Introduction
Breast cancer, one of the most common malignancies
among women (11.6% of total cancer cases in 2018), is a confounding
heterogeneous disease (1–3). Genetic mutations have been demonstrated
to serve a crucial role in the development and progression of
several types of cancer, including breast cancer (4–7).
Previous genome sequencing studies have indicated that clustered
mutations contribute to breast cancer progression (6,8), which
may explain the poor response to single target therapy.
Consequently, clustered mutations may pose as biomarkers for the
prognosis of breast cancer. Overall, it is critical to develop
novel prognostic biomarkers, in order to improve precision therapy
of breast cancer.
The Apolipoprotein B mRNA editing enzyme, catalytic
polypeptide-like (APOBEC) protein family is a large family of
evolutionarily conserved cytidine deaminases (9). APOBECs serve an important role in the
innate immune system, protecting against viral pathogens, such as
restriction of HIV-1 viral reverse transcription (10–14).
Notably, apolipoprotein B mRNA editing enzyme catalytic subunit 3B
(APOBEC3B), a member of the APOBEC family, has been demonstrated to
induce somatic mutations in several types of malignancies,
including breast cancer (15–21).
This suggests that APOBEC3B may be a key factor for clustered
mutations in cancer. A number of studies have demonstrated that
APOBEC3B mRNA expression levels are upregulated in breast cancer,
which is associated with the metastasis, endocrine therapy
resistance and poor prognosis of patients with estrogen receptor
positive (ER+) breast cancer (22–25);
however, to the best of our knowledge, the clinical relevance of
the APOBEC3B protein in breast cancer has not yet been determined.
Therefore, the present study aimed to investigate the role of
APOBEC3B protein expression in breast cancer. The results were
inconclusive with regards to the significance of APOBEC3B mRNA
expression levels in breast cancer, particularly for patients who
received endocrine therapy. Furthermore, related analyses were
performed especially in patients who received a different treatment
with publicly available data. As APOBEC3B is well-known for its
involvement in innate immunity and is also associated with tumor
infiltrating lymphocytes (TILs) (14,26),
associations between APOBEC3B and immune biomarkers of breast
cancer were also assessed.
Materials and methods
Patients and tissue samples
Clinicopathological data were collected from 120
female patients who received breast cancer surgery at The
Affiliated Hospital of Qingdao University (Qingdao, China) between
February 2008 and November 2010. The median age of the patients was
56 with an age range of 23–85 years. The patients were followed-up
from the date of diagnosis until May 2018 or mortality. Follow-up
began 6 months after surgery and was performed by outpatient
examination and/or by telephone every 3 months. The present study
was approved by the Ethics Committee of The Affiliated Hospital of
Qingdao University and all patients provided written informed
consent prior to enrolment in the study. TNM stage was classified
according to the American Joint Committee on Cancer guidelines, 7th
Edition (27).
Formalin-fixed and paraffin-embedded blocks of the
120 breast cancer samples were retrospectively collected and from
each block, a representative 1-mm-diameter core of tissue was
selected, re-arrayed and re-embedded in a recipient block to
prepare the tissue microarrays (TMAs). In order to ensure that
cancer tissue was collected, all slides were reviewed using
hematoxylin and eosin (H&E) staining. Hematoxylin staining was
performed for 4 min and eosin staining for 1 min at room
temperature. The most effective coring region was selected for TMA
construction. Additionally, the H&E sections of the recipient
blocks were observed following TMA construction, to confirm that
the selected region was contained within the cores. The 120 breast
cancer specimens were examined via immunohistochemistry for
APOBEC3B protein, as previously described (28). Scoring was based on the percentage of
stained tumor cells, as follows: 0–10%, negative (−); 11–25%,
slightly positive (+); 26–50%, moderately positive (++); and
51–100%, strongly positive (+++) (29). Cores were categorized into low
expression (− and +) and high expression (++ and +++) for further
analyses. All scoring was completed independently and blind by two
pathologists (Pathology Group of Breast Disease Center, the
Affiliated Hospital of Qingdao University) with agreement between
the two >90% of the time. Disagreements were resolved by
consensus. TILs were assessed using H&E stained sections and
the International TILs Working Group 2014 scoring standard was
implemented (30). Furthermore, TILs
were assessed independently and blind by the same investigators and
the mean values were used for analysis. The prognostic value of
APOBEC3B mRNA for breast cancer was determined using the
Kaplan-Meier Plotter online tool (http://kmplot.com). The cut-off value for APOBEC3B
mRNA expression levels was the median value. The patients with
similar prevalence to the SEER program in the Kaplan-Meier Plotter
online service were defined as ‘SEER similar’.
Reagents and antibodies
Monoclonal primary antibodies against APOBEC3B
(1:100; cat. no ab191695; Abcam) were used for IHC staining.
Anti-rabbit secondary antibodies were purchased from Abcam (cat. no
ab6112; Abcam) and a dilution of 1:500 was used for staining.
Immune cell infiltration analysis
RNA sequencing data and clinical data of 1,102
patients with breast cancer were downloaded from The Cancer Genome
Atlas (TCGA) database (https://portal.gdc.cancer.gov). Data on the abundance
of CD8 cells, CD4 T cells, CD8 T cells, neutrophils, macrophages
and dendritic cells per sample ID were obtained from Li et
al (31), which was accessible
through the Tumor Immune Estimation Response (TIMER) database
(http://cistrome.org/TIMER).
Statistical analysis
Statistical analyses were performed using SPSS
software (v.22.0; IBM Corp.). The χ2 test was used to assess the
association between APOBEC3B protein expression levels and
clinicopathological data. The Kaplan-Meier method and the
Cox-regression model were used for survival analysis. Differences
between two survival curves was assessed with log-rank test. All
analyses were two-sided. P<0.05 was considered to indicate a
statistically significant difference. Disease-free survival (DFS)
time was defined as the interval (in months) between the date of
breast surgery to first recurrence (loco-regional recurrence and/or
distant metastasis). Relapse-free survival (RFS) time was defined
as the interval (in months) between the date of breast surgery to
first loco-regional recurrence. Distant metastasis free survival
(DMFS) time was defined as the interval (in months) between the
date of breast surgery to first distant metastasis. Overall
survival (OS) time was defined as the interval (in months) between
the date of breast surgery and breast cancer-associated mortality.
The Kruskal-Wallis test was employed to compare the abundance of
immune cell infiltrates among the groups. The Dunn's test was used
as the post-hoc test following Kruskal-Wallis. The difference
between two groups was assessed with Mann-Whitney U test.
