Introduction
Pancreatic cancer (PC) is one of the most
devastating types of cancer worldwide. As the third leading cause
of cancer-associated mortality in the USA, PC was estimated to
cause 44,330 deaths in this country in 2018 (1). The number of PC cases is projected to
increase, and to become the second leading cause of
cancer-associated mortality before 2030 (2).
Due to the anatomical location of PC, it is usually
asymptomatic in the early stages of disease. However, without the
use of invasive procedures, current screening methods are unable to
detect early-stage PC (3), and these
procedures are difficult to perform at the early stage of disease.
Thus, the development of non-invasive detection methods, such as
liquid biopsy, has been increasingly emphasized in recent years,
and the identification of new biomarkers that can distinguish early
stage PC is urgently needed (4).
MicroRNAs (miRs) play critical roles in a number of
biological processes, such as cell proliferation, cell
differentiation and apoptosis, through their regulation of gene
expression by translational repression or mRNA degradation,
resulting in gene silencing. Recent studies have indicated that
miRs may also function as potential biomarkers for cancer detection
(5–8). Exosomes are a type of extracellular
vesicle, which are enclosed by a lipid bilayer that can contain
various components, including proteins, lipids, DNA, mRNAs, miRs
and non-coding RNAs (9). A number of
studies have demonstrated that miRs contained within exosomes
(exmiRs) play an important role in the development, metastasis and
drug resistance of tumors (10–12). The
lipid bilayer structure of exosomes prevents the enclosed miRs from
degradation, allowing the detection of accurate numbers of stable
exmiRs. Thus, isolating exosomes and quantifying the encapsulated
exmiRs has become a promising approach for the noninvasive
detection of cancer biomarkers (13).
miRs can be quantitatively measured in serum by
reverse transcription-quantitative (RT-q)PCR. However, this
approach is expensive and time-consuming (14). The tethered cationic lipoplex
nanoparticle (TCLN) biochip is a novel technology for the direct
analysis of exmiRs within a single exosome. Molecular beacons in
liposome nanoparticles serve as molecular probes for target miRs.
When serum is applied to the biochip, the molecular beacons
recognize target miRs in exosomes, and the interaction is read by a
total internal reflection fluorescence microscope (15). The copy numbers of the exmiRs can
then be obtained by the fluorescence signals of the molecular
beacons (MBs) (15). Wu et al
(13) demonstrated that the levels
of exmiRs detected by the TCLN biochip were consistent with the
widely used reverse transcription-quantitative PCR (RT-qPCR)
method. In addition, the TCLN biochip indicated a higher
sensitivity compared with RT-qPCR (13).
The aim of the present study was to evaluate the
expression levels of circulating exmiR-21, exmiR-10b and
exmiR-212-3p between patients with PC and healthy individuals using
a TCLN biochip and to determine whether these exmiRs can
distinguish early-stage PC.
Materials and methods
Patients and clinical samples
A total of 36 patients with PC and 65 healthy
individuals were enrolled for the present study between November
2017 and December 2018. The 36 patients with PC were divided into
two groups, early- and advanced-stage; patients with stages I and
II disease were categorized as the early stage group (n=19),
whereas patients with stages III and IV disease were categorized in
the advanced-stage group (n=17). All participants were from the Sir
Run Run Shaw hospital of Zhejiang University (Zhejiang, China). The
detailed characteristics of the patients are provided in Table I. All patients were newly diagnosed
with PC, which was confirmed by two blinded pathologists from the
Sir Run Run Shaw hospital, following surgery or ultrasound
endoscopic guided fine needle aspiration biopsy. The blood samples
of patients treated with chemotherapy, radiotherapy or
immunotherapy were excluded. Tumors were staged according to the
8th American Joint Committee on Cancer (AJCC) TNM Staging of
Pancreatic Cancer (16).
 | Table I.Characteristics of patients with
pancreatic cancer. |
Table I.
Characteristics of patients with
pancreatic cancer.
|
Characteristics | Cases, n (%) |
|---|
| Sex |
|
|
Female | 15 (41.7) |
|
Male | 21 (58.3) |
| Age (years) |
|
|
<60 | 8
(22.2) |
|
60–69 | 19 (52.8) |
|
≥70 | 9
(25.0) |
| Cigarette
smoking |
|
| No | 28 (77.8) |
|
Yes | 8
(22.2) |
| Alcohol
drinking |
|
| No | 7
(19.4) |
|
Yes | 29 (80.6) |
| Diabetes
mellitus |
|
| No | 33 (91.7) |
|
Yes | 3 (8.3) |
| ABO blood type |
|
| A | 13 (36.1) |
| B | 8
(22.2) |
| O | 12 (33.3) |
| AB | 3 (8.3) |
| Stage |
|
| I | 6
(16.7) |
| II | 13 (36.1) |
|
III | 5
(13.9) |
| IV | 12 (33.3) |
| BMI |
|
|
<19.57 | 8
(22.2) |
|
19.57–23.71 | 19 (52.8) |
|
>23.71 | 9
(25.0) |
The blood samples included in the present study were
collected in vacuum blood tubes with EDTA anticoagulant before
surgery and pharmacotherapy, and handled within 1 h following
collection. The blood samples were subjected to centrifugation at
2,795 × g for 10 min at 4°C. The plasma was then stored at −80°C.
Written informed consent was obtained from the participants before
sampling. The studies were conducted in accordance with the
International Ethical Guidelines for Biomedical Research Involving
Human Subjects (CIOMS), and the research protocols were approved by
the Clinical Research Ethics Committee of Sir Run Run Shaw Hospital
of Zhejiang University (Zhejiang, China)
Animal experiments
A total of 34 mice were obtained from the animal
unit of Zhejiang University (Zhejiang, China). BALB/C nude mice
(4–6 weeks old) were used in all experiments. All mice were female,
and their weight ranged from 20–26 g. The housing temperature was
from 22–24°C, and mice were given access to food and water. The air
remained clean by using ventilation devices for ~8–15 times per
hour, and the indoor lighting was controlled for 12 h of light and
dark. All animal experiments were performed in the animal unit of
Zhejiang University according to procedures authorized and
specifically approved by the institutional Ethical Committee.
Subcutaneous xenograft mouse model were established in 12 mice by
injecting 1×106 PANC-1-Luciferase cells (Stem Cell Bank,
Chinese Academy of Science) in the shoulders, via a left subcostal
incision, made in mice that were anesthetized with 3% isoflurane.
