Introduction
Hepatocellular carcinoma (HCC), representing 75–85%
of primary hepatic carcinoma, was the fourth most frequent cause of
cancer-associated mortality worldwide in 2018 (1). There were ~854,000 new liver cancer
cases and ~810,000 deaths globally in 2015 (2), with nearly equal rates of incidence and
mortality. Although, great advances have been made in surgical and
non-surgical treatments in the last decades, the outcome of HCC
remains poor (3) and >70% of
patients experience recurrence within 5 years (4,5). The
poor prognosis of HCC is primarily ascribed to delayed diagnosis
(6).
The causes of HCC occurrence are complex and involve
the accumulation of genetic and epigenetic changes (6). Cellular events are often accompanied by
upregulated expression of multiple factors (such as oncogenes) that
influence the survival of tumor cells by inhibiting apoptosis and
regulating the cell cycle (6).
Cytoskeleton-associated membrane protein 4 (CKAP4;
also referred to as P63), is a 63-kDa type II transmembrane protein
located in the endoplasmic reticulum (7,8). CKAP-4
plays a vital role in maintaining the structure of the endoplasmic
reticulum (8–10). Other studies have identified CKAP4 as
a cell surface receptor for surfactant protein A, antiproliferative
factor and Dickkopf 1 (DKK1) (11–14),
which are associated with tumor growth in cervical and bladder
cancer (12,13). As a novel DKK1 receptor that
stimulates tumor cell proliferation, CKAP4 is frequently
upregulated in pancreatic cancer, lung cancer and esophageal
squamous cell carcinoma (ESCC), and blocking CKAP4 activity via
knockdown or anti-CKAP4 antibodies can prevent xenograft tumor
formation (14–16). CKAP4 has also been demonstrated to be
upregulated in chronic lymphocytic leukemia (CLL) (17). Recently, exosomal CKAP4 was reported
to be a promising biomarker for pancreatic ductal adenocarcinoma
(PDAC) (18).
In 2013, Li et al (19) first revealed that CKAP4 was
associated with progression and metastasis in patients with HCC.
CKAP4 expression was demonstrated to be significantly higher in HCC
tissues compared with adjacent normal tissues, and was associated
with tumor size, intrahepatic metastasis, portal venous invasion
and tumor stage; furthermore, upregulated CKAP4 expression
increased overall survival (OS) and disease-free survival (DFS)
times in patients with HCC. Other studies have revealed that high
CKAP4 expression in HCC cells is associated with low proliferative
capacity and invasion potential via the suppression of epithelial
growth factor receptor (EGFR) signaling (20), which is involved in HCC occurrence
and development (6). The serum CKAP4
concentration in patients with HCC was significantly higher
compared with patients with chronic hepatitis B and cirrhosis, as
well as healthy controls, which suggests that CKAP4 is a promising
indicator for HCC diagnosis, especially when combined with
α-fetoprotein (AFP) (21). In
addition, upregulated CKAP4 expression was observed in HCC in a
proteomic study using isobaric tags for relative and absolute
quantification (22).
The above studies present contradictory results;
upregulated CKAP4 expression in HCC is reported to be associated
with low growth and favorable prognosis, but in other cancers,
CKAP4 expression appears to promote tumor growth and result in
worse prognosis. Additionally, an immunohistochemical study
comparing CKAP4 protein expression in cholangiocarcinoma and HCC
tissues demonstrated no CKAP4 protein expression in HCC (23).
In the present study, the association between CKAP4
mRNA expression and HCC was investigated using four CKAP4 datasets
of cohorts of patients with HCC from the Gene Expression Omnibus
(GEO) and The Cancer Genome Atlas (TCGA) databases. The data
obtained were used to determine the CKAP4 mRNA expression levels in
HCC tumor tissues and adjacent liver tissues, and their diagnostic
potential. Associations between CKAP4 mRNA expression and
clinicopathological characteristics and the prognosis of patients
with HCC were then analyzed.