Results
Clinicopathological characteristics of
patients with breast cancer
In total, 120 patients who received breast cancer
surgery at The Affiliated Hospital of Qingdao University were
evaluated in the present study. A total of four specimen were lost
during sectioning, thus 116 cases were analyzed. The
clinicopathological characteristics of the patients are presented
in Table I. The median diagnostic
age was 56 years (range, 23–85 years), and the majority of cases
were ductal carcinoma (n=87; 75.0%). The majority of patients
presented with histological grade 1 (n=52; 44.8%), followed by
grade 2 (n=50; 43.1%) and only two patients had T3 (1.7%; Table I). Regarding pathological
characteristics, the majority of patients were classified in stage
I (n=48; 41.4%) and stage II (n=56; 48.3%), and were both ER+
(n=86; 74.1%) and progesterone receptor (PR)+(n=73; 62.9%).
Regarding molecular subtypes, the majority of cases were
luminal-like(Luminal A, n=50, 43.1%; Luminal B, n=37, 31.9%),
whereas nine patients were human epidermal growth factor receptor-2
(HER2) positive (7.8%). After a median follow-up of 97 months
(range, 8.0–123.0 months), 15 patients had died and seven patients
had relapsed by the end of the present study (data not shown).
 | Table I.Clinicopathological characteristics
of patients with breast cancer (n=116). |
Table I.
Clinicopathological characteristics
of patients with breast cancer (n=116).
|
Characteristics | Patients, n | Percentage |
|---|
| Age at diagnosis,
years (median range) | 56 (23–85) |
|
| Age, years |
|
|
|
≤65 | 79 | 68.1 |
|
>65 | 37 | 31.9 |
| Overall survival,
months |
|
|
| Median,
range | 97 (8–123) |
|
| Number
of mortalities | 15 | 12.9 |
| Disease-free
survival, months |
|
|
| Median,
range | 91 (4–123) |
|
| Number
of recurrences | 22 | 19.0 |
| Histological
subtype |
|
|
|
IDC | 87 | 75.0 |
|
ILC | 12 | 10.3 |
|
Others | 17 | 14.7 |
| Menopausal
status |
|
|
|
Premenopausal | 47 | 40.5 |
|
Postmenopausal | 69 | 59.5 |
| Histological grade,
% |
|
|
| G1 | 52 | 44.8 |
| G2 | 50 | 43.1 |
| G3 | 14 | 12.1 |
| ER status |
|
|
|
Positive | 86 | 74.1 |
|
Negative | 30 | 25.9 |
| PR status |
|
|
|
Positive | 73 | 62.9 |
|
Negative | 43 | 37.1 |
| HER2 status |
|
|
|
Positive | 9 | 7.8 |
|
Negative | 107 | 92.2 |
| Ki67, % |
|
|
|
>14 | 68 | 58.6 |
|
≤14 | 48 | 41.4 |
| T category |
|
|
| T1 | 58 | 50.0 |
| T2 | 56 | 48.3 |
| T3 | 2 | 1.7 |
| LN status |
|
|
|
Negative | 81 | 69.8 |
|
Positive | 35 | 30.2 |
| Pathological
stage |
|
|
| I | 48 | 41.4 |
| II | 56 | 48.3 |
|
III | 12 | 10.3 |
| Molecular
subtype |
|
|
| Luminal
A | 50 | 43.1 |
| Luminal
B | 37 | 31.9 |
| HER2
amplification | 3 | 2.6 |
| Triple
negative | 26 | 22.4 |
| APOBEC3B protein
expression |
|
High | 66 | 56.9 |
|
Low | 50 | 43.1 |
APOBEC3B protein expression by TMA and
association with clinicopathological characteristics
APOBEC3B protein staining was performed in
accordance with a previous study on ovarian cancer (26) and was localized to both nuclear and
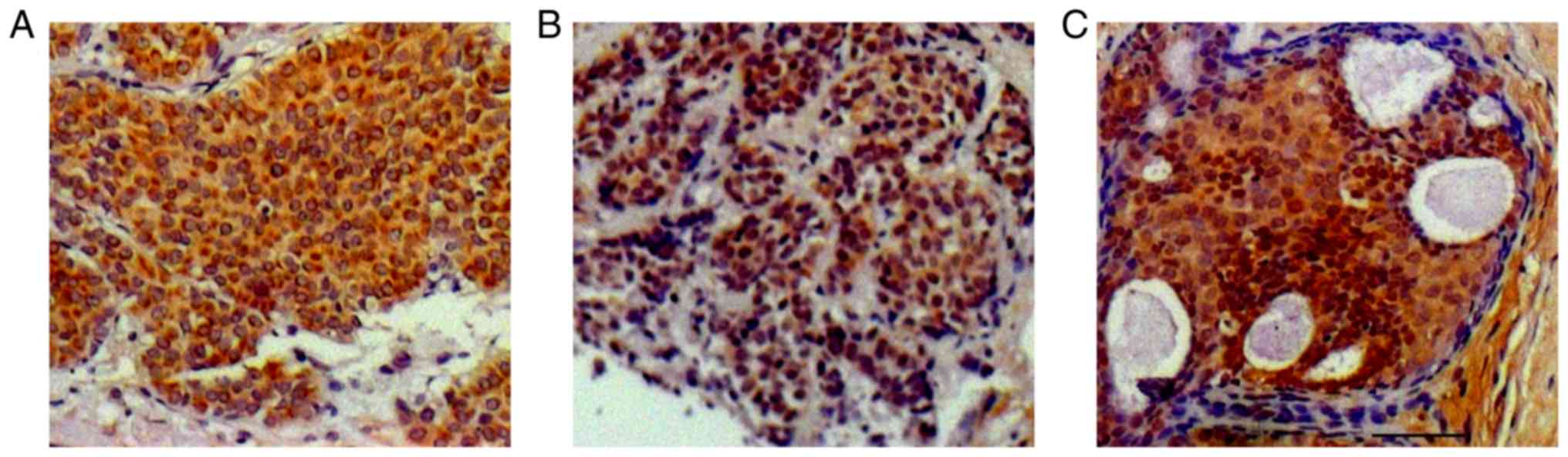
cytoplasmic compartments (Fig. 1).