The other 22 normal mice were used as the normal control and were
injected with the same volume of normal saline. Subsequently, the
blood of the 34 mice was collected, when the subcutaneous tumors in
the 12 ×enograft mice grew to a longitudinal diameter of 1 cm.
Blood was collected from the venous plexus of the eye socket, in
mice anesthetized with 3% isoflurane, and the plasma was then
separated by centrifugation at 2,795 × g for 10 min at 4°C. The
plasma was then stored at −80°C. All mice were sacrificed by
cervical dislocation under 3% isoflurane anesthesia, following the
collection of blood. The plasma samples of these mice were used for
further analysis.
Rationale for selecting exmiRs
As an oncomiR, a number of studies have demonstrated
that miR-21 plays an important role in cell proliferation,
migration, invasion and survival by regulating the cell cycle,
apoptosis and invasion-related genes (17–21).
Previous studies have proven that exosomal miR-21 is upregualted at
the early stage of lung cancer, breast cancer, liver cancer and
other malignant tumors (22–24).
Thus, exmiR-21 was considered as a potential
biomarker for the diagnosis of cancer in the present study.
However, to the best of our knowledge, few studies have focused on
the assocaition between exmiR-21 and PC, particularly at the early
stage of PC. miR-10b was associated with the invasive and
metastatic properties of various human cancer types. Kim et
al (25) identified that miR-10b
promotes breast cancer cell proliferation, migration and invasion
via the inhibition of transcription factor TBX5 expression, leading
to the repression of the tumor suppressor genes (dual specificity
tyrosine phosphorylation regulated kinase 1A and PTEN). Previous
studies regarded miR-10b as a potential diagnostic biomarker of
breast cancer, bladder cancer and lung cancer (26). Ayaz et al (27) reported that miR-212-3p was
specifically expressed in laryngeal squamous cell carcinoma.
Furthermore, a study revealed that miR-212-3p was upregulated in PC
tissue. However, to the best of our knowledge, few studies have
focused on exmiRs in the aforementioned cancer types. Given that,
exmiRs (particularly tumor-derived exmiRs) play an important role
in the early diagnosis of several types of cancer in humans.
Preparing cationic lipoplex
nanoparticles containing MBs
Firstly, miR-21-, miR-10b- and miR-212-3p-specific
probes with fluorescent signal markers were designed. BHQ1 and
6-carboxy-fluorescein were assembled at the 3′ and 5′ ends of the
probe, respectively. The center of the probe is a specially
designed single-stranded base sequence for identifying the target
RNA. The specific molecular probe is coated in the cationic
liposome nanoparticles and linked to the glass substrate to
construct a PC-associated miRNA liposome nanoparticle chip. The
probe sequences were as follows: MiR-21-MB,
5′-6FAM-TCAACATCAGTCTGATAAGCTATTATCAGACTGA-BHQ1-3′; miR-10b-MB,
5′-6FAM-ATACCACACAAATTCGGTTCTACAACCGAATTTGTG-BHQ1-3′;
miR-212-3p-MB,
5′-6FAM-CGGCCGTGACTGGAGACTGTTAAGTCTCCAGTCA-BHQ1-3′.
Exosome characterization and
quantification
A commercial exosome extraction kit (Total Exosome
Isolation kit; cat. no. 4484450; Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract mouse serum and human plasma
exosomes to detect their particle size and potential. The dynamic
light scattering (DLS) technology was used for detection, with the
use of a 640 nm laser and 30 frames per sec, which is also known as
Photon Correlation Spectroscopy (PCS) or quasi-elastic scattering,
is a method for measuring the variation of light intensity
fluctuations over time (28). The
Zetasizer Nano ZS90 instrument (Malvern Instruments, Inc.) was used
for particle size analysis. The DLS technology has the advantages
of accuracy, speediness and good repeatability, and has become a
more conventional characterization method in nanotechnology
(29). DLS is widely used, including
for particle science testing in the life science field; it is
mature, stable and credible. As a gold standard, it is recognized
by the US Bureau of Standards and incorporated into the US
measurement standard system. The calculation method of DLS should
be able to reflect the particle size distribution within the system
better.
Western blotting
Cells or exosomes were lysed with RIPA buffer
(Beyotime Institute of Biotechnology), supplemented with protease
and phosphatase inhibitor cocktail (Beyotime Institute of
Biotechnology) on ice for 15 min. The lysates were centrifuged at
12,000 × g for 5 min to pellet cell debris at 4°C. The protein
concentrations were measured by the BCA protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. Subsequently, the supernatant was added to SDS/PAGE
sample loading buffer (Beyotime Institute of Biotechnology) and
boiled for 10 min at 95°C. A total mass of 20 µg protein was run on
SDS-PAGE (10%) and transferred to polyvinylidene fluoride membrane
(Bio-Rad Laboratories, Inc.). The membranes were blocked for 60 min
in 5% (w/v) non-fat dry milk or bovine serum albumin for 1 h at
room temperature and incubated with primary antibodies overnight at
4°C. The next day, the membranes were washed thrice in
tris-buffered saline containing 0.1% (v/v) Tween-20 (TBST),
incubated with horseradish-peroxidase conjugated secondary
antibodies (Thermo Fisher Scientific, Inc.) for 1 h at room
temperature, and then washed thrice in TBST. Bands were visualized
using ChemiDoc Touch Imaging System (Bio-Rad Laboratories, Inc.)
and the band intensities were quantified using the Image Lab 5.2.1
software (Bio-Rad Laboratories, Inc.). The primary antibodies were:
Anti-CD9 (1:2,000; cat. no. ab92726; Abcam), anti-CD63 (1:1,000;
cat. no. ab134045; Abcam) and anti-heat shock protein 70 (HSP70;
1:1,000; cat. no. ab181606; Abcam). The secondary antibody was:
Goat anti-rabbit IgG (H+L) secondary antibody, HRP (1:10,000; cat.
no. 31460; Invitrogen; Thermo Fisher Scientific, Inc.)
Transmission electron microscopy
(TEM)
To evaluate the morphology of isolated exosomes, the
exosomes were observed under a transmission electron microscope.
Firstly, carbon-coated 400-mesh copper grids (Xinxing Bairui Co.
