Introduction
Renal cell carcinoma (RCC) accounts for
approximately 3% of all human malignancies and approximately 85% of
renal cancer (1). Clear cell renal
cell carcinoma (ccRCC) is responsible for approximately 80% of RCC
incidence and exhibits the highest mortality rate among all
urological malignancies (2,3). The 5-year survival rate of a major
percentage of patients diagnosed with metastatic ccRCC (>30%)
was below 20% (4). The successful
treatment for ccRCC, notably for metastatic ccRCC, is becoming
increasingly challenging for medical practitioners (5,6). The
discovery of novel tumor markers can improve the understanding,
diagnosis and treatment of ccRCC.
Gene associated with retinoid-interferon-induced
mortality-19 (GRIM-19) is a member of the GRIM family of proteins
(7,8). It is essential for the tumor cell death
induced by interferon-β and retinoic acid. As a subunit of
mitochondrial NADH: ubiquinone oxidoreductase (respiratory chain
complex I), GRIM-19 is critical for complex I assembly, activity
and mitochondrial membrane potential maintenance (9,10). Its
deficiency or mutation commonly increases the sensitivity of the
cells to their apoptotic stimuli (9,10).
GRIM-19 is well-known as a cell death regulatory gene widely
distributed in the cytoplasm, nucleus and mitochondria and has been
studied as a suppressor of various types of cancer (11–17).
GRIM-19 downregulation is associated with the development, invasion
and induction of apoptosis of lung cancer (11), glioma (12), cervical cancer (13), breast cancer (14), hepatocarcinoma (15,16) and
gastric cancer cells (17). GRIM-19
may be considered an important tumor marker.
The present study analyzed the mRNA and protein
expression levels and distribution of GRIM-19 in ccRCC tumor
tissues compared with those noted in the paracancerous non-tumor
tissues, and demonstrated that GRIM-19 deficiency promoted ccRCC
malignant progression by causing an increase in the tumor, lymph
nodes and metastasis (TNM) stage and the Fuhrman grade of the
tumors.
Materials and methods
Specimen collection from ccRCC
patients
A total of 93 paired-matched tumor and paracancerous
non-tumor renal tissues were obtained from ccRCC patients during
the period June 2015 and October 2016 at the Second Affiliated
Hospital of the Dalian Medical University, in Dalian, Liaoning,
China. Non-tumor renal tissues were obtained from a distance higher
than and/or equal to 5 (≥5) cm away from the edge of the resected
tumor. None of the patients received radiotherapy or chemotherapy
prior to surgery. The specimens were classified by the patient
clinicopathological parameters including age, gender, tumor
position, TNM stage and Fuhrman grade. A total of 74 and 19 paired
tissues were from male and female patients, whereas 43 and 50
paired tissues were from patients higher than 60 (>60) and lower
than and/or equal to 60 (≤60) years, respectively. A total of 51
and 42 paired tissues were obtained from the right and left side
kidneys of the patients, respectively. A total of 54, 16, 22 and 1
tumors were of T1, T2, T3 and T4 stage, respectively, whereas 53
and 40 tumors were of Fuhrman grade ≤2 and Fuhrman grade >2. The
histological subtypes and tumor stages were assessed by the 2016
WHO Classification of Tumors of the Urinary System of the Tumor,
Nodes and Metastasis system (18). A
total of 38 cases of paired surgical tissues were snap-frozen in
liquid nitrogen for RT-qPCR and western blotting analyses, whereas
55 paired tissues were fixed in 10% neutral-buffered formalin for
24 h at room temperature (RT) and embedded in paraffin for the
immunohistochemical (IHC) assay.
RT-qPCR assay
A piece of tissue (~50 mg) from each patient was cut
into 50 µm slices by a freezing microtome. Total RNA was extracted
using TRIzol reagent (Transgen Biotech Co., Ltd.) and reversely
transcribed into cDNA using EasyScript One-Step gDNA Removal and
cDNA Synthesis SuperMix kit (Transgen Biotech Co., Ltd.). RT-qPCR
was performed using Quantitect SYBR Green PCR kit (Transgen Biotech
Co., Ltd.) in a StepOnePlus PCR system (Thermo Scientific, Inc.).
The following conditions were used: Initial denaturation at 94°C
for 30 sec, followed by 45 cycles at 94°C for 5 sec, 60°C for 30
sec, 95°C for 15 sec, 60°C for 60 sec, 95°C for 30 sec and 60°C for
30 sec. GAPDH was used as the internal reference gene. The primer
sequences for GRIM-19 were the following: F:
5′-ACCGGAAGTGTGGGATACTG-3′, R: 5′-GCTCACGGTTCCACTTCATT-3′ and for
GAPDH F: 5′-AGAAGGCTGGGGCTCATTTG-3′, R:
5′-AGGGGCCATCCACAGTCTTC-3′. The 2−ΔΔCT method was used
for quantification (19).
GRIM-19 level in the tumorous tissue from one ccRCC patient
was strictly compared with and normalized against its level that
was defined as 1 (100%) in paired paracancerous normal tissue from
same patient. The amplification cycle times of the internal
reference in paired tumor and paracancerous tissues were separately
used for calculating the relative differential cycle time (ΔCT) and
relative differential expression level change (ΔΔCT) of
GRIM-19. The overall difference of GRIM-19 in ccRCC
patients' tumorous tissues with P below 0.05 was considered
statistically significant.