Of the 116 patients, 66 demonstrated high levels of APOBEC3B
protein expression (56.9%). The associations between APOBEC3B
protein expression levels and clinicopathological characteristics
are presented in Table II. APOBEC3B
protein expression was significantly associated with ER and PR
expression (both P<0.05), and with different subtypes of breast
cancer (P=0.045). However, no significant associations were
identified between APOBEC3B protein expression and other
clinicopathological characteristics such as age, histological
grade, tumor size and TNM stage. Notably, no significant
associations were identified between APOBEC3B protein expression
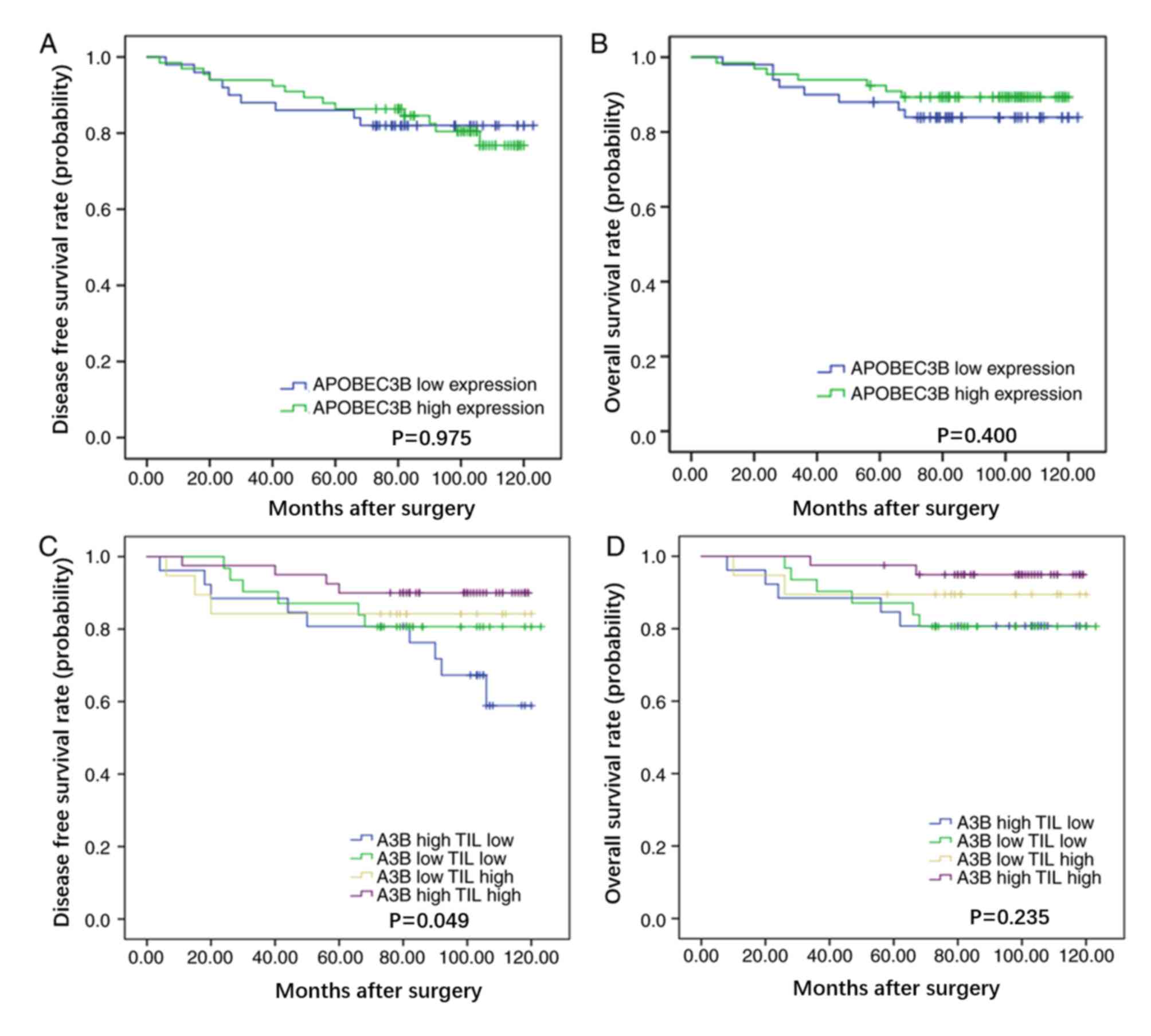
and DFS time (P=0.975) or OS time (P=0.400) in patients with breast
cancer (Fig. 2A and B,
respectively).
 | Table II.Associations between APOBEC3B protein
expression and clinicopathological characteristics of patients with
breast cancer. |
Table II.
Associations between APOBEC3B protein
expression and clinicopathological characteristics of patients with
breast cancer.
|
| APOBEC3B
expression, n |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | High | Low | χ2 | P-value |
|---|
| Age, years |
|
| 0.614 | 0.433 |
|
≤65 | 43 | 36 |
|
|
|
>65 | 23 | 14 |
|
|
| Histological
subtype |
|
| 2.520 | 0.284 |
|
IDC | 53 | 34 |
|
|
|
ILC | 6 | 6 |
|
|
|
Others | 7 | 10 |
|
|
| Menopause
status |
|
| 1.549 | 0.213 |
|
Premenopausal | 30 | 17 |
|
|
|
Postmenopausal | 36 | 33 |
|
|
| Histological
grade |
|
| 2.769 | 0.250 |
| G1 | 34 | 18 |
|
|
| G2 | 25 | 25 |
|
|
| G3 | 7 | 7 |
|
|
| ER status |
|
| 4.711 | 0.030 |
|
Positive | 54 | 32 |
|
|
|
Negative | 12 | 18 |
|
|
| PR status |
|
| 10.799 | 0.001 |
|
Positive | 50 | 23 |
|
|
|
Negative | 16 | 27 |
|
|
| HER2 status |
|
| 0.000 | 0.933 |
|
Positive | 5 | 4 |
|
|
|
Negative | 61 | 46 |
|
|
| Ki67, % |
|
| 1.194 | 0.274 |
|
>14 | 41 | 26 |
|
|
|
≤14 | 25 | 24 |
|
|
| T category |
|
| 2.577 | 0.276 |
| T1 | 34 | 24 |
|
|
| T2 | 30 | 26 |
|
|
| T3 | 2 | 0 |
|
|
| LN status |
|
| 0.139 | 0.709 |
|
Negative | 47 | 34 |
|
|
|
Positive | 19 | 16 |
|
|
| Pathological
stage |
|
| 1.315 | 0.518 |
| I | 29 | 19 |
|
|
| II | 29 | 27 |
|
|
|
III | 8 | 4 |
|
|
| Molecular
subtype |
|
| 8.050 | 0.045 |
| Luminal
A | 33 | 17 |
|
|
| Luminal
B | 23 | 14 |
|
|
| HER2
amplification | 1 | 2 |
|
|
| Triple
negative | 9 | 17 |
|
|
| iTILs |
|
| 0.185 | 0.667 |
|
High | 14 | 9 |
|
|
|
Low | 52 | 41 |
|
|
| sTILs |
|
| 5.564 | 0.018 |
|
High | 37 | 17 |
|
|
|
Low | 29 | 33 |
|
|
| TILs |
|
| 5.817 | 0.016 |
|
High | 40 | 19 |
|
|
|
Low | 26 | 31 |
|
|
APOBEC3B protein expression and
TILs
The association between APOBEC3B protein expression
and TILs was analyzed in the 116 patients with breast cancer, in
order to determine the role of APOBEC3B in the immune system. The
results demonstrated that high APOBEC3B protein expression was
associated with TILs (P=0.016), particularly TILs within the stroma
(P=0.018; Table II and Fig. S1). Subsequently, patients were
divided into four groups: i) High APOBEC3B protein expression and
high levels of TILs; ii) high APOBEC3B protein expression and low
levels of TILs; iii) low APOBEC3B protein expression and high
levels of TILs; and iv) low APOBEC3B protein expression and low
levels of TILs, in order to analyze the effects of APOBEC3B protein
expression and TILs on survival. The results demonstrated that
patients with high APOBEC3B protein expression and high levels of
TILs had improved DFS time (P=0.049), with a trend for improved OS
time (P=0.235), compared with patients with high APOBEC3B protein
expression and low levels of TILs (Fig.