Ltd.) were absorbed in a 20-µl aliquot of exosomes (~2 µg) for 2
min and allowed to dry at room temperature. Subsequently, the
exosomes were fixed in 1% (v/v) glutaraldehyde (EM-grade) for 5 min
at room temperature by placing drops carefully on the dried
preparation. Next, the grids were washed twice with water for 5 min
and then contrast-stained with 2% uranyl acetate for 10 min at room
temperature. Images were obtained by transmission electron
microscopy (Tecnai T10; FEI).
Statistical analysis
Statistical analyses were performed and graphs were
constructed using GraphPad Prism (version 7.0; GraphPad Software,
Inc.) and SPSS (version 24.0; IBM Corp.) statistical software. The
data are presented as the mean ± standard deviation. Significant
differences between the mean values of two groups were determined
using the Student's t-test. Significant differences between the
mean values of three or four groups were determined using the
one-way ANOVA, followed by Student-Newman-Keuls for further
multiple comparisons. Clinicopathological diagnoses were used as
the gold standard to assess diagnostic accuracy, using the receiver
operating characteristic (ROC) curves generated with the GraphPad
Prism 7.0 software. All diagnostic metrics (sensitivity and
specificity) were calculated using the standard formulae (30). P<0.05 was considered to indicate a
statistically significant difference.
Results
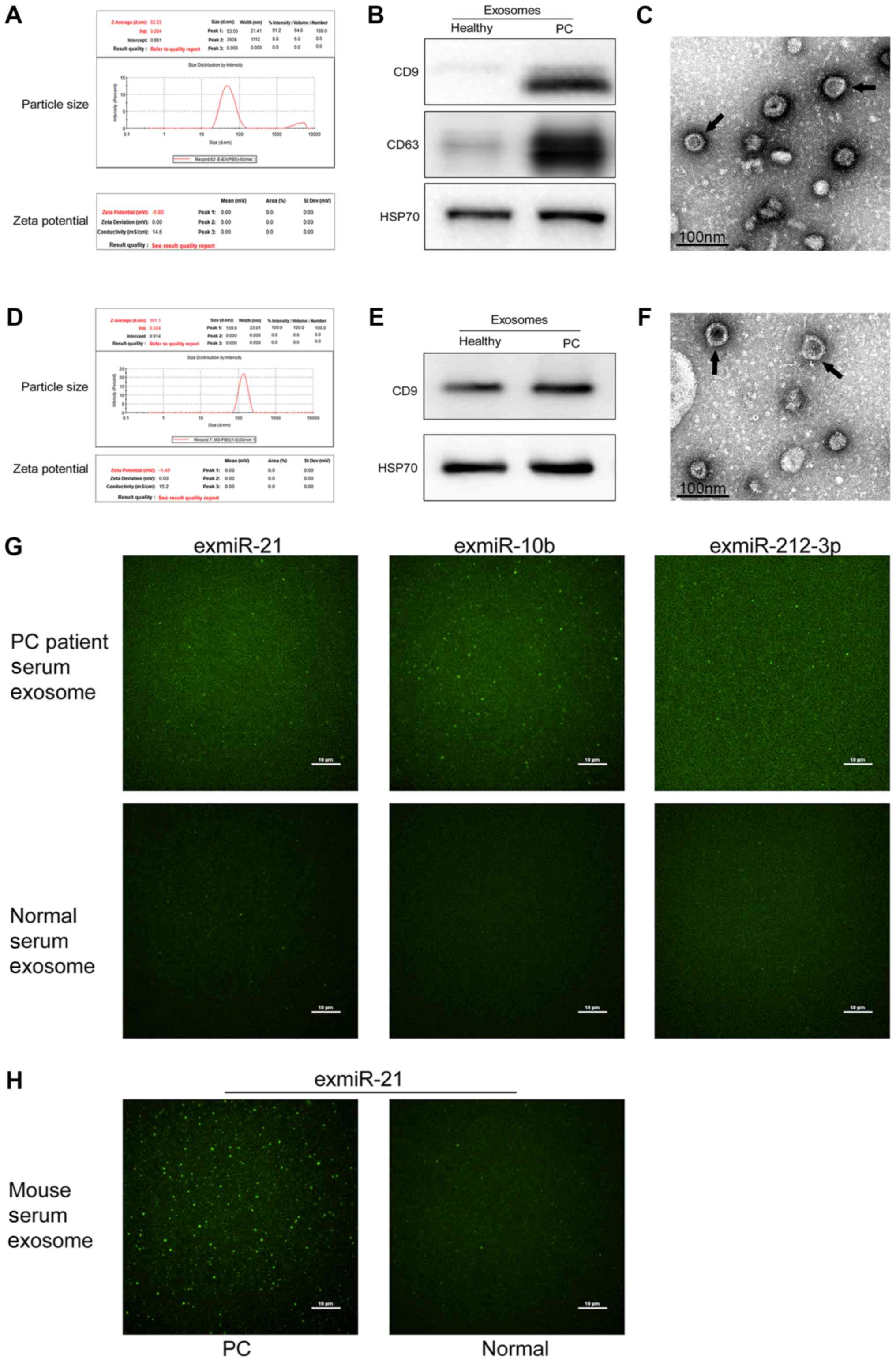
Exosome characterization
DLS was used to detect the size of particles. Based
on the DLS analysis, purified particles had a size of 53.5 nm in
human plasma samples and 139.9 nm in mouse plasma samples, which
was in accordance with the described view that exosomes are
spherical particles with sizes ranging between 30 and 150 nm
(31). The ζ potential values of
plasma exosomes from both humans and mice were negative, −5.80 and
−1.49 mV (Fig. 1A and D),
respectively. CD9, CD63 and HSP70 are considered as
exosomal-specific markers; the presence of these markers in the
purified particles was assessed to characterize the purified
particles as exosomes. Western blot analysis demonstrated that the
purified particles expressed these markers, and thus could be
considered as exosomes (Fig. 1B and
E). Additionally, electron micrographs showed that the
collected products had a distinctive cup shape (Fig. 1C and F). Overall, DLS, western blot
analyses and electron micrographs confirmed that the particles
tested were exosomes. Furthermore, as shown in Fig. 1G, the fluorescence intensity
distributions indicated that more exosomes with higher exmiR-21,
exmiR-10b and exmiR-212-3p expression were present in the samples
of patients compared with those from healthy controls. Furthermore,
murine PC exosomes revealed much higher fluorescence signals
compared with plasma exosomes from healthy mouse (Fig. 1H).