Materials and methods
Tissue sample data sources
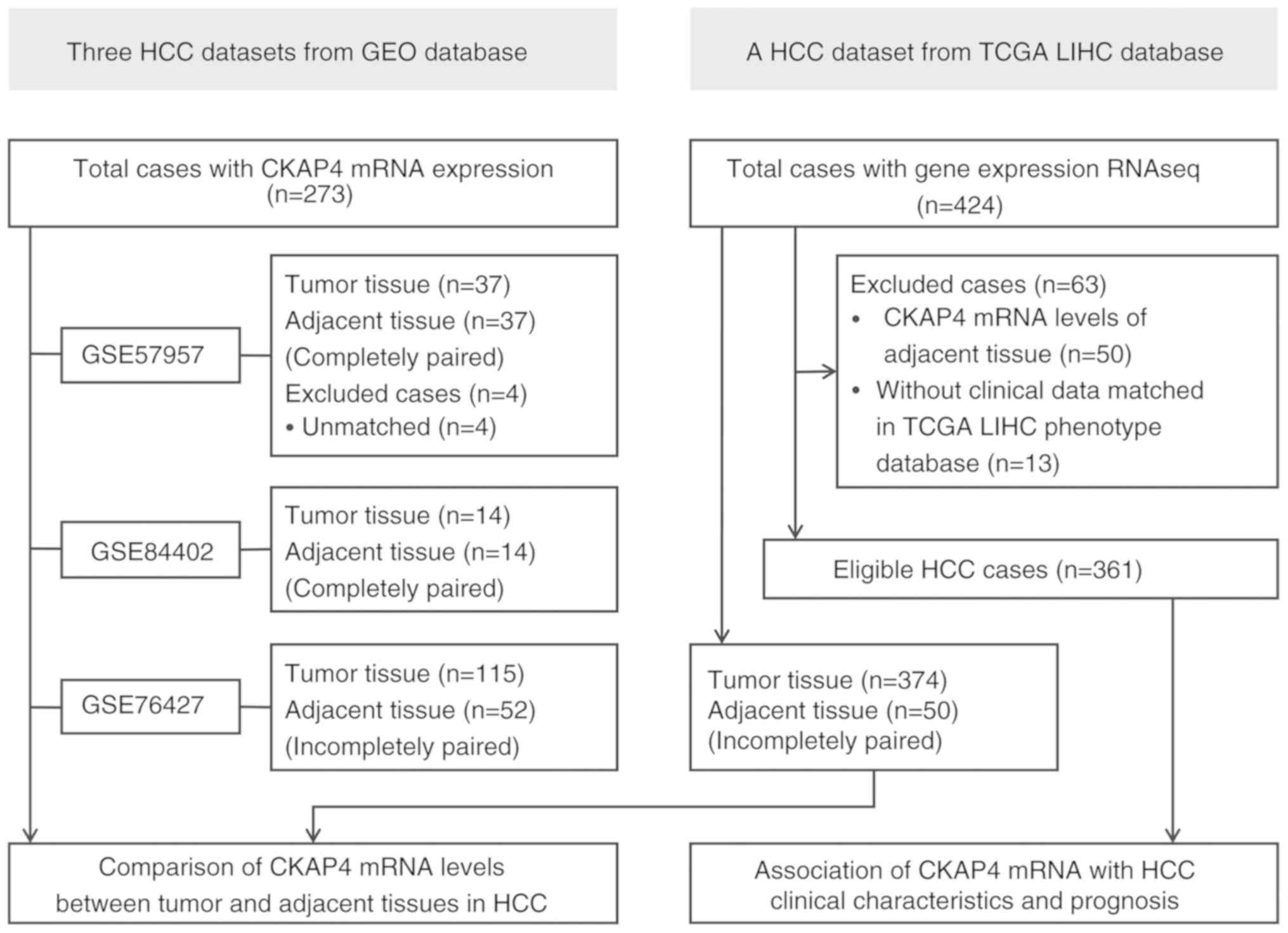
Three gene expression profile datasets of HCC tumor
and adjacent tissues, GSE76427 (24), GSE84402 (25) and GSE57957 (26), were downloaded from the GEO database
(https://www.ncbi.nlm.nih.gov/) (Fig. 1) and analyzed using software packages
in RStudio 3.6.1 version (https://rstudio.com/). The packages ‘illuminaHumanv4
db’ (https://bioconductor.org/packages/illuminaHumanv4 db/)
and ‘hgu133plus2 db’ (https://bioconductor.org/packages/hgu133plus2
db/) were used for gene annotation. The aforementioned packages
were used to convert gene IDs into gene symbols for subsequent
analysis. The packages ‘dplyr’ (http://dplyr.tidyverse.org/) and ‘tibble’ (http://tibble.tidyverse.org/) were used to preprocess
the data and the package ‘limma’ (https://bioconductor.org/packages/limma/) was used to
determine whether the data required normalization. Log2
normalization was performed for the extracted data. Detailed
information regarding tissue samples in the three datasets is
presented in Fig. 1.
In addition, a liver cancer (LIHC) dataset with
RNAseq data was downloaded from TCGA database (https://portal.gdc.cancer.gov/) and consisted of
HCC tumor (n=374) and adjacent tissue samples (n=50). LIHC
phenotype data were obtained from the GDC TCGA-LIHC cohort
(https://gdc.xenahubs.net/), which
contained 469 cases. All the data were processed using software
packages in the R program, including ‘dplyr’, ‘tidyr’ (http://tidyr.tidyverse.org/), ‘DESeq2’ (https://bioconductor.org/packages/DESeq2/) and ‘edgeR’
(https://bioconductor.org/packages/edgeR/).