2C and D, respectively). Multivariate analysis demonstrated
that patients in the subgroup with high APOBEC3B protein expression
and high levels of TILs exhibited improved DFS time compared with
patients with high APOBEC3B protein expression and low levels of
TILs [hazard ratio (HR)=0.65; 95% CI=0.45–0.95; P=0.024; Table III].
 | Table III.Univariate and multivariate analyses
of characteristics associated with DFS. |
Table III.
Univariate and multivariate analyses
of characteristics associated with DFS.
|
| DFS time |
|---|
|
|
|
|---|
|
Characteristics | HR (95% CI) | P-value |
|---|
| Univariate |
|
|
| Age,
years (≤65 vs. >65) | 1.95
(0.84–4.51) | 0.120 |
|
Histological subtype (IDC vs.
Others) | 0.84
(0.44–1.58) | 0.582 |
| Grade
(1 vs. 2/3) | 0.29
(0.11–0.73) | 0.009 |
| Tumor
size, cm (>5 vs. ≤5) | 4.04
(1.49–10.98) | 0.006 |
| Lymph
node (+ vs. -) | 3.00
(1.29–7.01) | 0.011 |
| TNM
stage (II/III vs. I) | 2.09
(1.13–3.87) | 0.019 |
| ER (+
vs. -) | 0.68
(0.28–1.67) | 0.402 |
| PR (+
vs. -) | 0.62
(0.26–1.43) | 0.260 |
| HER2 (+
vs. -) | 3.65
(1.22–10.92) | 0.020 |
| Ki67, %
(≤14 vs. >14) | 0.78
(0.33–1.87) | 0.581 |
| Molecular
subtype |
| 0.066 |
| Luminal
A vs. TNBC | 0.80
(0.29–2.21) | 0.671 |
| Luminal
B vs. TNBC | 0.43
(0.12–1.51) | 0.186 |
| HER2+
vs. TNBC | 4.28
(0.86–21.39) | 0.077 |
| TILs
(high vs. low) | 0.43
(0.18–1.05) | 0.064 |
| iTILs
(high vs. low) | 0.41
(0.10–1.76) | 0.232 |
| sTILs
(high vs. low) | 0.41
(0.16–1.04) | 0.061 |
|
APOBEC3B (high vs. low) | 0.99
(0.42–2.32) | 0.975 |
|
APOBEC3B and TILs (hl vs.
hh) | 0.27
(0.08–0.88) | 0.030 |
| Multivariate |
|
|
| Grade
(1 vs. 2/3) | 0.21
(0.08–0.56) | 0.002 |
| Tumor
size, cm (>5 vs. ≤5) | 6.25
(1.35–29.03) | 0.019 |
| Lymph
node (+ vs. -) | 4.05
(1.14–11.94) | 0.011 |
| TNM
stage (II/III vs. I) | 0.43
(0.07–2.51) | 0.346 |
| HER2 (+
vs. -) | 2.09
(0.56–7.73) | 0.271 |
|
APOBEC3B and TILs (hl vs.
hh) | 0.65
(0.45–0.95) | 0.024 |
APOBEC3B mRNA expression in breast
cancer
A total of 3,951 breast cancer cases were assessed
using the data in the Kaplan-Meier Plotter online tool. With the
median value set as the cut-off value, the results demonstrated
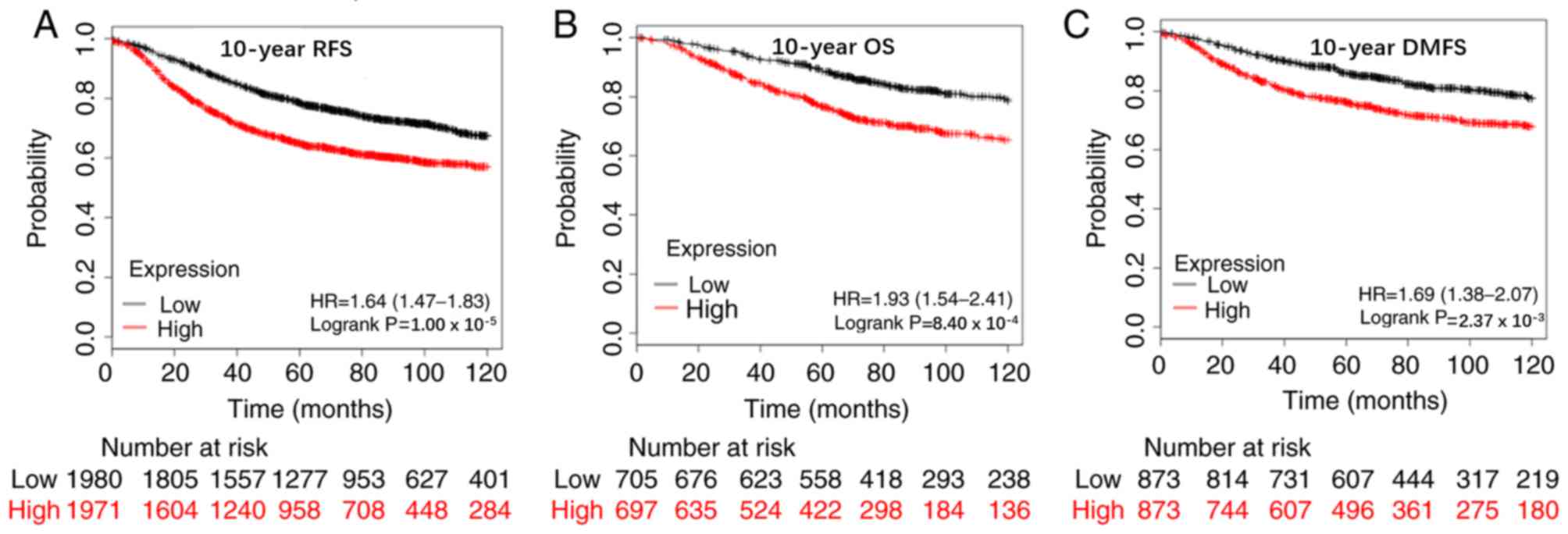
that patients with high APOBEC3B mRNA expression levels had poor
10-year relapse-free survival (RFS) rate (n=3,951; HR=1.64; 95%
CI=1.47–1.83; P<0.00001), OS rate (n=1,402; HR=1.93; 95%
CI=1.54–2.41; P=0.00084) and distant metastasis-free survival
(DMFS) rate (n=1,746, HR=1.69; 95% CI=1.38–2.07; P=0.00237;
Fig. 3). Analysis was also performed
in the patients who had a similar prevalence to the SEER
(Surveillance, Epidemiology, and End Results) database and similar
trends were observed for 10-year RFS rate (n=493; HR=1.47, 95%
CI=1.04–2.07; P=0.028), OS rate (n=301; HR=1.97; 95% CI=1.22–3.19;
P=0.0047) and DMFS rate (n=375; HR=1.54; 95% CI=1.02–2.31; P=0.036;
Fig. S2). Overall, these results
suggest that high APOBEC3B mRNA expression levels are associated
with poor prognosis of breast cancer.