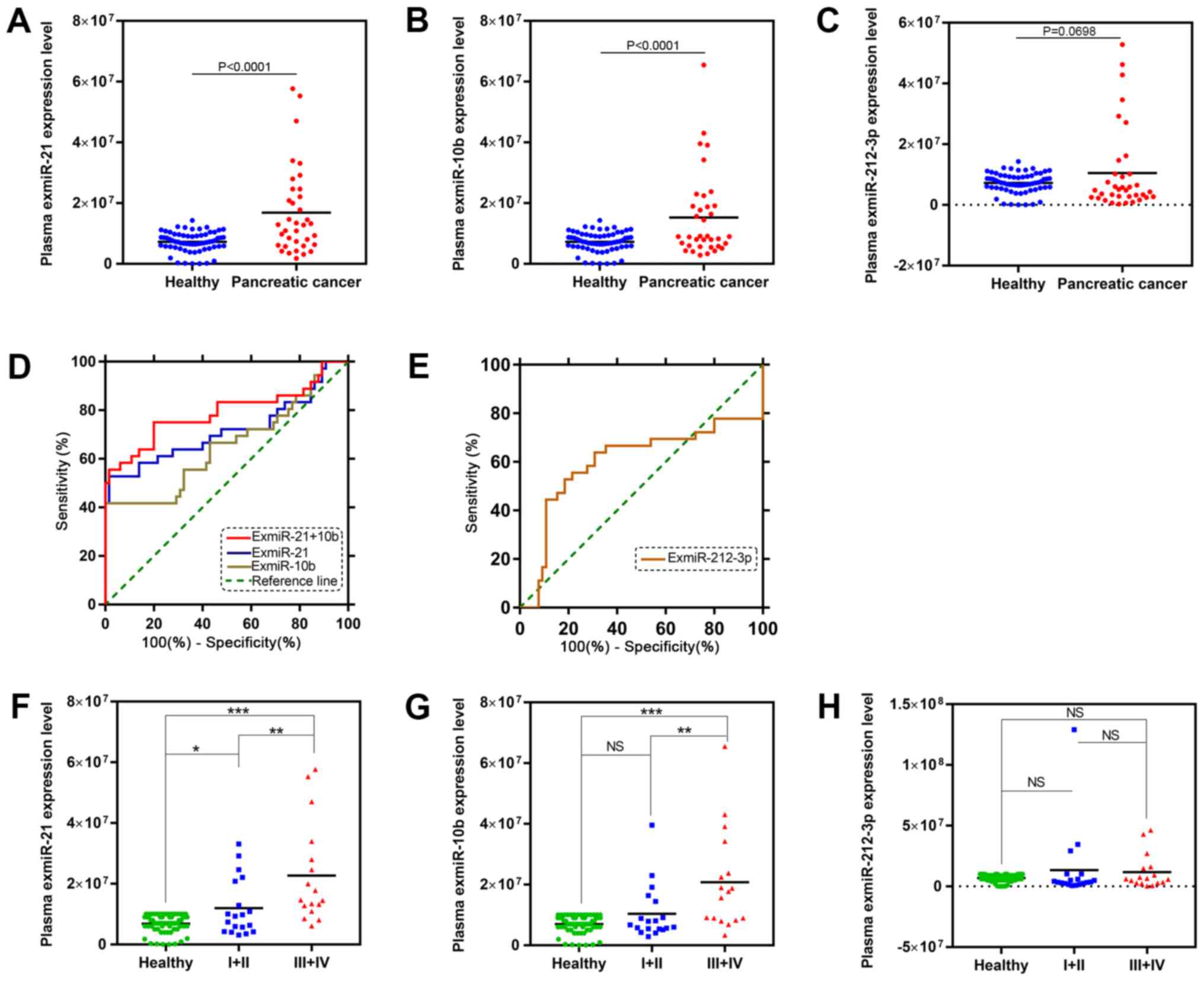
Differential exmiR expression in
patients with PC and in healthy controls
The expression levels of exmiR-21, exmiR-10b and
exmiR-212-3p were examined in the plasma of 36 patients with PC and
65 healthy individuals. The results showed that the expression
levels of exmiR-21 and exmiR-10b were significantly increased in
the plasma of patients with PC compared with the healthy control
group (Fig. 2A and B). However, no
significant difference (P=0.0698) was detected in the expression of
exmiR-212-3p between the two groups (Fig. 2C).
ROC curve analysis
In order to evaluate the diagnostic performance of
the three exmiRs, the ROC curve analysis was performed. The results
revealed that exmiR-21 and exmiR-10b, but not exmiR-212-3p, could
differentiate patients with PC from healthy individuals (Fig. 2D and E). The exmiR with the greatest
area under the curve (AUC) was exmiR-21 (P=0.0003; AUC, 0.7171);
the AUC of exmiR-10b was 0.6543 (P=0.0105). The ROC analysis
indicated that the diagnostic accuracy of exmiR-21 was superior to
the other miRs. ExmiR-21 and exmiR-10b were also combined in
further ROC analysis, which indicated that exmiR-10b could improve
the diagnostic performance of exmiR-21 (P<0.0001; AUC, 0.791;
Fig. 2).
