC, but a study conducted on a larger
population size would improve the validity and accuracy of this
hypothesis.
 | Table IV.Univariate and multivariate analysis
of recurrence or survival associated factors in 88 hepatocellular
carcinoma patients. |
Table IV.
Univariate and multivariate analysis
of recurrence or survival associated factors in 88 hepatocellular
carcinoma patients.
|
| Recurrence | Survival |
|---|
|
|
|
|
|---|
|
| Univariate
analysis | Multivariate
analysis | Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|
|---|
|
Characteristics | HR, 95% CI | P-value | HR, 95% CI | P-value | HR, 95% CI | P-value | HR, 95% CI | P-value |
|---|
| Sex, female vs.
male | 1.030
(0.372–2.856) | 0.954 |
|
| 23.029
(0.009–57589.1) | 0.432 |
|
|
| Age, ≤50 vs. >50
years | 1.333
(0.734–2.422) | 0.345 |
|
| 1.200
(0.446–3.226) | 0.718 |
|
|
| Tumor size, ≤5 vs.
>5 cm | 2.229
(1.302–3.818) | 0.003b | 1.580
(0.857–2.912) | 0.143 | 2.458
(0.954–6.337) | 0.063 |
|
|
| Tumor number,
single vs. multiple | 1.692
(0.870–3.290) | 0.121 |
|
| 1.571
(0.442–5.584) | 0.485 |
|
|
| Tumor
encapsulation, present vs. absent | 0.392
(0.228–0.676) | 0.001b | 0.535
(0.302–0.950) | 0.033a | 0.291
(0.111–0.761) | 0.012a |
0.560(0.198–1.580) | 0.273 |
| Microvascular
invasion, yes vs. no | 2.177
(1.249–3.795) | 0.006b | 1.412
(0.772–2.582) | 0.263 | 2.954
(1.038–8.410) | 0.042a | 1.338
(0.413–4.338) | 0.627 |
| AFP, ≤20 vs. >20
ng/ml | 1.032
(0.605–1.763) | 0.907 |
|
| 0.604
(0.233–1.565) | 0.299 |
|
|
| HBsAg, negative vs.
positive | 1.866
(0.743–4.689) | 0.184 |
|
| 1.861
(0.416–8.328) | 0.416 |
|
|
| HBV-DNA, negative
vs. positive | 0.691
(0.402–1.186) | 0.180 |
|
| 0.933
(0.369–2.360) | 0.883 |
|
|
| TNM stage, I–II vs.
III–IV | 1.246
(0.731–2.126) | 0.419 |
|
| 0.755
(0.282–2.021) | 0.576 |
|
|
| Liver cirrhosis, no
vs. yes | 1.500
(0.844–2.667) | 0.167 |
|
| 1.397
(0.509–3.830) | 0.516 |
|
|
| BCLC stage, 0+A vs.
B+C | 0.356
(0.196–0.646) | 0.001b | 0.523
(0.261–1.047) | 0.067 | 0.224
(0.083–0.605) | 0.003b | 0.224
(0.079–0.636) | 0.005b |
| CLEC4M, low vs.