Western blotting
Approximately 50 mg of tissue from each patient was
washed with PBS, grounded in liquid nitrogen in a mortar covered
with aluminum foil into powder and suspended in 450 µl of ice-cold
RIPA solution. The tissue mixture was grounded using a pestle and
suspended thrice for 10 min each time on ice. The total protein was
collected from the supernatant by centrifugation at 12,879 × g at
4°C for 15 min. The protein concentration was determined by the
Bradford assay. Equal amounts of protein from each group were
boiled for 5 min and separated by PAGE using 10% SDS gels. The
protein bands were transferred onto nitrocellulose membranes (PALL
Co., Ltd.), blocked in 5% (w/v) skimmed milk (Becton, Dickinson) in
TBST for 3 h at RT and incubated with primary antibodies against
GRIM-19 (1:1,000; 10986-1-AP; Proteintech Group, Inc.) and GAPDH
(1:2,000; 10494-1-AP; Proteintech Group, Inc.) at 4°C overnight.
The membranes were washed with TBST thrice for 10 min each time,
incubated with the secondary antibody IgG (1:2,000; SA00001-2;
Proteintech Group, Inc.) for 3 h at RT, and washed with TBST for an
additional three times (10 min each time). The protein bands were
visualized by ECL (K-12045-D50; Advansta) and analyzed by the
Bio-Rad ChemiDoc™ MP system (Bio-Rad, Laboratories, Inc.). GRIM-19
protein expression level in the tumorous tissue from one ccRCC
patient was strictly compared with and normalized against its level
that was defined as 1 (100%) in paired paracancerous normal tissue
from same patient. Its level change with P-value less than 0.05 was
regarded as statistically significant.
IHC assay. Paraffin-embedded tumor and
non-tumor renal tissues were cut into 4-µm slices
The slides were boiled in citrate buffer in a
micro-wave oven with high power for 4 min and cooled down at RT.
This step was repeated four times. Following washing with PBS three
times for 5 min each time, the slices were blocked in 3%
H2O2 for 20 min and in 10% non-immune goat
serum (ZSGB-BIO, Co., China) for 15 min at RT. The samples were
incubated with GRIM-19 antibody (1:200) at 4°C overnight, placed at
37°C for 30 min and treated with biotin-streptavidin HRP detection
kit (ZSGB-BIO, Co.). The images were developed using
3,3′-diamino-benzidine kit (ZSGB-BIO, Co.) under an upright light
BX3-CBH microscope (Olympus, Co.).
The classification of the IHC staining was assessed
by the immunoreaction intensity and the DAB positive staining
quantity of the tumor cells (20).
The immunoreaction intensity was classified as 0 (negative), 1
(weak), 2 (moderate) and 3 (strong). The DAB staining quantity of
each sample was rated as 0 (none), 1 (1-10% positive cells/field),
2 (10-50%), 3 (51-75%) and 4 (>76%). Their multiplication scores
ranged from 0–2, 3–5, 6–8 and 9–12, and were considered as negative
(−), weak (+), moderate (++) and strong (+++) with regard to
GRIM-19 expression levels, respectively.
Data processing and statistical
analysis
Experimental data were expressed as mean ± standard
deviation. The data were processed by SPSS 17.0 Software (SPSS,
Inc.). The differential expression between tumor and paired
non-tumor tissues was analyzed by the paired t-test and the one-way
ANOVA (and nonparametric) analysis with the post hoc Brown-Forsythe
test. The expression levels of GRIM-19 were compared with the
patient clinicopathological parameters by the χ2 or
Fisher's exact tests. Significant differences were considered for
P<0.05.