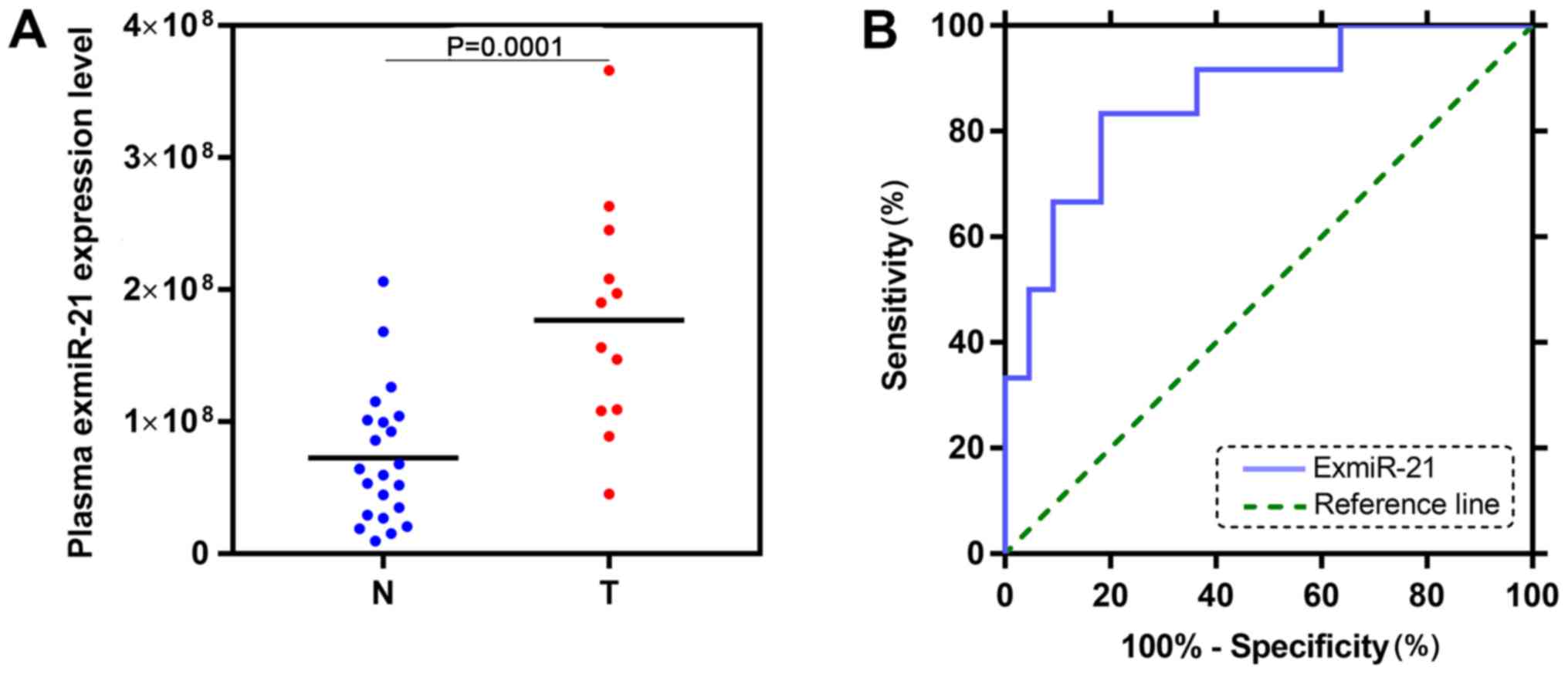
ExmiR-21 was significantly upregulated
in mice with PC and is a sensitive diagnostic marker
The expression of exmiR-21 was also examined using
the TCLN biochip in the plasma of 12 mice with PC and 22 healthy
mice. The level of exmiR-21 in the plasma of mice with PC was
significantly higher (P=0.0001) compared with the healthy controls
(Fig. 3A). The AUC of exmiR-21 in
mice was 0.8636 (P=0.0005; Fig.
3B).
ExmiR-21 was significantly higher in
patients with early-stage PC compared with healthy controls
The 36 patients with PC were stratified to examine
the associations between the plasma levels of the three exmiRs and
the PC stage, based on TNM staging. Detecting PC at an early stage
(stages I or II) (32) may provide a
window of opportunity when the disease can be adequately treated
and stopped (33). The levels of
exmiR-21 were significantly different in the early-stage group
compared with the healthy control and patients with advanced-stage
PC (Fig. 2F). In contrast, the
expression levels of exmiR-10b and exmiR-212-3p showed no
significant differences between the early-stage and the control
groups (Fig. 2G and H).
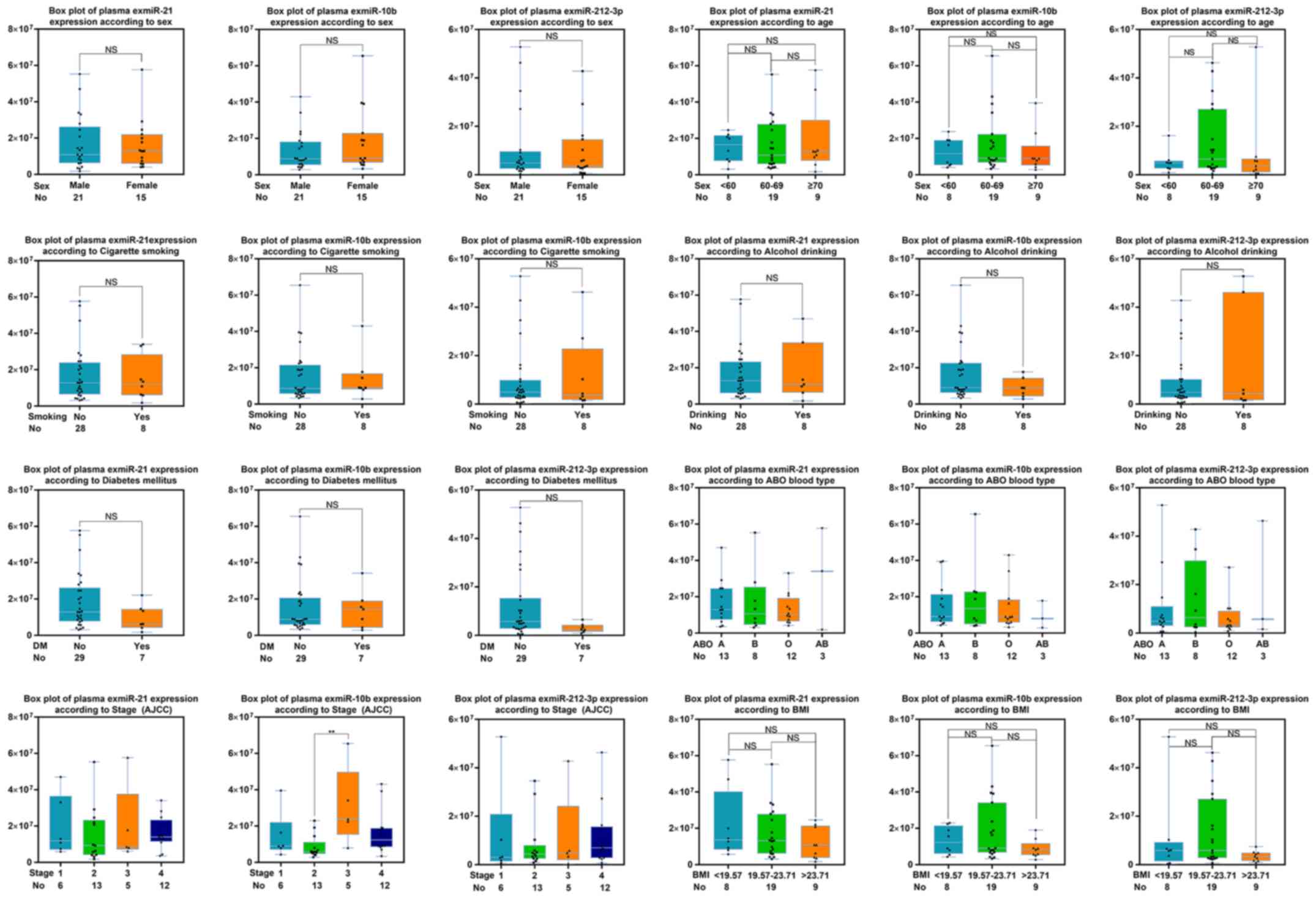
Evaluating levels of the selected
exmiRs with clinical parameters of patients with PC
Additionally, various clinical characteristics of
the 36 patients with PC, such as age, sex, history of smoking or
drinking, diabetes mellitus, blood type, cancer stage and body mass
index were analyzed. However, the levels of the three exmiRs showed
no significant differences according to these characteristics
(Fig. 4).