high | 2.738
(1.588–4.720) |
<0.001c | 1.889
(1.056–3.381) | 0.032a | 4.659
(1.778–12.207) | 0.002b | 3.928
(1.360–11.347) | 0.011a |
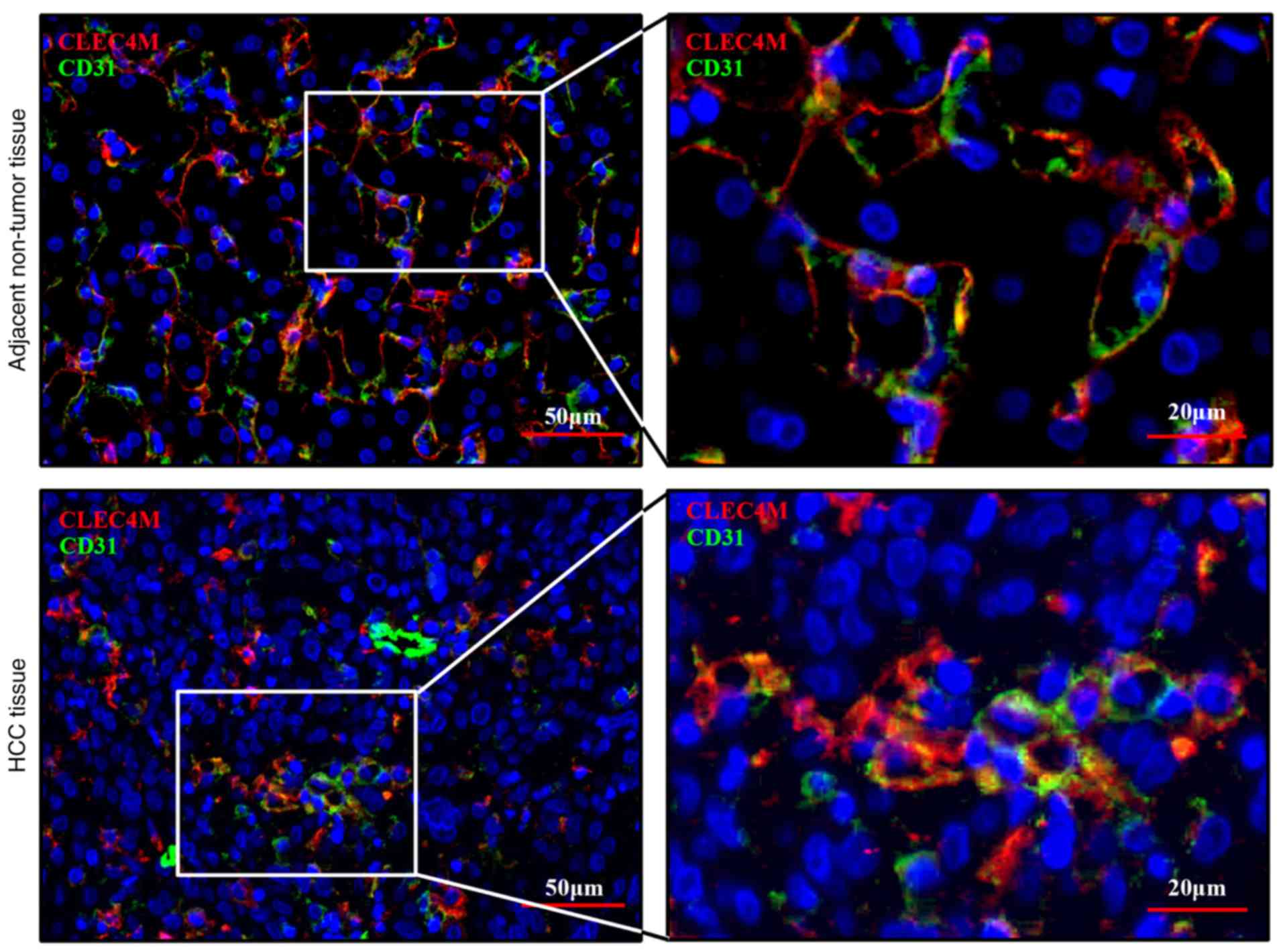
CLEC4M is specifically expressed in
sinusoidal endothelial cells
Immunohistochemical staining illustrated that
low-positive staining of CLEC4M was visualized in HCC tissues. The
expression level of CLEC4M in HCC cells was also investigated. CD31
is a biomarker that is commonly used to label endothelial cells
(27). Thus, co-localization
staining, with CLEC4M and CD31 antibodies, was performed using the
pathological tissue sections. The results of the current study
indicated that CLEC4M is expressed in the same cells in which
CD31-positive staining was observed (Fig. 4), which suggests that CLEC4M is
specifically expressed in sinusoidal endothelial cells (but not in
HCC cells), consistent with the results of a previous study
(28).