Log2(count+1) normalization of the CKAP4 mRNA expression
data was achieved with ‘DESeq2’ and ‘vst’. Finally, 361 HCC cases
with matched CKAP4 mRNA and clinical data were evaluated (Fig. 1).
Presentation of CKAP4 mRNA levels and
the diagnostic value of CKAP4 mRNA in patients with HCC
Differences in CKAP4 mRNA levels between tumor and
adjacent normal tissues in the four datasets are presented as
expression heatmaps and violin plots. The software package
‘pheatmap’ (https://CRAN.R-project.org/package=pheatmap) in the R
program was used to plot the heatmaps and the packages
‘ggstatsplot’ (https://indrajeetpatil.github.io/ggstatsplot/) and
‘ggpubr’ (https://rpkgs.datanovia.com/ggpubr/) were used to plot
the violin plots. The diagnostic value of the CKAP4 mRNA levels in
HCC was evaluated using a receiver operating characteristic (ROC)
curve.
Analysis of the association between
CKAP4 mRNA levels and clinicopathological characteristics of
patients with HCC
Using patient samples with complete clinical data
from TCGA-LIHC dataset, the association between CKAP4 mRNA levels
in tumor tissues and clinical characteristics was analyzed. The
patients were divided into high- and low-level CKAP4 mRNA groups
using the median CKAP4 mRNA level as the cut-off value, and
differences in clinical parameters were compared between the 2
groups. In addition, the ‘ggboxplot’ function in the ‘ggpubr’
package of the R program was used to present and compare
differences in CKAP4 mRNA expression based on clinical parameters,
demonstrating significant differences between the initial high- and
low-level CKAP4 mRNA groups.
Survival analyses associated with
CKAP4 mRNA levels in patients with HCC
Survival analyses were performed in the cohort of
361 patients with HCC from TCGA-LIHC dataset. In this analysis,
cases with a follow-up time of <30 days after curative surgical
resection were excluded. Univariate Cox regression analysis, was
used to evaluate the hazard ratios (HRs), confidence intervals (CI)
and P-values of CKAP4 mRNA and the clinicopathological parameters
for OS and DFS. However, multivariate Cox regression analysis was
not conducted owing to an insufficient number of cases with
complete data. Kaplan-Meier survival analysis of the predictive
power of CKAP4 mRNA levels for OS and DFS of patients with HCC
after hepatectomy was performed using the packages ‘survival’
(https://github.com/therneau/survival/) and ‘survminer’
(https://rpkgs.datanovia.com/survminer/) in R. CKAP4
mRNA expression levels were compared among patients with different
survival times and among patients with different recurrence times
after hepatectomy using the function ‘ggboxplot’ in the ‘ggpubr’
packages in R software.
Statistical analysis
Data are presented as the mean ± SD, median and
interquartile range (IQR), or constituent ratio, according to the
data type. The paired t-test (for groups with completely paired
tumor and adjacent samples), unpaired t-test (for independent
groups) and χ2 test were used to compare CKAP4 mRNA
levels between two groups and calculated using R. One-way ANOVA
followed by Bonferroni's post-hoc test was used for comparing
differences in the CKAP4 mRNA levels between multiple groups, and
calculated using SPSS Statistics software v.24.0 (IBM Corp.). Ridit
analysis and the Mann-Whitney U test were used to compare
differences in clinicopathological parameters between the low- and
high-level CKAP4 mRNA groups, and calculated using SPSS v.24.0.
Kaplan-Meier curves were generated and compared using the
Tarone-Ware test (for survival plots where crossover between the
groups is observed) or log-rank test in R. Univariate Cox
regression analysis was used to calculate HRs and their 95% CIs, as
well as the associated P values. A two-sided P<0.05 was
considered to indicate a statistically significant difference.