Discussion
One of the most important causes of the high
mortality of patients with PC is the lack of robust and sensitive
biomarkers for early diagnosis. The majority of patients with PC
are diagnosed at a stage too late for surgical treatment (32). Thus, the development of new
diagnostic methods that can detect early-stage PC is necessary to
improve the survival of patients with PC.
The present study demonstrated that exmiR-21 and
exmiR-10b were expressed at significantly higher levels in patients
with PC compared with the healthy controls. Among the three exmiRs,
exmiR-21 demonstrated the optimal diagnostic performance and, when
combined with exmiR-10b, the diagnostic performance of exmiR-21
increased. The plasma of 34 mice with PC were also examined, which
demonstrated elevated levels of exmiR-21 expression compared with
controls. Furthermore, the expression of the three exmiRs in
patients with early- or late-stage PC was also investigated in the
present study. The results indicated that exmiR-21 level may
predict the stage of PC and suggested that elevated exmiR-21
expression may serve as a potential biomarker for the early
diagnosis of PC.
Numerous studies have reported an association
between the overexpression of miR-21 and miR-10b in the plasma of
patients with PC (34,35). miR-21 and miR-10b have been
associated with multiple cancer types, such as gastric and
esophageal cancer (36), lung cancer
(37) and breast cancer (38). However, few studies have examined
exmiRs in plasma samples of patients with PC. Several studies have
reported that exmiR-21 (4,39) and exmiR-10b (39,40) were
significantly upregulated in patients with PC, and the results of
the present study are consistent with these findings. The
diagnostic performance of these exmiRs were also compared, which
revealed that exmiR-21 showed the best diagnostic performance among
these three exmiRs. Furthermore, the diagnostic performance of
exmiR-21 was increased when combined with exmiR-10b. Wu et
al (41) reported that
miR-212-3p plays an important role in the development of gastric
cancer. MiR-212-3p is one of the top differentially expressed miRs
in PC (42), and a previous study
confirmed that miR-212-3p is expressed at higher levels in PC
tissues compared with controls (42). Furthermore, patients with PC and
higher miR-212-3p expression had a decreased overall survival
compared with those with low miR-212-3p expression. Therefore,
miR-212-3p was selected as a candidate miR for analysis in the
present study. However, no significant difference was observed in
the expression of miR-212-3p between patients with PC and healthy
controls, and exmiR-212-3p exhibited the poorest diagnostic
performance among the three exmiRs.
The results indicated that exmiR-21 could
distinguish early-stage patients from both healthy individuals and
patients with advanced-stage PC, indicating that exmiR-21 showed
potential diagnostic value for early-stage PC.
Although the etiology of PC remains largely unclear,
cigarette smoking, obesity, history of drinking alcohol and
diabetes mellitus have been associated with an increased risk of PC
(43). These risk factors were thus
examined for any effect on the expression of the exmiRs in patients
with PC in the present study. The expression levels of exmiRs were
evaluated in subgroups, according to age, gender, body mass index,
blood type (44), history of
smoking, history of drinking and diabetes mellitus in the 36
patients with PC. No differences were observed in the levels of
exmiRs in any of the subgroups. Further studies with a larger
number of cases are vital to evaluate the expression levels of
these exmiRs based on clinical parameters.
To the best of our knowledge, the present study is
the first to use the TCLN biochip to detect the expression levels
of these candidate exmiRs in patients with PC, healthy individuals
and mice, with the aim of investigating the diagnostic value of
plasma exmiRs in PC. Compared with RTq-PCR, which involves
cumbersome and time-consuming methods for exosome miR extraction
and detection, the TCLN biochip is a novel and efficient method of
detecting RNA targets for the early diagnosis of cancer (6). Using the TCLN biochip, the levels of
exmiRs can be easily determined in various cancer types. In the
present study, the utility of using the TCLN biochip to evaluate
exmiR-21 levels was demonstrated to distinguish patients with
early-stage PC from healthy individuals. Although further
large-scale studies are required to validate the present findings,
these results suggest that this method will increase the number of
patients with PC diagnosed at the early-stages, giving the
opportunity to be treated with surgery and prevent disease
progression.
The present study has several limitations. Due to
the limited number of patients with PC in the study, a training and
validation cohort could not be set. However, the plasma levels of
exmiR-21 in 34 mice were validated. The present study also only
compared the expression levels of the three exmiRs in patients with
PC and healthy individuals; benign pancreatic disease could be a
valuable inclusion (such as chronic pancreatitis, pancreatic
neuroendocrine neoplasms or cystic tumors). Bloomston et al
(45) suggested that miR-21 was
uniquely upregulated in patients with PC compared with normal
individuals and patients with chronic pancreatitis. Abue et
al (46) reported that the
miR-483-3p and miR-21 expression levels were significantly higher
in PDAC than in normal individuals and patients with intraductal
papillary mucinous neoplasm. Additionally, the present study only
compared the preoperative patients with healthy individuals. Future
studies should examine the levels of these exmiRs before and after
treatment, such as surgery, radiotherapy, chemotherapy and
molecular targeted therapy. As aforementioned, further studies with
larger samples of both patients with PC and healthy individuals
will be important to verify the reliability of the diagnostic
performance of exmiR-21. Furthermore, other exmiRs that may have
higher sensitivity and specificity in PC could also be considered.