Results
GRIM-19 mRNA downregulation is
associated with ccRCC clinical progression
The alterations in GRIM-19 mRNA levels were
evaluated in paired tumor and paracancerous non-tumor tissues from
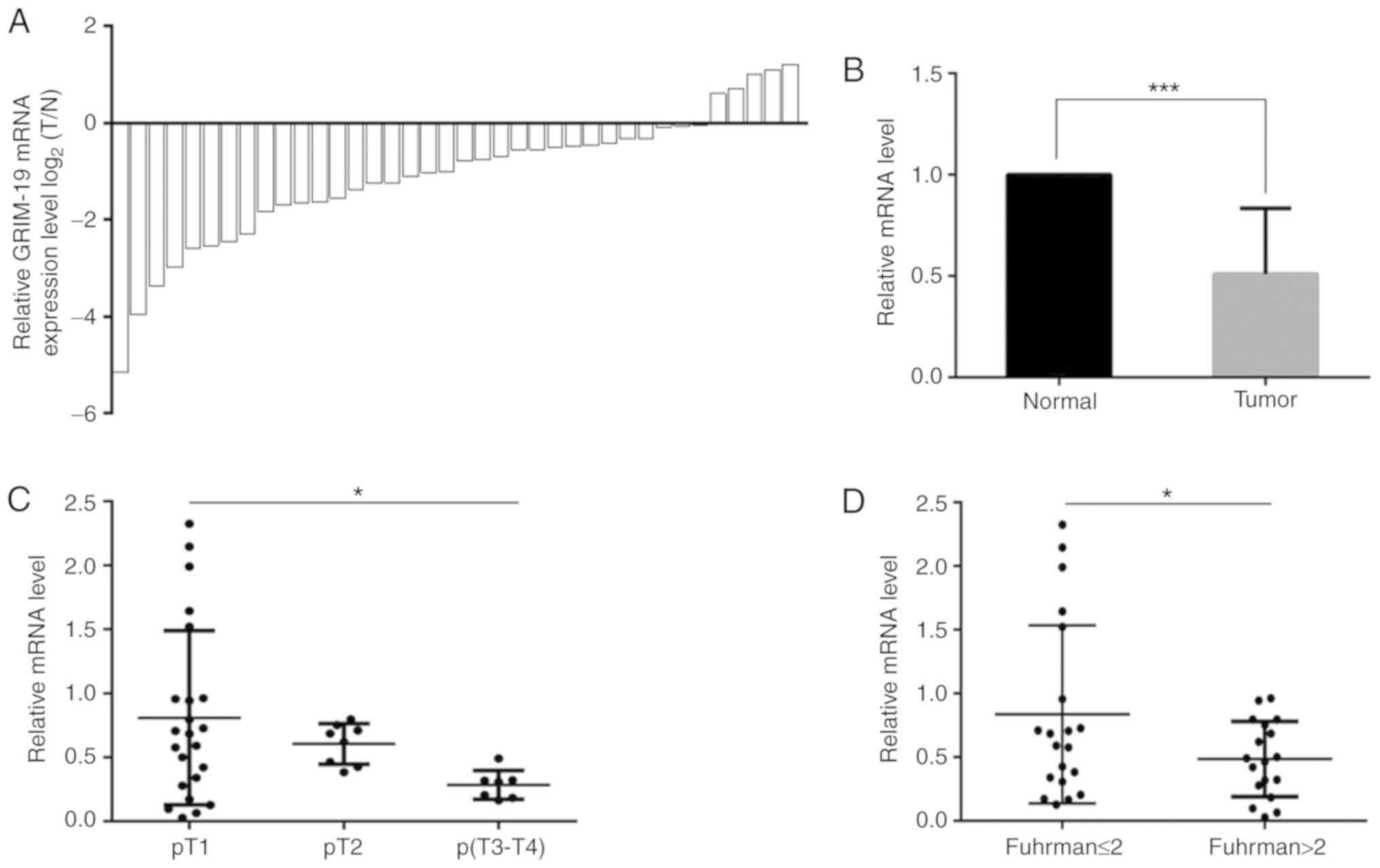
38 patients by RT-qPCR. GRIM-19 was downregulated in the
majority (33/38) of the tumor tissues (Fig. 1A) with an overall decrease of 49.1%
(P=0.0007, Fig. 1B). GRIM-19
downregulation was inversely associated with the T stage and the
Fuhrman grade of the tumors, while it was not associated with the
patient age (P=0.07), gender (P=0.3) and tumor location (tumor from
left or right kidney, P=0.1, Table
I). The GRIM-19 levels noted in the T2 and T3-T4 tumor
tissues were decreased by 25.2 and 64.8%, respectively compared
with those noted in the T1 stage samples (Fig. 1C, Table
I, P=0.02). GRIM-19 levels in tumors with Fuhrman grade
lower than and/or equal to 2 (≤2) were 72.4% higher than those of
tumors with Fuhrman grade higher than 2 (>2, Fig. 1D, Table
I, P=0.03). These findings suggested that GRIM-19 deficiency
promoted ccRCC progression.
 | Table I.Clinical association of GRIM-19 mRNA
and protein alterations with clear cell renal cell carcinoma. |
Table I.
Clinical association of GRIM-19 mRNA
and protein alterations with clear cell renal cell carcinoma.