Previous studies have reported that high APOBEC3B
mRNA expression levels indicate a worse survival in ER+ patients
(24–26). The present study analyzed different
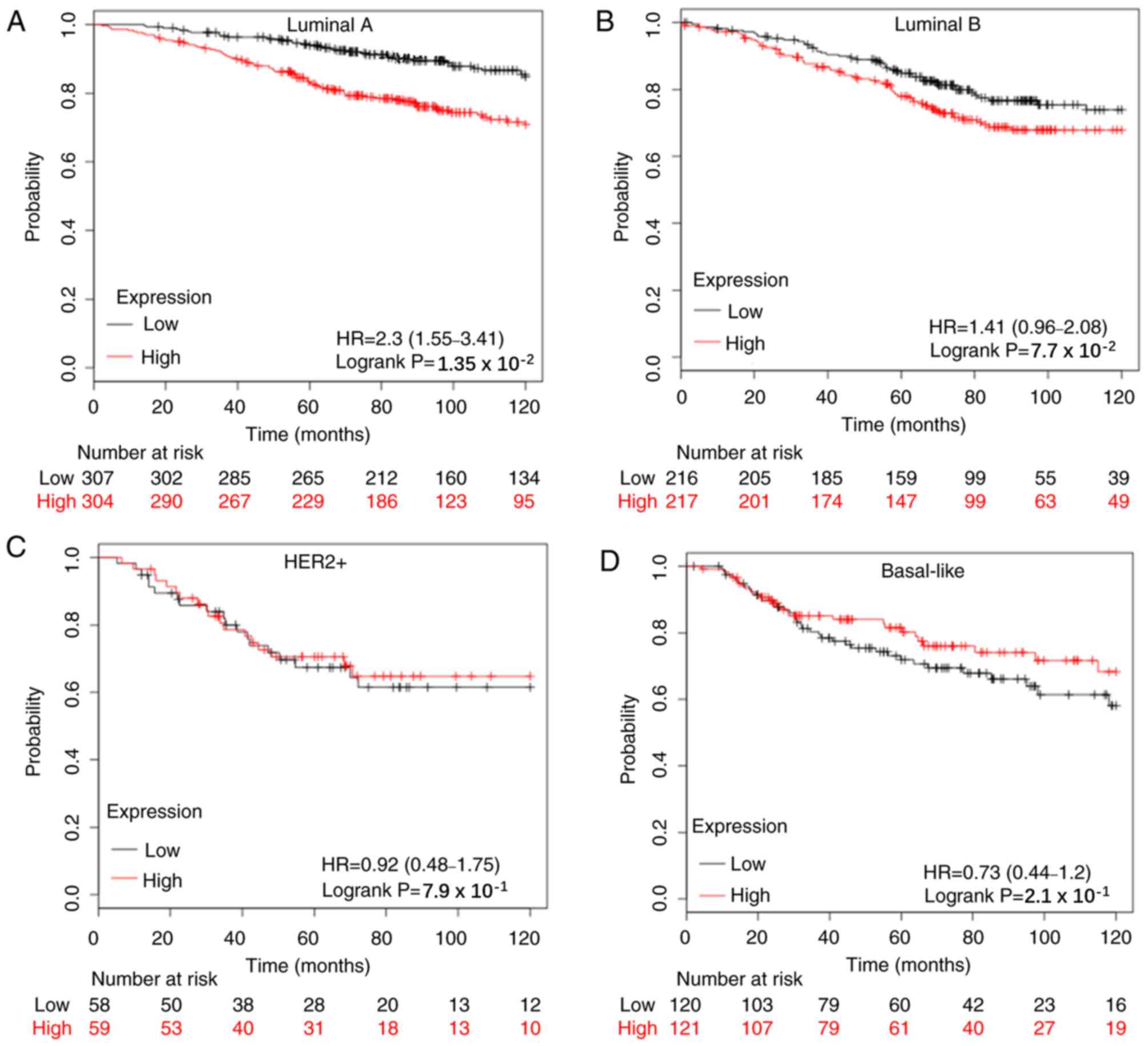
subtypes of breast cancer and the results demonstrated that the
prognostic value of APOBEC3B mRNA expression was significantly
associated with the Luminal A subtype in 10-year-RFS rate (n=1,933;
HR=1.68; 95% CI=1.41–2.01; P=0.00084; Fig. S3A), 10-year-OS rate (n=611; HR=2.3;
95% CI=1.55–3.41; P=0.0135; Fig. 4A)
and 10-year-DMFS rate (n=965; HR=1.67; 95% CI=1.22–2.27; P=0.0011;
Fig. S4A). Conversely, APOBEC3B
mRNA expression was only significantly associated with the Luminal
B subtype in 10-year-RFS rate (n=1,149; HR=1.32; 95% CI=1.08–1.60;
P=0.0056; Fig. S3B).