Therefore, RNA sequencing-based screening to identify PC-specific
exmiRs will be conducted, in order to focus on exmiRs with the
highest levels of upregulation, for further analysis.
The results of the present study demonstrated that
plasma levels of exmiR-21 and exmiR-10b were increased in patients
with PC compared with controls. ROC analyses demonstrated that
exmiR-21 had the best diagnostic performance among the three
exmiRs. ExmiR-21 was also found to discriminate patients with
early-stage PC from both healthy controls and patients with
advanced PC. These findings suggest that evaluating exmiR-21 using
the TCLN biochip may be a useful non-invasive strategy for
diagnosing early stage PC.
Acknowledgements
The authors would like to acknowledge Hangzhou
Dixiang Co. Ltd. (Hangzhou, China) for their support with the
tethered cationic lipoplex nanoparticle biochip technology.
Funding
The present study was supported by the Major Project
of Medical Science and Technology of Zhejiang Province (grant no.
WKI-ZJ-1824); the Foundation Project for Medical Science and
Technology of Zhejiang province (grant no. 2018KY102); the Major
Research Project of Science Technology Department of Zhejiang
Province (grant no. 2019C03048).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
LC and SJ designed the study, performed the
experiments, wrote the manuscript and prepared the figures. XP and
GD were involved in the study design, and performed the experiments
and data analysis. SZ collected the samples and characteristics of
healthy individuals and patients. MW assisted with performing the
experiment and verifying the results. All authors reviewed the
manuscript, and approved the final version of the published
manuscript.
Ethics approval and consent to
participate
The research protocol was reviewed and approved by
the Research Ethics Committee of Sir Run Run Shaw Hospital, School
of Medicine, Zhejiang University (approval; no. 20170222-16). All
participants or their guardians provided written informed consent
for scientific research statement. All animal experiments were
carried out in the animal unit of Zhejiang University according to
procedures authorized and specifically approved by the
institutional Ethical Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu H, Li T, Du Y and Li M: Pancreatic
cancer: Challenges and opportunities. BMC Med. 16:2142018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goto T, Fujiya M, Konishi H, Sasajima J,
Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, et
al: An elevated expression of serum exosomal microRNA-191, - 21,
−451a of pancreatic1 neoplasm is considered to be efficient
diagnostic marker. BMC Cancer. 18:1162018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin Y, Lin Z, Fang Z, Li H, Zhi X and
Zhang Z: Plasma MicroRNA-34a as a potential biomarker for early
diagnosis of esophageal cancer. Clin Lab. 65:2019. View Article : Google Scholar
|
|
6
|
Fadaka AO, Pretorius A and Klein A:
Functional prediction of candidate MicroRNAs for CRC management
using in silico approach. Int J Mol Sci. 20:E51902019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matsuzaki J and Ochiya T: Circulating
microRNAs and extracellular vesicles as potential cancer
biomarkers: A systematic review. Int J Clin Oncol. 22:413–420.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kawaguchi T, Komatsu S, Ichikawa D,
Tsujiura M, Takeshita H, Hirajima S, Miyamae M, Okajima W, Ohashi
T, Imamura T, et al: circulating MicroRNAs: A next-generation
clinical biomarker for digestive system cancers. Int J Mol Sci.
17:E14592016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Salehi M and Sharifi M: Exosomal miRNAs as
novel cancer biomarkers: Challenges and opportunities. J Cell
Physiol. 233:6370–6380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Zhao F, Xiao Z and Yao L: Exosomal
microRNA-205 is involved in proliferation, migration, invasion, and
apoptosis of ovarian cancer cells via regulating VEGFA. Cancer Cell
Int. 19:2812019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin
YF, Yuan Y and Zhuang SM: Hepatoma cell-secreted exosomal
microRNA-103 increases vascular permeability and promotes
metastasis by targeting junction proteins. Hepatology.
68:1459–1475. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li XJ, Ren ZJ, Tang JH and Yu Q: Exosomal
MicroRNA MiR-1246 promotes cell proliferation, invasion and drug
resistance by targeting CCNG2 in breast cancer. Cell Physiol
Biochem. 44:1741–1748. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Y, Kwak KJ, Agarwal K, Marras A, Wang
C, Mao Y, Huang X, Ma J, Yu B, Lee R, et al: Detection of
extracellular RNAs in cancer and viral infection via tethered
cationic lipoplex nanoparticles containing molecular beacons. Anal
Chem. 85:11265–11274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee LJ, Yang Z, Rahman M, Ma J, Kwak KJ,
McElroy J, Shilo K, Goparaju C, Yu L, Rom W, et al: Extracellular
mRNA detected by tethered lipoplex nanoparticle biochip for lung
adenocarcinoma detection. Am J Respir Crit Care Med. 193:1431–1433.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu J, Kwak KJ, Shi J, Yu B, Sheng Y and
Lee LJ: Overhang molecular beacons encapsulated in tethered
cationic lipoplex nanoparticles for detection of single-point
mutation in extracellular vesicle-associated RNAs. Biomaterials.