|
|
| Level changes and
clinical significance |
|---|
|
|
|
|
|---|
| Parameter | Patients, n | Relative mRNA
levela | P-value | Relative protein
levela | P-value |
|---|
| Sex |
|
| 0.300 |
| 0.591 |
| Male | 31 | 0.715 |
| 0.231 |
|
|
Female | 7 | 0.466 |
| 0.119 |
|
| Age, years |
|
| 0.073 |
| 0.293 |
|
≤60 | 22 | 0.834 |
| 0.192 |
|
|
>60 | 16 | 0.443 |
| 0.116 |
|
| Tumor location |
|
| 0.129 |
| 0.541 |
| Left
kidney | 17 | 0.825 |
| 0.241 |
|
| Right
kidney | 21 | 0.543 |
| 0.119 |
|
| T stage |
|
| 0.023 |
| 0.003 |
|
pT1 | 23 | 0.809 |
| 0.200 |
|
|
pT2 | 7 | 0.605 |
| 0.089 |
|
|
pT3-4b | 8 | 0.285 |
| 0.033 |
|
| Fuhrman grade |
|
| 0.035 |
| 0.009 |
|
≤2c | 20 | 0.836 |
| 0.197 |
|
|
>2d | 18 | 0.485 |
| 0.086 |
|
GRIM-19 protein downregulation in
tumor tissues exhibits a negative correlation with ccRCC clinical
aggressiveness
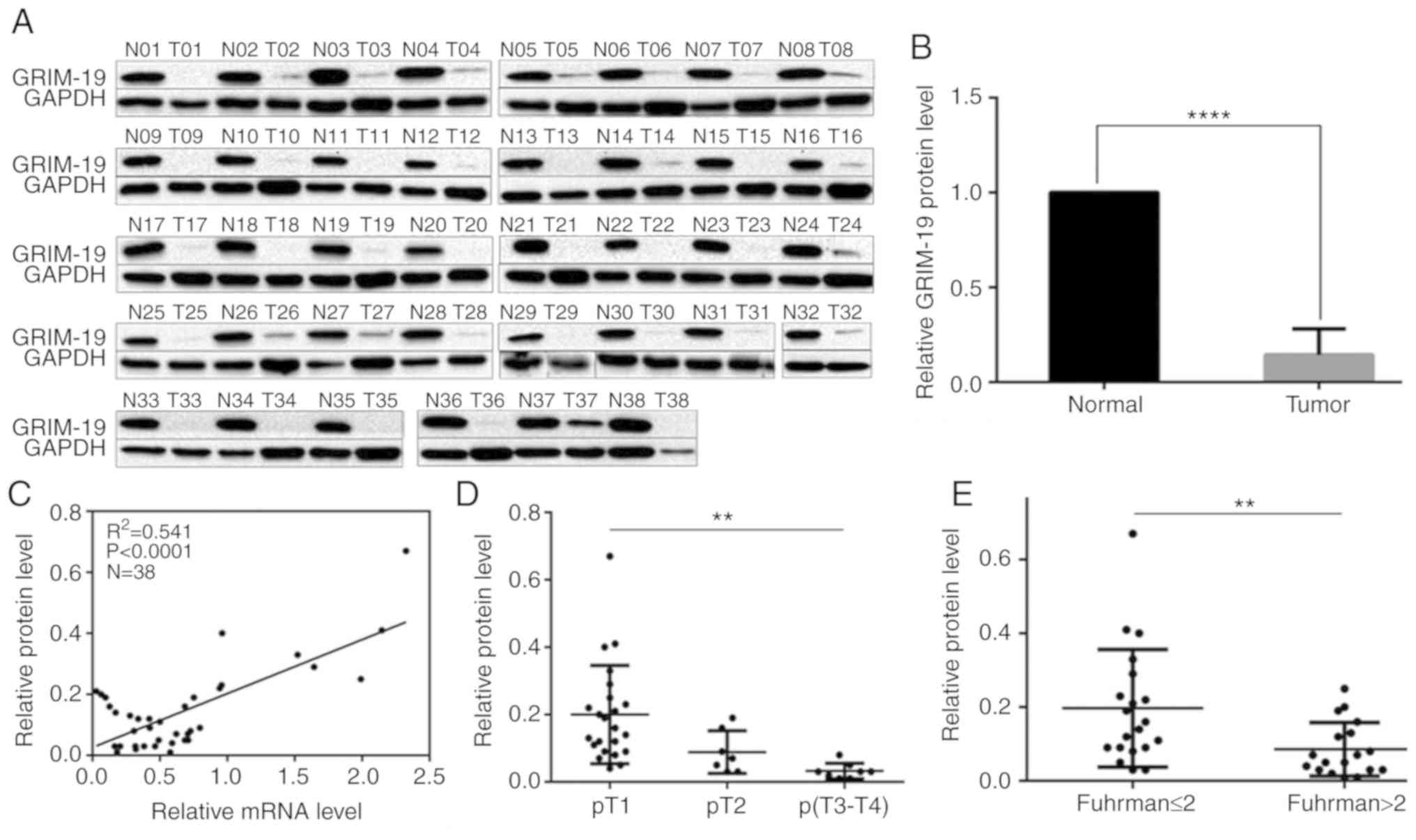
Western blotting indicated that GRIM-19 protein
levels were ubiquitously suppressed in each tumor tissue compared
with the corresponding paired non-tumor tissues (Fig. 2A). GRIM-19 levels in tumor tissues
increased from 1 to 67% compared with those in paired non-tumor
tissues (Fig 2B, P=0.0001). An
overall decrease of approximately 85.6% was noted (Fig. 2B, P=0.0001). A positive correlation
was noted between GRIM-19 protein and mRNA downregulation (Fig. 2C, P<0.0001). The downregulation
noted in the protein levels was inversely associated with the
increase in the tumor stage and in the Fuhrman grade (Table I). GRIM-19 protein levels were
decreased by 55.0 and 83.5% in T2 and T3-4 tumors, respectively
compared with the levels noted in T1 tumors (Fig. 2D, Table
I, P=0.003). GRIM-19 protein levels were decreased by 56.4% in
tumors with Fuhrman grade higher than 2 (>2) compared with those
of tumors with Fuhrman grade lower than and/or equal to 2 (≤ 2)
(Fig. 2E, Table I, P=0.009). This evidence was
consistent with the aforementioned findings. No significant
differences were noted with regard to the reduction in the GRIM-19
protein expression levels measured in tumor tissues from female and
male patients (48.5% reduction, Table
I, P=0.591). Similar findings were noted in the tissues from
patients higher and lower than, and/or equal to 60 years (>60 or
≤60 years) (39.6% lower, Table I,
P=0.293). Furthermore, no significant differences were noted with
regard to the reduction in the GRIM-19 protein expression levels in
tissues from patients with right and left kidneys (50.6% lower,
Table I, P=0.541). The
downregulation of the mRNA and protein levels of GRIM-19 was
concordantly associated with ccRCC aggressiveness.