Results
CKAP4 mRNA levels are elevated in
tumor tissues compared with adjacent liver tissues in patients with
HCC
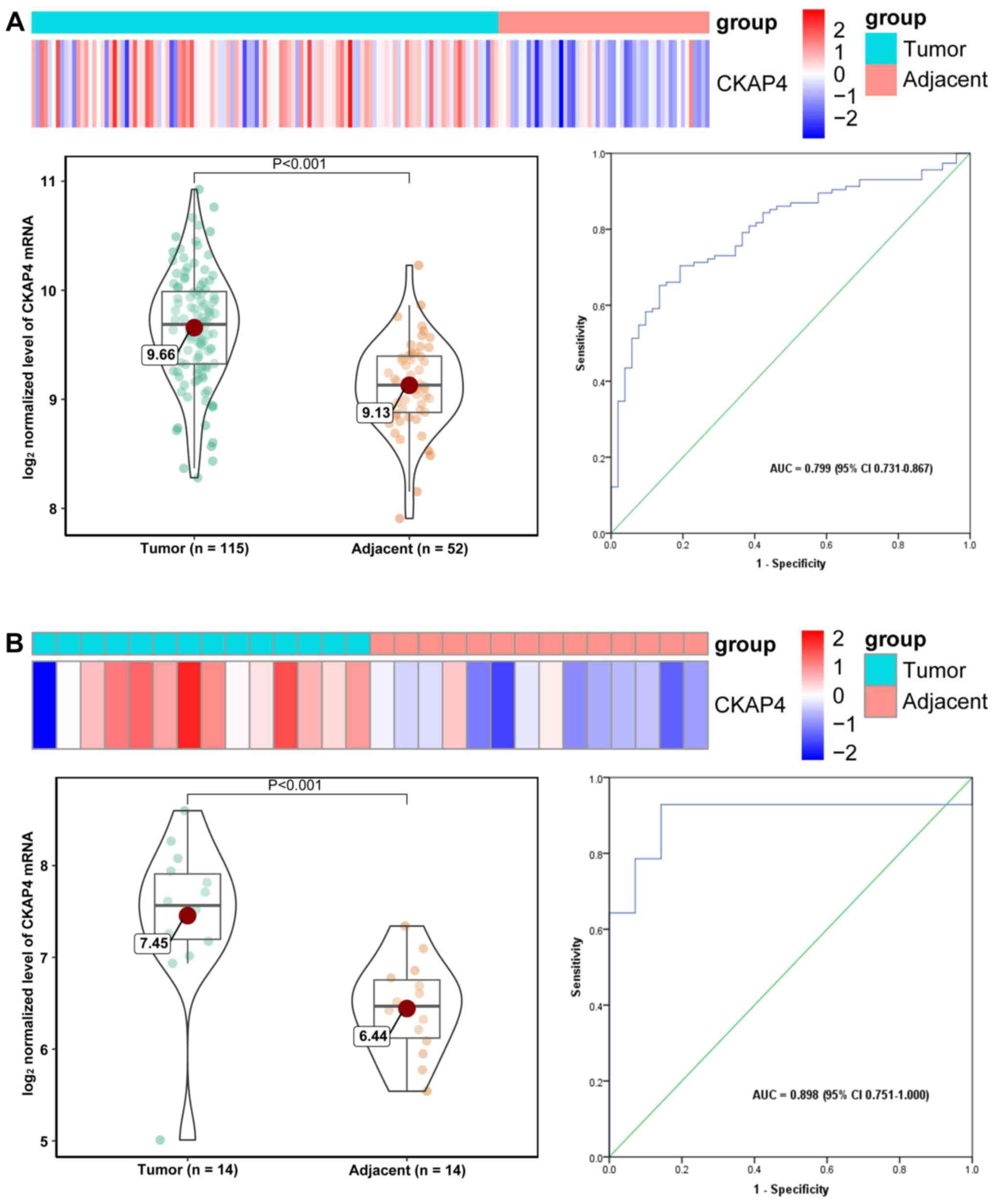
Heat maps and violin plots were used to intuitively
display CKAP4 mRNA expression in tumor and adjacent liver tissues,
and areas under ROC curves were used to evaluate the diagnostic
value for HCC. Heat maps of CKAP4 mRNA levels in all four datasets
demonstrate elevated (‘hot’) expression in tumor tissues and
reduced (‘cold’) expression in adjacent liver tissues (Fig. 2A-D). Compared with adjacent liver
tissues, tumor tissues from patients with HCC exhibit significantly
upregulated CKAP4 mRNA levels (following normalization);
furthermore, CKAP4 mRNA demonstrated moderate power for the
diagnosis of HCC in all of the datasets, with AUCs ranging from
0.799–0.898, indicating its potential diagnostic value in HCC
(Fig. 2A-D).