183:20–29. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC cancer staging manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
MiR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hong L, Han Y, Zhang Y, Zhang H, Zhao Q,
Wu K and Fan D: MicroRNA-21: A therapeutic target for reversing
drug resistance in cancer. Expert Opin Ther Targets. 17:1073–1080.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sheth S, Jajoo S, Kaur T, Mukherjea D,
Sheehan K, Rybak LP and Ramkumar V: Resveratrol reduces prostate
cancer growth and metastasis by inhibiting the
Akt/MicroRNA-21pathway. PLoS One. 7:e516552012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiong B, Cheng Y, Ma L and Zhang C: MiR-21
regulates biological behavior through the PTEN/PI-3 K/Akt signaling
pathway in human colorectal cancer cells. Int J Oncol. 42:219–228.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qian X, Ren Y, Shi Z, Long L, Pu P, Sheng
J, Yuan X and Kang C: Sequence-dependent synergistic inhibition of
human glioma cell lines by combined temozolomide and miR-21
inhibitor gene therapy. Mol Pharm. 9:2636–2645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lai X and Friedman A: Exosomal miRs in
lung cancer: A mathematical model. PLoS One. 11:e01677062016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bica-Pop C, Cojocneanu-Petric R, Magdo L,
Raduly L, Gulei D and Berindan-Neagoe I: Overview upon miR-21 in
lung cancer: Focus on NSCLC. Cell Mol Life Sci. 75:3539–3551. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang M, Ji S, Shao G, Zhang J, Zhao K,
Wang Z and Wu A: Effect of exosome biomarkers for diagnosis and
prognosis of breast cancer patients. Clin Transl Oncol. 20:906–911.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim J, Siverly AN, Chen D, Wang M, Yuan Y,
Wang Y, Lee H, Zhang J, Muller WJ, Liang H, et al: Ablation of
miR-10b suppresses oncogene-induced mammary tumorigenesis and
metastasis and reactivates tumor-suppressive pathways. Cancer Res.
76:6424–6435. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jin X, Chen Y, Chen H, Fei S, Chen D, Cai
X, Liu L, Lin B, Su H, Zhao L, et al: Evaluation of tumor-derived
exosomal miRNA as potential diagnostic biomarkers for early-stage
non-small cell lung cancer using next-generation sequencing. Clin
Cancer Res. 23:5311–5319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ayaz L, Görür A, Yaroğlu HY, Ozcan C and
Tamer L: Differential expression of microRNAs in plasma of patients
with laryngeal squamous cell carcinoma: Potential early-detection
markers for laryngeal squamous cell carcinoma. J Cancer Res Clin
Oncol. 139:1499–1506. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stetefeld J, McKenna SA and Patel TR:
Dynamic light scattering: A practical guide and applications in
biomedical sciences. Biophys Rev. 8:409–427. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fischer K and Schmidt M: Pitfalls and
novel applications of particle sizing by dynamic light scattering.
Biomaterials. 98:79–91. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Parikh R, Mathai A, Parikh S, Chandra
Sekhar G and Thomas R: Understanding and using sensitivity,
specificity and predictive values. Indian J Ophthalmol. 56:45–50.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Simpson RJ, Lim JW, Moritz RL and
Mathivanan S: Exosomes: Proteomic insights and diagnostic
potential. Expert Rev Proteomics. 6:267–283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Root A, Allen P, Tempst P and Yu K:
Protein biomarkers for early detection of pancreatic ductal
adenocarcinoma: Progress and challenges. Cancers (Basel).
10:E672018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lennon AM, Wolfgang CL, Canto MI, Klein
AP, Herman JM, Goggins M, Fishman EK, Kamel I, Weiss MJ, Diaz LA,
et al: The early detection of pancreatic cancer: What will it take
to diagnose and treat curable pancreatic neoplasia? Cancer Res.
74:3381–3389. 2018. View Article : Google Scholar
|
|
34
|
Duell EJ, Lujan-Barroso L, Sala N, Deitz
McElyea S, Overvad K, Tjonneland A, Olsen A, Weiderpass E, Busund
LT, Moi L, et al: Plasma microRNAs as biomarkers of pancreatic
cancer risk in a prospective cohort study. Int J Cancer.
141:905–915. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ouyang H, Gore J, Deitz S and Korc M:
MicroRNA-10b enhances pancreatic cancer cell invasion by
suppressing TIP30 expression and promoting EGF and TGF-β actions.
Oncogene. 36:49522017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jamali L, Tofigh R, Tutunchi S, Panahi G,
Borhani F, Akhavan S, Nourmohammadi P, Ghaderian SMH, Rasouli M and
Mirzaei H: Circulating microRNAs as diagnostic and therapeutic
biomarkers in gastric and esophageal cancers. J Cell Physiol.
233:8538–8550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shan X, Zhang H, Zhang L, Zhou X, Wang T,
Zhang J, Shu Y, Zhu W, Wen W and Liu P: Identification of four
plasma microRNAs as potential biomarkers in the diagnosis of male
lung squamous cell carcinoma patients in China. Cancer Med.
7:2370–2381. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matamala N, Vargas MT, González-Cámpora R,
Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G,
Yanowsky K, Calvete-Candenas J, et al: Tumor microRNA expression
profiling identifies circulating microRNAs for early breast cancer
detection. Clin Chem. 61:1098–1106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lai X, Wang M, McElyea SD, Sherman S,
House M and Korc M: A microRNA signature in circulating exosomes is
superior to exosomal glypican-1 levels for diagnosing pancreatic
cancer. Cancer Lett. 393:86–93. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Joshi GK, Deitz-McElyea S, Liyanage T,
Lawrence K, Mali S, Sardar R and Korc M: Label-free
nanoplasmonic-based short noncoding RNA sensing at attomolar
concentrations allows for quantitative and highly specific assay of
MicroRNA-10b in biological fluids and circulating exosomes. ACS
Nano. 9:11075–11089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu R, Li F, Zhu J, Tang R, Qi Q, Zhou X,
Li R, Wang W, Hua D and Chen W: A functional variant at miR-132-3p,
miR-212-3p, and miR-361-5p binding site in CD80 gene alters
susceptibility to gastric cancer in a Chinese Han population. Med
Oncol. 31:602014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu Z, Zhou L, Ding G and Cao L:
Overexpressions of miR-212 are associated with poor prognosis of
patients with pancreatic ductal adenocarcinoma. Cancer Biomark.
18:35–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Maisonneuve P and Lowenfels AB: Risk
factors for pancreatic cancer: A summary review of meta-analytical
studies. Int J Epidemiol. 44:186–198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li X, Xu H and Gao P: ABO blood group and
diabetes mellitus influence the risk for pancreatic cancer in a
population from China. Med Sci Monit. 24:9392–9398. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bloomston M, Frankel WL, Petrocca F,
Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C and Croce
CM: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Abue M, Yokoyama M, Shibuya R, Tamai K,
Yamaguchi K, Sato I, Tanaka N, Hamada S, Shimosegawa T, Sugamura K
and Satoh K: Circulating miR-483-3p and miR-21 is highly expressed
in plasma of pancreatic cancer. Int J Oncol. 46:539–547. 2015.
View Article : Google Scholar : PubMed/NCBI
|