Similar analyses were performed for patients who did
or did not receive systemic therapy. The results for patients that
did not receive treatment were as follows: 10-year-RFS rate
(n=1,010; HR=1.58; 95% CI=1.26–1.98; P<0.05) and 10-year-OS rate
(n=382; HR=1.98; 95% CI=1.20–3.28; P=0.0068; Fig. S5). The results for patients that
received endocrine therapy were as follows: 10-year-RFS rate
(n=929; HR=1.35; 95% CI=1.04–1.77; P=0.026) and 10-year-OS rate
(n=133; HR=2.95; 95% CI=1.36–6.38; P=0.0039), and those that
received tamoxifen: 10-year-RFS rate (n=740; HR=1.37; 95%
CI=1.00–1.87; P=0.0047) and 10-year-OS rate (n=130; HR=2.61; 95%
CI=1.19–5.74; P=0.013; Fig. S6).
Overall, patients with high APOBEC3B mRNA expression levels were
revealed to have worse 10-year-RFS and OS rates. APOBEC3B mRNA
expression has no significant effect on 10-year-RFS rate (n=798;
HR=1.28; 95% CI=0.97–1.67; P=0.075) and 10-year-OS rate (n=300;
HR=0.99; 95% CI=0.61–1.62; P=0.98; Fig.
S6) for patients with chemotherapy.
APOBEC3B mRNA and the immune
system
The median cut-off value was used in the TCGA
database to determine the association between APOBEC3B mRNA
expression levels and immune-related genes. The results
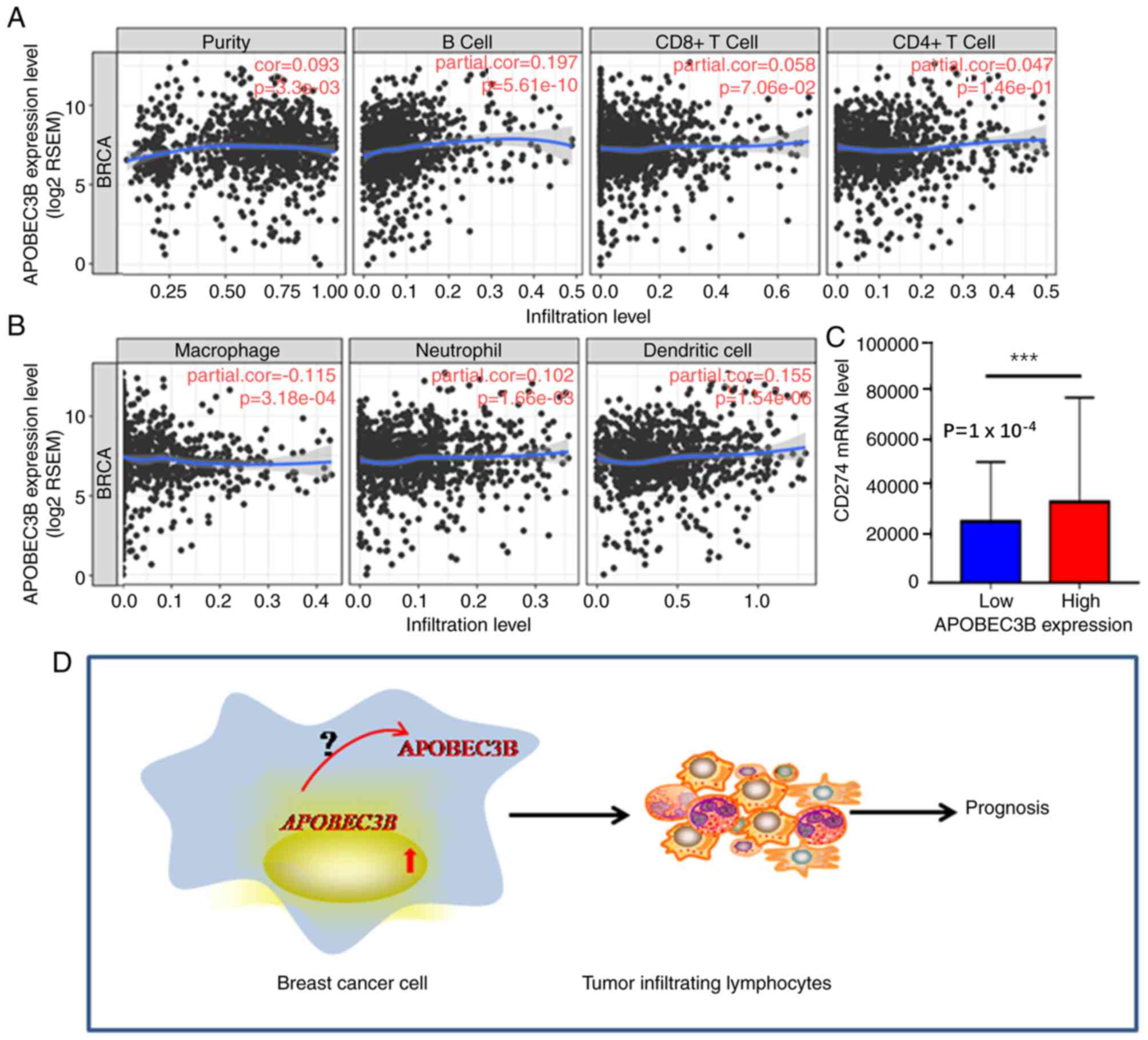
demonstrated that APOBEC3B mRNA expression levels were positively
associated with B-cells (r=0.197; P<0.05) and dendritic cells
(r=0.155; P<0.05); however, a negative trend association was
demonstrated with macrophages (r=−0.115; P>0.05; Fig. 5A and B). High APOBEC3B mRNA
expression levels were also indicated to be associated with
programmed cell death-ligand 1,PD-L1 (CD274) expression (P=0.0001;
Fig. 5C), which is a target for
immunotherapy. Overall, these results suggest that APOBEC3B
expression may be a novel predictor of immunotherapy response.
Discussion
Breast cancer is the most common malignancy in women
as well as the leading cause of cancer-associated mortality in
women worldwide (1). In 2018, there
were 2,088,849 newly-diagnosed cases of breast cancer and 626,679
cancer-associated mortalities worldwide (1). Despite advancements made in the
development of novel treatments, and improved survival of patients
with breast cancer, the number of reported cases of recurrence
and/or metastasis remains high. Previous studies have highlighted
breast cancer as a heterogeneous disease, which is comprised of
several somatic mutations, including those in PI3KCA, P53 and ESR1
(2,3,6,2,8).
Discovering novel biomarkers for these mutations may prove
beneficial in the development of treatments for breast cancer.
APOBEC3B has been demonstrated to induce somatic mutations, and
thus may be a useful biomarker (15–21).