The expression levels and protein
distribution of GRIM-19 are associated with ccRCC clinical
characteristics
GRIM-19 levels and protein localization in patient
tumor and paracancerous non-tumor tissues were analyzed by
immunohistochemistry. The expression of GRIM-19 was mainly present
in the cytoplasm (Fig. 3). GRIM-19
levels were highly elevated in normal kidney epithelial cells,
tubular epithelial cells and collecting duct epithelial cells
(Figs. 3-1A-B, 2A-B). This protein was not expressed in the
nuclei of kidney glomerular cells (Fig.
3). The following immunoreactivity scores were noted in 61.8%
(34/55), 27.3% (15/55), 5.45% (3/55) and 5.45% (3/55) of tumor
tissues: -, +, ++ and +++. The same immunoreactivity expression
pattern (−, +, ++ and +++) was noted in 0% (0/40), 25% (10/40), 35%
(14/40) and 40% (16/40) of paracancerous non-tumor tissues
(Table II, P<0.0001). GRIM-19
was undetected in 61.8% (34/55) of tumor tissues, while, it was
present in all (40/40) the paracancerous tissues. The percentage of
tumor tissues that exhibited ++ and +++ GRIM-19 immunoreactivity
was 10.9% (6/55), which was 64.1% lower than that of the
paracancerous non-tumor tissues (75%, 30/40). GRIM-19 expression
levels were downregulated or undetectable in tumor tissues compared
with those noted in paracancerous tissues (Figs. 3-1C-D, 2C-D, Figs.
3-1A-B, 2A-B). GRIM-19 levels
were decreased or undetectable in tissues that exhibited higher
tumor stage compared with the corresponding levels noted in low
stage tumor tissues (Figs. 3-1C-D,
3-2C-D). Accordingly, the structural
arrangements of the kidney tubules and collecting duct became loose
(Fig. 3-1C-D) or collapsed (Fig. 3-2C-D). Further analysis indicated
that GRIM-19 downregulation was inversely associated with the
increase in the tumor stage (P=0.013) and in the tumor Fuhrman
grade (P=0.026), while it was not associated with the patient
gender, age and tumor location (Table
II). These findings were consistent with the RT-qPCR and
Western blotting results.
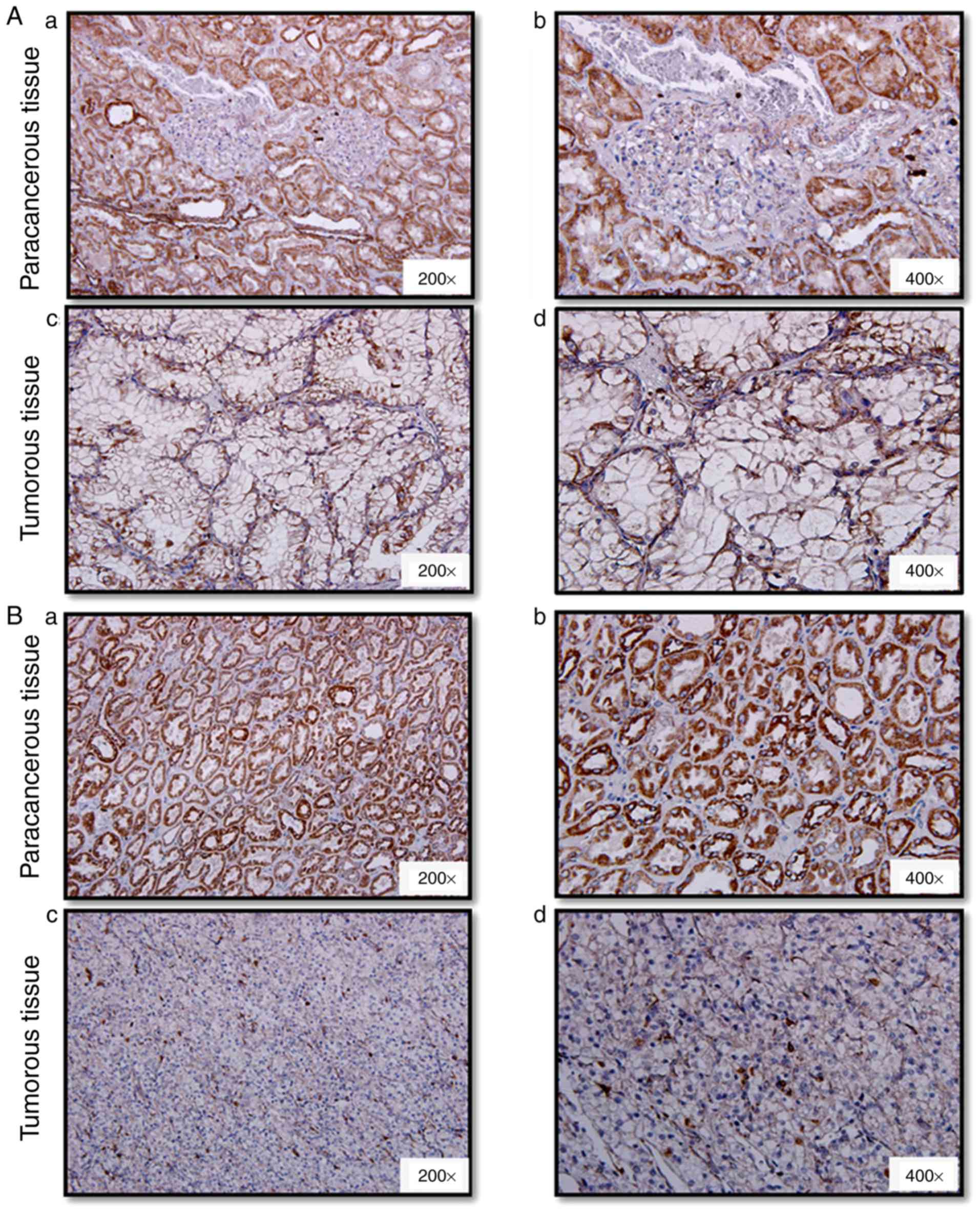 | Figure 3.Immunohistochemistry evaluation of
GRIM-19 levels and protein distribution in ccRCC tumor and
paracancerous tissues. Representative images of GRIM-19 expression
in the paracancerous region, (Aa and Ab) and the tumor region
(well-differentiated, Ac and Ad) from one tumor sample of Fuhrman
grade 2; Paracancerous tissues (Ba and Bb) and tumor tissues
(poorly differentiated, Bc and Bd) from one patient with Fuhrman