Discussion
Tumor recurrence and metastasis are major
contributors to the high mortality rate of patients with HCC
(29). The discovery of sensitive
and reliable biomarkers for the early diagnosis of HCC represents
an effective strategy to reduce the rate of recurrence and
metastasis, and improves prognosis. The clinical significance and
biological function of CLEC4M have been well researched in colon
(12) and gastric cancer (13). However, the influence that CLEC4M
exerts on HCC progression is not yet well characterized. In the
present study, CLEC4M expression was shown to be downregulated in
HCC tissues compared with adjacent non-tumor tissues. Furthermore,
the association between CLEC4M expression and certain
clinicopathological features was investigated, including tumor
size, extent of encapsulation, microvascular invasion,
differentiation and tumor recurrence or metastasis. Higher CLEC4M
expression in HCC tumor tissues correlated with shorter
postoperative RFS and OS times in patients with HCC. Additionally,
moderate- and strong-positive staining of CLEC4M was identified to
be an independent prognostic indicator of RFS and OS in patients
with HCC (following hepatectomy). The results of the present study
systematically highlight CLEC4M as an effective biomarker that may
predict the prognosis of patients with HCC.
CLEC4M is a C-type lectin that is primarily
expressed in sinusoidal endothelial cells of the healthy liver, as
demonstrated in the non-tumor section of Fig. 1E. Notably, CLEC4M expression has also
been observed in the HCC tissues of patients that have experienced
recurrence or metastasis. Additionally, high CLEC4M expression
significantly correlated with microvascular invasion and tumor
metastasis, which suggests that CLEC4M may promote angiogenesis.
Multiple studies have provided indirect evidence supporting the
present results. For example, Borentain et al (30) demonstrated that the inhibition of
E-selectin suppressed hepatocellular carcinoma growth via the
impairment of tumor angiogenesis. Moreover, DC-SIGN, which is
highly homologous to CLEC4M, interacted with the Lewis X residues
of carcinoembryonic antigen-related cell adhesion molecule 1,
resulting in angiogenesis (31,32).
Thus, an investigation into the influence of CLEC4M on the
angiogenesis of HCC tissues should be performed in future
experiments to further prove that CLEC4M play important roles in
metastasis and invasion of HCC tissues. In summary, the present
data indicate that CLEC4M is implicated in the progression of HCC,
in a similar manner to its association with colon and gastric
cancer.
RT-qPCR determined that the expression of CLEC4M was
significantly downregulated in tumor tissues, compared with
non-tumor tissues. This appeared to contradict the fact that
patients with HCC and high CLEC4M expression in tumor tissues
typically exhibited shorter OS and RFS times. This may be
attributable to the fact that CLEC4M was specifically expressed in
sinusoidal endothelial cells, even in HCC tissues (Fig. 4), consistent with previous studies
(33,34). Additionally, in tissues containing
many endothelial cells, the staining of CLEC4M appears stronger.
Liver sinusoids consist of a line of sinusoidal endothelial liver
cells and Kupffer cells, providing oxygen and nutrients to
hepatocytes and forming a distribution network throughout the liver
(27,35,36).
Additionally, CLEC4M can bind to intercellular adhesion molecule 3
(ICAM3; 28), resulting in the activation and recruitment of
ICAM3-positive T cells and initiating an immune response to
pathogens or cancer cells (37).
Thus, a microenvironment with a low expression level of CLEC4M and
incomplete microvasculature may favor early tumor development, in
association with proliferation of tumor cells and escaping from
immune surveillance in HCC cells. Subsequently, a gradual increase
in the genesis of hepatic sinusoids and the surrounding vasculature
may provide sufficient nutrition and oxygen proportional to the
growth of the tumor, whilst allowing it time to adapt to the immune
pressures of the host environment. Furthermore, it has been
demonstrated that CLEC4M enhances the mobility and invasiveness of
tumor cells in gastric and colon cancer (13,21).
Additionally, high CLEC4M expression in HCC tissues is associated
with a poorer prognosis, which is consistent with previous
literature on lung (38) and
cervical cancer (39). Therefore, it
is hypothesized that an increase in CLEC4M expression proportional
to microvascular development may be beneficial to the growth and
metastasis of HCC cells. This may also explain the correlation
between moderate or strong-positive staining of CLEC4M in cancer
tissues, and the high risk of recurrence and metastasis.