Furthermore, previous studies have reported that APOBEC3B mRNA
expression is upregulated in cancer tissues compared with normal
tissues, including breast cancer (12–17,20).
APOBEC3B mRNA expression has also been associated with poorer RFS
and OS rates in ER+ breast cancer (22–25). The
present study analyzed APOBEC3B protein expression in patients with
breast cancer and investigated its association with the immune
system.
The results demonstrated that APOBEC3B protein
levels were similar to mRNA levels, and were associated with ER and
PR status. However, the results did not detect an association
between APOBEC3B protein levels and DFS or OS time, which differs
from previous studies (26,32). This may be due to the fact that
APOBEC3B mRNA and protein levels are not consistent in breast
cancer and that APOBEC3B has different roles depending on its
surroundings. Rüder et al (26) reported that patients with high-grade
serous ovarian carcinoma that are negative for both APOBEC3B mRNA
and protein expression, were associated with poor progression-free
survival (PFS) time; however, in multivariate analysis, only
APOBEC3B mRNA expression was demonstrated to be an independent
factor for PFS time. Furthermore, only APOBEC3B cytoplasmic
staining indicated a trend towards improved PFS time (26). The difference in results compared
with previous studies may be due to the small sample size used in
the present study. Future studies are required with larger sample
sizes, in order to confirm these observations. Analyses using
publicly available data demonstrated that APOBEC3B mRNA expression
levels were associated with the prognosis of Luminal breast cancer,
particularly the Luminal A subtype. This was the case whether the
patient received endocrine therapy or no treatment; however, it was
not the case for patients treated using chemotherapy. Taken
together, the results suggest that the prognostic value of APOBEC3B
mRNA expression levels is valuable in low-risk patients. Consistent
with the findings of the present study, previous studies have
demonstrated that APOBEC3B mRNA expression levels are associated
with the survival of ER+ and lymph node negative patients (23,24).
Furthermore, overexpression of APOBEC3B in MCF7 has been reported
to induce tamoxifen resistance (25), which may explain the poor prognosis
of these patients. A previous study demonstrated that high APOBEC3B
mRNA expression indicates an improved response rate to neoadjuvant
chemotherapy in breast cancer, particularly triple negative breast
cancer; however, no significant effect was observed on the
prognosis (33). This suggests that
APOBEC3B mRNA expression levels may serve a predictive role for
chemotherapy sensitivity in some breast cancer subtypes, however,
future studies are required to determine the exact predictive value
of APOBEC3B.
APOBEC3B has been reported to serve vital roles in
the immune system (13,22,26),
which include TILs. TILs play important roles in the prediction and
prognosis in breast cancer. Moreover, immunotherapy maybe effective
in breast cancer. However, to the best of our knowledge, no study
has analyzed the association between APOBEC3B expression and TILs
in breast cancer. The results demonstrated that APOBEC3B protein
expression levels were positively associated with lymphocytes in
breast cancer tissue, particularly lymphocytes within the stroma.
Such associations have been demonstrated to improve the survival
and pathological complete response in breast cancer (34). The precursor study to the present
study reported that different types of lymphocyte serve varying
roles in the prediction and prognosis of breast cancer (34,35).
Despite failure to detect an association between APOBEC3B protein
expression levels and lymphocyte subtypes, the present study
demonstrated that high APOBEC3B protein expression was associated
with increased TIL levels. Furthermore, survival analysis indicated
that high APOBEC3B protein expression and high levels of TILs were
associated with improved DFS, as an independent factor, compared
with the high APOBEC3B protein expression and low levels of TILs.
It is plausible that APOBEC3B and TILs may be associated with the
active immune status contribute to cancer control, thus making the
APOBEC3B protein a notable marker for tumor immune status.
The association between APOBEC3B mRNA expression
levels and different subtypes of lymphocyte was also assessed using
TCGA and TIMER databases. The results were similar to those of
previous studies on ovarian and lung cancer (13,26),
demonstrating that high APOBEC3B mRNA and protein expression were
associated with TILs in breast cancer, which may contribute to the
prognosis of these patients. However, the key factors which are
attributed to the translation and location of the APOBEC3B protein,
its direct interaction with lymphocytes and the resulting prognosis
remain unclear (Fig. 5D), thus
further studies are required. Furthermore, APOBEC3B mRNA expression
has been reported to be associated with an immune therapeutic
response in lung cancer, suggesting its potential as a biomarker
(13). Although the predictive value
of APOBEC3B, with regards to immune therapy, was not assessed in
the present study, the results demonstrated that APOBEC3B mRNA
expression was positively associated with PD-L1 expression, which
is a known biomarker for immune therapy. This indicates that
APOBEC3B may be a promising marker for immunotherapy.
Overall, the results of the present study
demonstrated that APOBECB3B mRNA and protein expression levels
served varied roles in breast cancer progression. APOBEC3B mRNA
expression indicated worse prognosis for patients with breast
cancer, particularly patients with the luminal A subtype, while
both APOBEC3B protein and mRNA expression were associated with some
immune system cells and may have the potential to be developed as a
novel prognostic marker for patients with breast cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Doctoral Funding of Shandong Province (grant nos.
ZR2017BH061 and ZR2019BH013), the National Natural Science
Foundation of China (grant nos. 81572616 and 81772845), the Qingdao
Science and Technology Innovation Plan (grant no. 17-1-1-54-jch),
the postdoctoral funding of Qingdao City, youth funding of Qingdao
University (Clinical medicine + X) and The Affiliated Hospital of
Qingdao University.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YM designed the present study, analyzed the data and
drafted the initial manuscript. ML, GN, YoW and JC acquired the
clinicopathological data and performed TMA construction. YuW, WC,
XL and XW performed the experiments. YZ interpreted the data and HW
designed and supervised the present study. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
All procedures performed in the present study were
in accordance with the Ethical Standards of the Institutional and
National Research Committee and with The Helsinki Declaration. The
present study was approved by the Ethics Committee of the
Affiliated Hospital of Qingdao University (approval no. QYFY WZLL
25616) and written informed consent was obtained from all patients
prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stephens PJ, Tarpey PS, Davies H, Van Loo
P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell
GR, et al: The landscape of cancer genes and mutational processes
in breast cancer. Nature. 486:400–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas N: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Greenman C, Stephens P, Smith R, Dalgliesh
GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C,
et al: Patterns of somatic mutation in human cancer genomes.