grade 3. The images corresponding to Aa, Ac, Ba and Bc, and to Ab,
Ad, Bb and Bd were obtained at ×200 and ×400 magnification,
respectively. GRIM-19, gene associated with
retinoid-interferon-induced mortality-19; ccRCC, clear cell renal
cell carcinoma. |
 | Table II.Immunohistochemistry assay of GRIM-19
association with clear cell renal cell carcinoma clinicopathology
parameters. |
Table II.
Immunohistochemistry assay of GRIM-19
association with clear cell renal cell carcinoma clinicopathology
parameters.
|
|
| GRIM-19 positive
immunoreactivity |
|---|
|
|
|
|
|---|
| Parameter | Specimen, n
(n=55) | Positive, n (%)
(n=21) | Negative, n (%)
(n=34) | P-value |
|---|
| Sex |
|
|
| 0.750 |
|
Male | 43 | 17 (81.0) | 26 (76.5) |
|
|
Female | 12 | 4
(19.0) | 8
(23.5) |
|
| Age, years |
|
|
| 0.269 |
|
≤60 | 28 | 13 (61.9) | 15 (44.1) |
|
|
>60 | 27 | 8
(38.1) | 19 (55.9) |
|
| Tumor position |
|
|
| 0.610 |
| Left
kidney | 34 | 13 (61.9) | 21 (61.8) |
|
| Right
kidney | 21 | 8
(38.1) | 13 (38.2) |
|
| T stage |
|
|
| 0.013 |
|
T1-T2 | 40 | 19 (90.5) | 21 (61.8) |
|
| T3 | 15 | 2 (9.5) | 13 (38.2) |
|
| Fuhrman grade |
|
|
| 0.026 |
| ≤2 | 33 | 17 (81.0) | 16 (47.1) |
|
|
>2 | 22 | 4
(19.0) | 18 (52.9) |
|
In conclusion, the present study explored the
dysregulation in GRIM-19 expression levels with the clinical
development and progression of ccRCC by RT-qPCR, Western blotting
and IHC assays. GRIM-19 acted as a tumor suppressor in ccRCC. Its
deficiency was associated with an increase in the tumor stage and
in the Fuhrman tumor grade of ccRCC.
Discussion
The contribution of GRIM-19 in the regulation of
tumor growth has been recently recognized. GRIM-19 is commonly
regarded as a tumor suppressor in certain cancer types. The
suppression of GRIM-19 expression promoted lung cancer metastasis
(11) and enhanced glioma cell
proliferation and migration (12).
Accordingly, GRIM-19 overexpression suppressed cell proliferation
of cervical cancer, breast cancer, glioma, hepatocarcinoma and
gastric cancer by inducing the apoptotic cascade (12–17).
This protein may be considered an important tumor marker. Proteomic
analysis indicated that GRIM-19 was downregulated in RCC (21). A recent study highlighted that
GRIM-19 was downregulated in RCC tissues and that it could inhibit
tumor growth by decreasing cell proliferation and inducing
apoptosis (22). The present study
was specifically designed to explore the contribution of GRIM-19
deficiency in the clinical progression of ccRCC, which is
considered the most severe type of RCC.