In conclusion, the current study demonstrated that
expression levels of CLEC4M in HCC tissues may be an effective
indicator of HCC progression, and may represent a potential target
for therapeutic development.
Acknowledgements
The authors would like to thank Dr Bin Wang and Dr
Mojiao Liu, The Department of Pathology, Mengchao Hepatobiliary
Hospital of Fujian Medical University, for their technical help.
The results shown in this study are in part based upon data
generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81602102 and grant
no. 81672376), the Natural Science Foundation of Fujian Province
(grant no. 2016J01417 and 2017J01266), the Young and Middle-aged
Talent Training Project of Fujian provincial health and Family
Planning Commission (grant no. 2018-ZQN-76; 2018-ZQN-37; and
2016-1-44), the Joint Funds for the Innovation of Science and
Technology of Fujian province (grant no. 2017Y9116 and 2017Y9041),
the Scientific Foundation of Fuzhou City (grant no. 2015-S-143-19)
and the Startup Fund for scientific research, Fujian Medical
University (grant no. 2018QH1194).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XLL, YCW and JFL were responsible for research
creation and design, and provided study material. LHC provided the
pathological sections and perform the immunochemical score together
with LPL. LPL, KK, BXZ, LLW, CLZ and FW collected, assembled the
clinical and follow-up data. NSL and XYZ analysed and interpreted
the data. LPL and YCW drafted and finalized the manuscript. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All the experiments were approved by the Ethics
committee of Mengchao Hepatobiliary Hospital of Fujian Medical
University (Fujian, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
CLEC4M
|
C-type lectin domain family 4 member
M
|
|
RFS
|
recurrence free survival
|
|
OS
|
overall survival
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu ZX, Huang JW, Liao MH and Zeng Y:
Treatment strategy for hepatocellular carcinoma in China:
Radiofrequency ablation versus liver resection. Jpn J Clin Oncol.
46:1075–1080. 2016.PubMed/NCBI
|
|
3
|
Zhao LY, Huo RR, Xiang X, Torzilli G,
Zheng MH, Yang T, Liang XM, Huang X, Tang PL, Xiang BD, et al:
Hepatic resection for elderly patients with hepatocellular
carcinoma: A systematic review of more than 17,000 patients. Expert
Rev Gastroenterol Hepatol. 12:1059–1068. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y, Wang M, Cui C, Zhang L, Liao F, Li
H and Wu X: Significance of combined tests of serum golgi
glycoprotein 73 and other biomarkers in diagnosis of small primary
hepatocellular carcinoma. Cancer Biomark. 15:677–683. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson PJ: The role of serum
alpha-fetoprotein estimation in the diagnosis and management of
hepatocellular carcinoma. Clin Liver Dis. 5:145–159. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amr KS, Elmawgoud Atia HA, Elazeem
Elbnhawy RA and Ezzat WM: Early diagnostic evaluation of miR-122
and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis.
4:215–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ding D, Yao Y, Zhang S, Su C and Zhang Y:
C-type lectins facilitate tumor metastasis. Oncol Lett. 13:13–21.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dambuza IM and Brown GD: C-type lectins in
immunity: Recent developments. Curr Opin Immunol. 32:21–27. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ferroni P, Roselli M, Spila A,
D'Alessandro R, Portarena I, Mariotti S, Palmirotta R, Buonomo O,
Petrella G and Guadagni F: Serum sE-selectin levels and
carcinoembryonic antigen mRNA-expressing cells in peripheral blood
as prognostic factors in colorectal cancer patients. Cancer.
116:2913–2921. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo RM, Zhao CB, Li P, Zhang L, Zang SH
and Yang B: Overexpression of CLEC18B associates with the
proliferation, migration, and prognosis of glioblastoma. ASN Neuro.