Association between CKAP4 mRNA levels
and clinical characteristics of patients with HCC
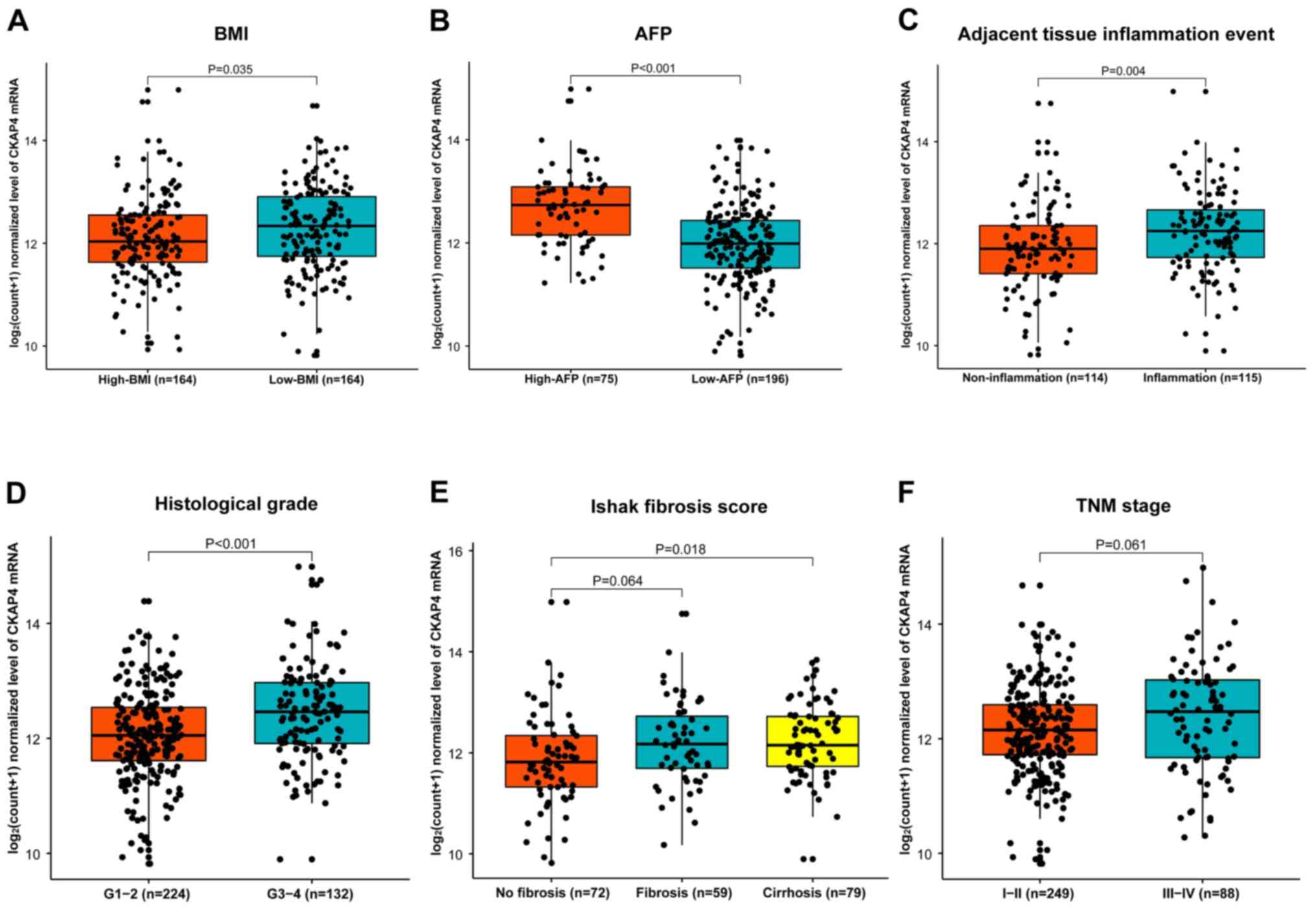
Clinicopathological and demographic characteristics
were obtained for the 361 patients with HCC from TCGA database, and
are summarized in Table I. Body mass
index (BMI) (P=0.005), AFP level (P<0.001), tumor histological
grade (P<0.001), adjacent liver tissue inflammation event
(P<0.001), Ishak fibrosis score (P=0.035), and American Joint
Committee on Cancer (AJCC) tumor node metastasis (TNM) stage
(P=0.014) were associated with CKAP4 mRNA levels, as they
significantly differed between the high- and low-level CKAP4 mRNA
groups (Table I). The CKAP4 mRNA
levels were further compared between subgroups for each of these
parameters (Fig. 3), and the results
demonstrated that CKAP4 mRNA levels in tumor tissues were
significantly different between the subgroups for all five
parameters, except for AJCC TNM stage (Fig. 3).
 | Table I.Differences in clinicopathological and
demographic characteristics between high- and low-level CKAP4 mRNA
groups in patients with HCC. |
Table I.
Differences in clinicopathological and
demographic characteristics between high- and low-level CKAP4 mRNA
groups in patients with HCC.