The results suggested that GRIM-19 mRNA and protein
levels were lower in tumor tissues from ccRCC patients compared
with the levels noted in the paired paracancerous non-tumor renal
tissues. The findings were similar to those noted from other
reports that confirmed that the expression levels of GRIM-19 were
reduced or totally suppressed in a number of human tumor types,
such as lung cancer, cervical cancer, hepatocarcinoma, gastric
cancer and glioma. GRIM-19 deficiency was associated with poor
differentiation, lymph node metastasis and vascular invasion of
colorectal cancer (23). Moreover,
GRIM-19 deficiency enhanced the progression of lung cancer
(11) and glioma (12). Its downregulation led to high
clinical stage, increased volume of ascites and increased size of
primary tumor lesion, while it was also associated with reduced
survival rate of epithelial ovarian carcinoma patients (24). Downregulation of GRIM-19 closely
correlated with high histological grade in HCC. The association of
GRIM-19 levels with the clinicopathological features of ccRCC is
unclear. The present study demonstrated that GRIM-19 deficiency was
associated with high TNM stage and Fuhrman grade of ccRCC
tissues.
GRIM-19 expression is differentially distributed in
various organelles depending on the tumor cell and tissue type. It
was expressed mainly in the nucleus of HeLa cells (13) and in the cytoplasm of hepatocarcinoma
cells (15). Fan et al
(11) demonstrated that GRIM-19 was
primarily localized in the cytoplasm of inflamed lung tissues,
whereas it demonstrated nuclear distribution in the nuclei of lung
cancer cells. GRIM-19 was only detected in the cytoplasm of
colorectal cancer cells compared with its expression in the
corresponding normal cells, where it was localized in both
cytoplasmic and nuclear regions (23). The present study indicated that
GRIM-19 was highly expressed in normal kidney epithelial, renal
tubular epithelial and collecting duct epithelial cells, while it
was not expressed in the nuclei and glomerular cells of
paracancerous tissues. GRIM-19 expression levels were undetectable
in tumor tissues compared with those noted in paracancerous
tissues. The loss or reduction of GRIM-19 was more evident in tumor
tissues of higher stage than tumor tissues of low stage, which
resulted in enhanced structural loss or collapse of the kidney
tubules and the collecting duct.
Signal transducer and activator of transcription 3
(STAT3) is a latent cytoplasmic transcription factor that plays
important roles in cell growth, reduction of apoptosis and cell
transformation. Accumulating evidence has suggested that GRIM-19 is
a negative regulator of STAT3, repressing STAT3
transcriptional activity and its downstream target gene expression.
In human cervical cancer, the restoration of GRIM-19 levels
reestablishes the control over STAT3-dependent gene expression and
affects tumor growth in vivo (13). The downregulation of GRIM-19
expression correlates with hyperactivation of STAT3 in HCC
(15). GRIM-19 expression sensitizes
BGC-803 gastric cancer cells to radiation possibly via the
suppression of STAT3 accumulation (17). Overexpression of GRIM-19 is
associated with hyperactivation of STAT3-induced gene expression in
renal carcinoma (22).
Downregulation of GRIM-19 correlated with STAT3 overexpression in
tumor samples from breast cancer patients (25). This finding provides additional
evidence for unraveling the exact mechanism of action of GRIM-19 in
ccRCC.
In summary, the present study suggests that GRIM-19
acts as a tumor suppressor in ccRCC. Its deficiency promotes ccRCC
aggressiveness. The overall GRIM-19 levels were dramatically
decreased and exerted an effect on the relaxed and/or collapsed
structures of the kidney tubules and the collecting duct in the
tumor tissues from ccRCC patients. GRIM-19 deficiency is a
potential indicator for ccRCC malignant transformation.
In future study on GRIM-19, we will follow up on the
lifespan of the patients. and consider for getting the tissue array
of ccRCC patients with survival times and rates. At the same time,
we will investigate the association of GRIM-19 deficiency with the
malignant progression, especially, the survival time for ccRCC
patients. Surely enough, more investigations are worth doing for
validating and confirming the role of GRIM-19 as a reliable
biomarker, surgical and therapeutic target in clinical significance
for ccRCC.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from National
Natural Science Foundation of China (grant nos. 81672737 and
81272186).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NY and XF performed the experiments and wrote
manuscript draft. SJ collected the specimens and extracted proteins
from specimens. WS designed the experiment and validated
clinicopathological diagnosis of patients. MZS and SL designed and
supervised the study and corrected the manuscript.
Ethics approval and consent to
participate
Tissue use and the study protocol were approved by
the Medical Ethics Committee of Dalian Medical University, and
granted by the informed consent from all patients.
Patient consent for publication
Written informed consents were obtained from all
individual participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barata PC and Rini BI: Treatment of renal
cell carcinoma: Current status and future directions. CA Cancer J
Clin. 67:507–524. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Meerleer G, Khoo V, Escudier B, Joniau
S, Bossi A, Ost P, Briganti A, Fonteyne V, Van Vulpen M, Lumen N,
et al: Radiotherapy for renal-cell carcinoma. Lancet Oncol.
15:e170–e177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oosterwijk E, Rathmell WK, Junker K,
Brannon AR, Pouliot F, Finley DS, Mulders PF, Kirkali Z, Uemura H
and Belldegrun A: Basic research in kidney cancer. Eur Urol.