10:17590914187819492018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Gisbergen KP, Aarnoudse CA, Meijer GA,
Geijtenbeek TB and van Kooyk Y: Dendritic cells recognize
tumor-specific glycosylation of carcinoembryonic antigen on
colorectal cancer cells through dendritic cell-specific
intercellular adhesion molecule-3-grabbing nonintegrin. Cancer Res.
65:5935–5944. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zuo Y, Ren S, Wang M, Liu B, Yang J, Kuai
X, Lin C, Zhao D, Tang L and He F: Novel roles of liver sinusoidal
endothelial cell lectin in colon carcinoma cell adhesion, migration
and in-vivo metastasis to the liver. Gut. 62:1169–1178. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Zhang Q, Zhang M, Yuan M, Wang Z,
Zhang J, Zhou X, Zhang Y, Lin F, Na H, et al: DC-SIGNR by
influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4
in gastric cancer liver metastasis. Mol Cancer. 16:782017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Coupland LA, Chong BH and Parish CR:
Platelets and P-selectin control tumor cell metastasis in an
organ-specific manner and independently of NK cells. Cancer Res.
72:4662–4671. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yasmin-Karim S, King MR, Messing EM and
Lee YF: E-selectin ligand-1 controls circulating prostate cancer
cell rolling/adhesion and metastasis. Oncotarget. 5:12097–12110.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pohlmann S, Soilleux EJ, Baribaud F,
Leslie GJ, Morris LS, Trowsdale J, Lee B, Coleman N and Doms RW:
DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds
to human and simian immunodeficiency viruses and activates
infection in trans. Proc Natl Acad Sci USA. 98:2670–2675. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Soilleux EJ, Morris LS, Leslie G, Chehimi
J, Luo Q, Levroney E, Trowsdale J, Montaner LJ, Doms RW, Weissman
D, et al: Constitutive and induced expression of DC-SIGN on
dendritic cell and macrophage subpopulations in situ and in vitro.
J Leukoc Biol. 71:445–457. 2002.PubMed/NCBI
|
|
18
|
da Silva RC, Segat L, Zanin V, Arraes LC
and Crovella S: Polymorphisms in DC-SIGN and L-SIGN genes are
associated with HIV-1 vertical transmission in a Northeastern
Brazilian population. Hum Immunol. 73:1159–1165. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen PC, Chuang PK, Chen CH, Chan YT, Chen
JR, Lin SW, Ma C, Hsu TL and Wong CH: Role of N-linked glycans in
the interactions of recombinant HCV envelope glycoproteins with
cellular receptors. ACS Chem Biol. 9:1437–1443. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Op den Brouw ML, de Jong MA, Ludwig IS,
van der Molen RG, Janssen HL, Geijtenbeek TB and Woltman AM:
Branched oligosaccharide structures on HBV prevent interaction with
both DC-SIGN and L-SIGN. J Viral Hepat. 15:675–683. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Na H, Liu X, Li X, Zhang X, Wang Y, Wang
Z, Yuan M, Zhang Y, Ren S and Zuo Y: Novel roles of DC-SIGNR in
colon cancer cell adhesion, migration, invasion, and liver
metastasis. J Hematol Oncol. 10:282017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cai Z, Zeng Y, Xu B, Gao Y, Wang S, Zeng
J, Chen L, Huang A, Liu X and Liu J: Galectin-4 serves as a
prognostic biomarker for the early recurrence/metastasis of
hepatocellular carcinoma. Cancer Sci. 105:1510–1517. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Remmele W, Hildebrand U, Hienz HA, Klein
PJ, Vierbuchen M, Behnken LJ, Heicke B and Scheidt E: Comparative
histological, histochemical, immunohistochemical and biochemical
studies on oestrogen receptors, lectin receptors, and Barr bodies
in human breast cancer. Virchows Arch A Pathol Anat Histopathol.