Nature. 446:153–158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nik-Zainal S, Alexandrov LB, Wedge DC, Van
Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J,
Stebbings LA, et al: Mutational processes molding the genomes of 21
breast cancers. Cell. 149:979–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Børresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smith HC, Bennett RP, Kizilyer A,
McDougall WM and Prohaska KM: Functions and regulation of the
APOBEC family of proteins. Semin Cell Dev Biol. 23:258–268. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Refsland EW and Harris RS: The APOBEC3
family of retroelement restriction factors. Curr Top Microbiol
Immunol. 371:1–27. 2013.PubMed/NCBI
|
|
11
|
Harris RS and Liddament MT: Retroviral
restriction by APOBEC proteins. Nat Rev Immunol. 4:868–877. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ng JCF, Quist J, Grigoriadis A, Malim MH
and Fraternali F: Pan-cancer transcriptomic analysis dissects
immune and proliferative functions of APOBEC3 cytidine deaminases.
Nucleic Acids Res. 47:1178–1194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang S, Jia M, He Z and Liu XS: APOBEC3B
and APOBEC mutational signature as potential predictive markers for
immunotherapy response in non-small cell lung cancer. Oncogene.
37:3924–3936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wen WX, Soo JS, Kwan PY, Hong E, Khang TF,
Mariapun S, Lee CS, Hasan SN, Rajadurai P, Yip CH, et al: Germline
APOBEC3B deletion is associated with breast cancer risk in an Asian
multi-ethnic cohort and with immune cell presentation. Breast
Cancer Res. 18:562016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Burns MB, Lackey L, Carpenter MA, Rathore
A, Land AM, Leonard B, Refsland EW, Refsland EW, Kotandeniya D,
Tretyakova N, et al: APOBEC3B is an enzymatic source of mutation in
breast cancer. Nature. 494:366–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Middlebrooks CD, Banday AR, Matsuda K,
Udquim KI, Onabajo OO, Paquin A, Figueroa JD, Zhu B, Koutros S,
Kubo M, et al: Association of germline variants in the APOBEC3
region with cancer risk and enrichment with APOBEC-signature
mutations in tumors. Nat Genet. 48:1330–1338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Roberts SA, Lawrence MS, Klimczak LJ,
Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL,
Saksena G, et al: An APOBEC cytidine deaminase mutagenesis pattern
is widespread in human cancers. Nat Genet. 45:970–976. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harris RS: Molecular mechanism and
clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer.
Breast Cancer Res. 17:82015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Delahanty R, Guo X, Zheng W and
Long J: Integrative genomic analysis reveals functional
diversification of APOBEC gene family in breast cancer. Hum
Genomics. 9:342015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kuong KJ and Loeb LA: APOBEC3B mutagenesis
in cancer. Nat Genet. 45:964–965. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanu N, Cerone MA, Goh G, Zalmas LP,
Bartkova J, Dietzen M, McGranahan N, Rogers R, Law EK, Gromova I,
et al: DNA replication stress mediates APOBEC3 family mutagenesis
in breast cancer. Genome Biol. 17:1852016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cescon DW, Haibe-Kains B and Mak TW:
APOBEC3B expression in breast cancer reflects cellular
proliferation, while a deletion polymorphism is associated with
immune activation. Proc Natl Acad Sci USA. 112:2841–2846. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sieuwerts AM, Willis S, Burns MB, Look MP,
Meijer-Van Gelder ME, Schlicker A, Heideman MR, Jacobs H, Wessels
L, Leyland-Jones B, et al: Elevated APOBEC3B correlates with poor
outcomes for estrogen-receptor positive breast cancers. Horm
Cancer. 5:405–413. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsuboi M, Yamane A, Horiguchi J, Yokobori
T, Kawabata-Iwakawa R, Yoshiyama S, Rokudai S, Odawara H, Tokiniwa
H, Oyama T, et al: APOBEC3B high expression status is associated
with aggressive phenotype in Japanese breast cancers. Breast
Cancer. 23:780–788. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Law EK, Sieuwerts AM, LaPara K, Leonard B,
Starrett GJ, Molan AM, Temiz NA, Vogel RI, Meijer-van Gelder ME,
Sweep FC, et al: The DNA cytosine deaminase APOBEC3B promotes
tamoxifen resistance in ER-positive breast cancer. Sci Adv.
2:e16017372016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rüder U, Denkert C, Kunze CA, Jank P,
Lindner J, Jöhrens K, Kulbe H, Sehouli J, Dietel M, Braicu E and
Darb-Esfahani S: APOBEC3B protein expression and mRNA analyses in
patients with high-grade serous ovarian carcinoma. Histol
Histopathol. 34:405–417. 2018.PubMed/NCBI
|
|
27
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC cancer staging manual. 7th. Springer
(ed.); New York: pp. 345–376. 2010
|
|
28
|
Chang W, Gao X, Han Y, Du Y, Liu Q, Wang
L, Tan X, Zhang Q, Liu Y, Zhu Y, et al: Gene expression
profling-derived immunohistochemistry signature with high
prognostic value in colorectal carcinoma. Gut. 63:1457–1467. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao XH, Liu QZ, Chang W, Xu XD, Du Y, Han
Y, Liu Y, Yu ZQ, Zuo ZG, Xing JJ, et al: Expression of ZNF148 in
diferent developing stages of colorectal cancer and its prognostic
value: A large Chinese study based on tissue microarray. Cancer.
119:2212–2222. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Salgado R, Denkert C, Demaria S, Sirtaine
N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL,
Penault-Llorca F, et al: The evaluation of tumor-infiltrating
lymphocytes (TILs) in breast cancer: Recommendations by an
International TILs Working Group 2014. Ann Oncol. 26:259–271. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li B, Severson E, Pignon JC, Zhao H, Li T,
Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al: Comprehensive
analyses of tumor immunity: Implications for cancer immunotherapy.
Genome Biol. 17:1742016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du Y, Tao X, Wu J, Yu H, Yu Y and Zhao H:
APOBEC3B up-regulation independently predicts ovarian cancer
prognosis: A cohort study. Cancer Cell Int. 18:782018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fujiki Y, Yamamoto Y, Sueta A,
Yamamoto-Ibusuki M, Goto-Yamaguchi L, Tomiguchi M, Takeshita T and
Iwase H: APOBEC3B gene expression as a novel predictive factor for
pathological complete response to neoadjuvant chemotherapy in
breast cancer. Oncotarget. 9:30513–30526. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mao Y, Qu Q, Chen X, Huang O, Wu J and
Shen K: The Prognostic value of tumor-infiltrating lymphocytes in
breast cancer: A Systematic review and meta-analysis. PLoS One.
11:e01525002016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mao Y, Qu Q, Zhang Y, Liu J, Chen X and
Shen K: The value of tumor infiltrating lymphocytes for predicting
response to neoadjuvant chemotherapy in breast cancer: A systemic
review and meta-analysis. PLoS One. 9:e01151032014. View Article : Google Scholar
|