60:622–633. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ficarra V, Guillè F, Schips L, de la
Taille A, Prayer Galetti T, Tostain J, Cindolo L, Novara G,
Zigeuner R, Bratti E, et al: Proposal for revision of the TNM
classification system for renal cell carcinoma. Cancer.
104:2116–2123. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan M, Zhang L, Wu Y, Gao L, Yang W, Li J,
Chen Y and Jin X: Increased expression of kindlin-2 is correlated
with hematogenous metastasis and poor prognosis in patients with
clear cell renal cell carcinoma. FEBS Open Bio. 6:660–665. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soultati A, Stares M, Swanton C, Larkin J
and Turajlic S: How should clinicians address intratumour
heterogeneity in clear cell renal cell carcinoma? Curr Opin Urol.
25:358–366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kalvakolanu DV: The GRIMs: A new interface
between cell death regulation and interferon/retinoid induced
growth suppression. Cytokine Growth Factor Rev. 15:169–194. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chidambaram NV, Angell JE, Ling W, Hofmann
ER and Kalvakolanu DV: Chromosomal localization of human GRIM-19, a
novel IFN-beta and retinoic acid-activated regulator of cell death.
J Interferon Cytokine Res. 20:661–665. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu H and Cao X: GRIM-19 is essential for
maintenance of mitochondrial membrane potential. Mol Biol Cell.
19:1893–1902. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fearnley IM, Carroll J, Shannon RJ,
Runswick MJ, Walker JE and Hirst J: GRIM-19, a cell death
regulatory gene product, is a subunit of bovine mitochondrial
NADH:ubiquinone oxidoreductase (complex I). J Biol Chem.
276:38345–38348. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan XY, Jiang ZF, Cai L and Liu RY:
Expression and clinical significance of GRIM-19 in lung cancer. Med
Oncol. 29:3183–3189. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Hao H, Zhao S, Liu Q, Yuan Q, Ni
S, Wang F, Liu S, Wang L and Hao A: Downregulation of GRIM-19
promotes growth and migration of human glioma cells. Cancer Sci.
102:1991–1999. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou Y, Li M, Wei Y, Feng D, Peng C, Weng
H, Ma Y, Bao L, Nallar S, Kalakonda S, et al: Down-regulation of
GRIM-19 expression is associated with hyperactivation of
STAT3-induced gene expression and tumor growth in human cervical
cancers. J Interferon Cytokine Res. 29:695–703. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Angell JE, Lindner DJ, Shapiro PS, Hofmann
ER and Kalvakolanu DV: Identification of GRIM-19, a novel cell
death-regulatory gene induced by the interferon-beta and retinoic
acid combination, using a genetic approach. J Biol Chem.
275:33416–33426. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li F, Ren W, Zhao Y, Fu Z, Ji Y, Zhu Y and
Qin C: Downregulation of GRIM-19 is associated with hyperactivation
of p-STAT3 in hepatocellular carcinoma. Med Oncol. 29:3046–3054.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kong D, Zhao L, Du Y, He P, Zou Y, Yang L,
Sun L, Wang H, Xu D, Meng X, et al: Overexpression of GRIM-19, a
mitochondrial respiratory chain complex I protein, suppresses
hepatocellular carcinoma growth. Int J Clin Exp Pathol.
7:7497–7507. 2014.PubMed/NCBI
|
|
17
|
Bu X, Zhao C, Wang W and Zhang N: GRIM-19
inhibits the STAT3 signaling pathway and sensitizes gastric cancer
cells to radiation. Gene. 512:198–205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bezhanova SD: Tumors of the kidney. The
new 2016 WHO classification of tumors of the genitourinary system.
Arkh Patol. 79:48–52. 2017.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu L and Yang W: Criteria for judging the
results of immunohistochemistry. Chin Oncol. 6:31996.
|
|
21
|
Alchanati I, Nallar SC, Sun P, Gao L, Hu
J, Stein A, Yakirevich E, Konforty D, Alroy I, Zhao X, et al: A
proteomic analysis reveals the loss of expression of the cell death
regulatory gene GRIM-19 in human renal cell carcinomas. Oncogene.
25:7138–7147. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui L, Meng Q, Wen J, Yan Z, Gao Z, Tian
Y, Xu P, Lian P and Yu H: The effect of a gene associated with
retinoid-interferon-induced mortality 19 (GRIM-19) on STAT3-induced
gene expression in renal carcinoma. J Biochem. 164:285–294. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hao M, Shu Z, Sun H, Sun R, Wang Y, Liu T,
Ji D and Cong X: GRIM-19 expression is a potent prognostic marker
in colorectal cancer. Hum Pathol. 46:1815–1820. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shao YP, Cheng XD and Wan XY: Expression
and clinical significance of GRIM-19 in epithelial ovarian
carcinoma. Zhonghua Fu Chan Ke Za Zhi. 47:751–755. 2012.(In
Chinese). PubMed/NCBI
|
|
25
|
Zhou T, Chao L, Rong G, Wang C, Ma R and
Wang X: Down-regulation of GRIM-19 is associated with STAT3
overexpression in breast carcinomas. Hum Pathol. 44:1773–1779.
2013. View Article : Google Scholar : PubMed/NCBI
|