|
|
| CKAP4 mRNA group |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Patients, n | Low-level | High-level | P-value |
|---|
| Age, (years) n,
median (IQR) | 360 | 181, 62.0 | 179, 60.0 | 0.119 |
|
|
| (53.0–69.0) | (51.0–68.0) |
|
| Sex, n (%) | 361 |
|
| 0.085 |
|
Male | 244 | 130 (53.3) | 114 (46.7) |
|
|
Female | 117 | 51
(43.6) | 66
(56.4) |
|
| Ethnicity, n
(%) | 349 |
|
| 0.071 |
|
Asian | 156 | 68
(43.6) | 88
(56.4) |
|
|
White | 176 | 97
(55.1) | 79
(44.9) |
|
| Black
or African American | 17 | 8
(47.1) | 9
(52.9) |
|
| History of risk
factors, n (%) | 342 |
|
| 0.803 |
| Alcohol
consumption | 66 | 36
(54.5) | 30
(45.5) |
|
|
Hepatitis B | 94 | 41
(43.6) | 53
(56.4) |
|
|
Hepatitis C | 51 | 27
(52.9) | 24
(47.1) |
|
|
Non-alcoholic fatty liver
disease | 17 | 9
(52.9) | 8
(47.1) |
|
|
Other | 28 | 12
(42.9) | 16
(57.1) |
|
|
None | 86 | 46
(53.5) | 40
(46.5) |
|
| Family history of
cancer, n (%) | 312 |
|
| 0.196 |
| No | 203 | 98
(48.3) | 105 (51.7) |
|
|
Yes | 109 | 61
(56.0) | 48
(44.0) |
|
| Body mass index,
kg/m2, n, median (IQR) | 328 | 167, 25.3 | 161, 23.6 | 0.005 |
|
|
| (22.4–29.85) | (21.2–27.5) |
|
| α-fetoprotein,
ng/ml, n, median (IQR) | 271 | 7
(3–47) | 35 (8–2466) | <0.001 |
| α-fetoprotein,
ng/ml, n (%) | 271 |
|
| <0.001 |
|
<200 | 196 | 121 (61.7) | 75
(38.3) |
|
|
≥200 | 75 | 21
(28.0) | 54
(72.0) |
|
| Adjacent liver
tissue inflammation event, n (%) | 229 |
|
| <0.001 |
| No | 114 | 78
(68.4) | 36
(31.6) |
|
|
Yes | 115 | 52
(45.2) | 63
(54.8) |
|
| Tumor histological
grade, n (%) | 356 |
|
| <0.001 |
| G1 | 53 | 35
(66.0) | 18
(34.0) |
|
| G2 | 171 | 93
(54.4) | 78
(45.6) |
|
| G3 | 121 | 47
(38.8) | 74
(61.2) |
|
| G4 | 11 | 3
(27.3) | 8
(72.7) |
|
| Vascular invasion,
n (%) | 305 |
|
| 0.215 |
|
None | 200 | 108 (54.0) | 92
(46.0) |
|
|
Micro | 89 | 42
(47.2) | 47
(52.8) |
|
|
Macro | 16 | 7
(43.8) | 9
(56.2) |
|
| Ishak fibrosis
score, n (%) | 210 |
|
| 0.035 |
| 0, no
fibrosis | 72 | 50
(69.4) | 22
(30.6) |
|
| 1-4,
fibrosis | 59 | 29
(49.2) | 30
(50.8) |
|
| 5-6,
cirrhosis | 79 | 41
(51.9) | 38
(48.1) |
|
| AJCC TNM stage, n
(%) | 337 |
|
| 0.014 |
| I | 167 | 92
(55.1) | 75
(44.9) |
|
| II | 82 | 38
(46.3) | 44
(53.7) |
|
|
III | 84 | 35
(41.7) | 49
(58.3) |
|
| IV | 4 | 0
(0.0) | 4
(100.0) |
|
| Recurrence type, n
(%) | 152 |
|
| 0.400 |
|
Extrahepatic | 39 | 21 (53.8) | 18 (46.2) |
|
|
Intrahepatic | 64 | 32 (50.0) | 32 (50.0) |
|
|
Locoregional | 49 | 22 (44.9) | 27 (55.1) |
|
High CKAP4 mRNA levels are associated
with poor prognosis in patients with HCC
Using univariate Cox regression analysis,
associations between CKAP4 mRNA levels in tumor tissues and the
clinicopathological parameters considered as risk factors for the
survival of HCC patients were evaluated, and the results are
summarized in Table II. High CKAP4
mRNA levels, macrovascular invasion and TNM III–IV stage are risk
factors for both OS and DFS in patients with HCC (Table II). BMI was a protective factor for
DFS and hepatitis B was a protective factor for OS and DFS with
reference to alcohol consumption (Table
II).
 | Table II.Associations of CKAP4 mRNA level and
other metrics with OS and DFS in patients with HCC. |
Table II.
Associations of CKAP4 mRNA level and
other metrics with OS and DFS in patients with HCC.