409:127–147. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Speir ML, Zweig AS, Rosenbloom KR, Raney
BJ, Paten B, Nejad P, Lee BT, Learned K, Karolchik D, Hinrichs AS,
et al: The UCSC Genome Browser database: 2016 update. Nucleic Acids
Res. 44:D717–D725. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Crouch EE and Doetsch F: FACS isolation of
endothelial cells and pericytes from mouse brain microregions. Nat
Protoc. 13:738–751. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lai WK, Sun PJ, Zhang J, Jennings A, Lalor
PF, Hubscher S, McKeating JA and Adams DH: Expression of DC-SIGN
and DC-SIGNR on human sinusoidal endothelium: A role for capturing
hepatitis C virus particles. Am J Pathol. 169:200–208. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen Y, Gao SG, Chen JM, Wang GP, Wang ZF,
Zhou B, Jin CH, Yang YT and Feng XS: Risk factors for the long-term
efficacy, recurrence, and metastasis in small hepatocellular
carcinomas. Cell Biochem Biophys. 72:627–631. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Borentain P, Carmona S, Mathieu S, Jouve
E, El-Battari A and Gerolami R: Inhibition of E-selectin expression
on the surface of endothelial cells inhibits hepatocellular
carcinoma growth by preventing tumor angiogenesis. Cancer Chemother
Pharmacol. 77:847–856. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bogoevska V, Horst A, Klampe B, Lucka L,
Wagener C and Nollau P: CEACAM1, an adhesion molecule of human
granulocytes, is fucosylated by fucosyltransferase IX and interacts
with DC-SIGN of dendritic cells via Lewis × residues. Glycobiology.
16:197–209. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Koch AE, Halloran MM, Haskell CJ, Shah MR
and Polverini PJ: Angiogenesis mediated by soluble forms of
E-selectin and vascular cell adhesion molecule-1. Nature.
376:517–519. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bashirova AA, Geijtenbeek TB, van
Duijnhoven GC, van Vliet SJ, Eilering JB, Martin MP, Wu L, Martin
TD, Viebig N, Knolle PA, et al: A dendritic cell-specific
intercellular adhesion molecule 3-grabbing nonintegrin
(DC-SIGN)-related protein is highly expressed on human liver
sinusoidal endothelial cells and promotes HIV-1 infection. J Exp
Med. 193:671–678. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Falkowska E, Durso RJ, Gardner JP, Cormier
EG, Arrigale RA, Ogawa RN, Donovan GP, Maddon PJ, Olson WC and
Dragic T: L-SIGN (CD209L) isoforms differently mediate
trans-infection of hepatoma cells by hepatitis C virus
pseudoparticles. J Gen Virol. 87:2571–2576. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brunt EM, Gouw AS, Hubscher SG, Tiniakos
DG, Bedossa P, Burt AD, Callea F, Clouston AD, Dienes HP, Goodman
ZD, et al: Pathology of the liver sinusoids. Histopathology.
64:907–920. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Patten DA, Wilson GK, Bailey D, Shaw RK,
Jalkanen S, Salmi M, Rot A, Weston CJ, Adams DH and Shetty S: Human
liver sinusoidal endothelial cells promote intracellular crawling
of lymphocytes during recruitment: A new step in migration.
Hepatology. 65:294–309. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Koppel EA, van Gisbergen KP, Geijtenbeek
TB and van Kooyk Y: Distinct functions of DC-SIGN and its
homologues L-SIGN (DC-SIGNR) and mSIGNR1 in pathogen recognition
and immune regulation. Cell Microbiol. 7:157–165. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu X, Zhang H, Su L, Yang P, Xin Z, Zou
J, Ren S and Zuo Y: Low expression of dendritic cell-specific
intercellular adhesion molecule-grabbing nonintegrin-related
protein in lung cancer and significant correlations with brain
metastasis and natural killer cells. Mol Cell Biochem. 407:151–160.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Jiang Y, Yuan M, Chen C, Wang K,
Zhang Q, Zuo Y and Ren S: Overexpression of dendritic cell-specific
intercellular adhesion molecule-3-grabbing nonintegrin-related
protein in cervical cancer and correlation with squamous cell
carcinoma antigen. Oncol Lett. 14:2813–2821. 2017. View Article : Google Scholar : PubMed/NCBI
|