|
| Cox regression for
OS | Cox regression for
DFS |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| CKAP4 mRNA |
|
|
|
|
|
Low-level | Reference |
| Reference |
|
|
High-level | 1.494
(1.044–2.138) | 0.028 | 1.616
(1.022–2.555) | 0.040 |
| Age, years | 1.007
(0.993–1.021) | 0.307 | 1.005
(0.978–1.023) | 0.595 |
| Sex |
|
|
|
|
|
Male | Reference |
| Reference |
|
|
Female | 0.810
(0.562–1.167) | 0.257 | 0.744
(0.471–1.177) | 0.207 |
| Race |
|
|
|
|
|
Asian | Reference |
| Reference |
|
|
White | 1.304
(0.885–1.919) | 0.179 | 0.995
(0.615–1.609) | 0.984 |
| Black
or African American | 1.365
(0.540–3.455) | 0.511 | NA |
|
| History of risk
factors |
|
|
|
|
| Alcohol
consumption | Reference |
| Reference |
|
|
Hepatitis B | 0.258
(0.136–0.487) | <0.001 | 0.471
(0.226–0.985) | 0.045 |
|
Hepatitis C | 0.854
(0.474–1.538) | 0.600 | 0.580
(0.269–1.251) | 0.165 |
|
Non-alcoholic fatty liver
disease | 0.403
(0.123–1.325) | 0.135 | 0.787
(0.236–2.628) | 0.698 |
|
Other | 0.615
(0.282–1.345) | 0.224 | 0.695
(0.300–1.610) | 0.396 |
|
None | 1.069
(0.667–1.712) | 0.782 | 1.260
(0.702–2.262) | 0.439 |
| Family history of
cancer |
|
|
|
|
| No | Reference |
| Reference |
|
|
Yes | 1.157
(0.793–1.693) | 0.454 | 1.314
(0.822–2.10) | 0.255 |
| Body mass
index | 0.967
(0.934–1.002) | 0.063 | 0.947
(0.907–0.989) | 0.014 |
| α-fetoprotein,
ng/ml |
|
|
|
|
|
<200 | Reference |
| Reference |
|
|
≥200 | 1.123
(0.689–1.829) | 0.642 | 1.818
(0.935–3.535) | 0.078 |
| Adjacent liver
tissue inflammation event |
|
|
|
|
| No | Reference |
| Reference |
|
|
Yes | 1.242
(0.752–2.052) | 0.397 | 0.927
(0.504–1.704) | 0.806 |
| Tumor histological
grade |
|
|
|
|
| G1 | Reference |
| Reference |
|
| G2 | 1.220
(0.710–2.096) | 0.471 | 1.325
(0.664–2.645) | 0.425 |
|
G3-G4 | 1.195
(0.681–2.098) | 0.535 | 1.789
(0.891–3.592) | 0.102 |
| Vascular
invasion |
|
|
|
|
|
None | Reference |
| Reference |
|
|
Micro | 1.366
(0.858–2.175) | 0.189 | 1.159
(0.610–2.203) | 0.652 |
|
Macro | 2.575
(1.215–5.457) | 0.014 | 3.104
(1.178–8.181) | 0.022 |
| Ishak fibrosis
score |
|
|
|
|
| 0, No
fibrosis | Reference |
| Reference |
|
| 1-4,
Fibrosis | 0.749
(0.381–1.472) | 0.403 | 0.589
(0.267–1.300) | 0.190 |
| 5-6,
Cirrhosis | 0.732
(0.396–1.356) | 0.321 | 0.565
(0.267–1.198) | 0.137 |
| AJCC TNM stage |
|
|
|
|
| I | Reference |
| Reference |
|
| II | 1.573
(0.938–2.640) | 0.086 | 1.304
(0.636–2.673) | 0.468 |
|
III–IV | 3.209
(2.079–4.954) | <0.001 | 3.838
(2.138–6.891) | <0.001 |
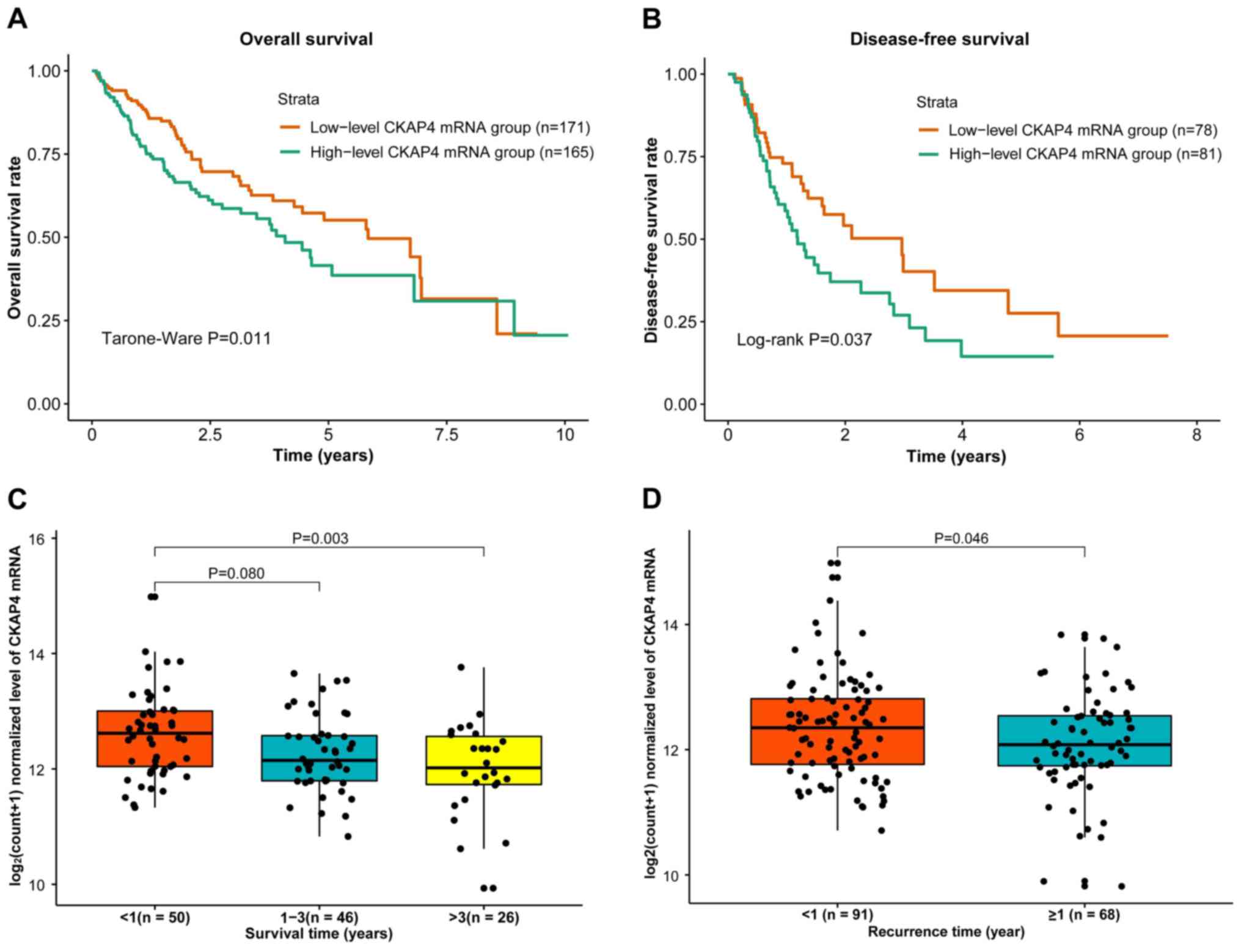
HCC patient survival was compared between the high-
and low-level CKAP4 mRNA groups and the results demonstrated
significantly lower OS and DFS probabilities in the high-level
group (Fig. 4A and B). Additionally,
CKAP4 mRNA levels were significantly different between the patients
with survival times of <1 year and >3 years (P=0.003;
Fig. 4C), and between the patients
with a recurrence times <1 year and ≥1 year (P=0.046; Fig. 4D), indicating that higher CKAP4 mRNA
levels are associated with poor prognosis.
Discussion
In the present study, CKAP4 mRNA levels in tumor and
adjacent liver tissues from patients with HCC were compared, their
associations with clinicopathological parameters were analyzed, and
their potential diagnostic and prognostic values for HCC were
evaluated. The present study demonstrated that CKAP4 mRNA levels
were significantly upregulated in tumor tissues [with a moderate
area under curve (AUC) for HCC diagnosis], and that this was
significantly associated with poor clinicopathological parameters
(lower BMI, higher AFP, adjacent liver tissue inflammation event,
poor tumor histological grade, higher Ishak fibrosis score, and
advanced AJCC TNM stage) and with poor prognosis in patients with
HCC (lower OS and DFS survival probabilities, shorter survival and
recurrence times after tumor resection).
Data were extracted from four independent HCC
datasets from the GEO and TCGA databases, and the CKAP4 mRNA levels
in tumor and adjacent liver tissues from patients with HCC were
compared. All four datasets showed consistent results, indicating
that CKAP4 mRNA levels were significantly higher in tumor tissues
compared with adjacent liver tissues. These results confirm the
upregulation of CKAP4 in tumor tissues in accordance with previous
results found in HCC (19) and other
types of cancer, including cervical cancer, bladder cancer, lung
cancer, pancreatic cancer, ESCC, CLL, PDAC and intrahepatic
cholangiocellular carcinoma (12–18,27).
CKAP4 upregulation in HCC tissues makes it a
potential diagnostic and prognostic biomarker in HCC. In the
present study, CKAP4 mRNA provided moderate power for the diagnosis
of HCC, with AUCs ranging from 0.799–0.898 for HCC diagnosis. To
the best of our knowledge, similar studies involving the diagnostic
evaluation of CKAP4 mRNA for HCC have not been conducted; however,
Wang et al (21) reported
that the serum concentration of CKAP4 protein was significantly
elevated in patients with HCC compared with healthy controls, and
with patients with chronic hepatitis B and cirrhosis; the study
also revealed that 86% of AFP-negative patients with HCC displayed
elevated serum CKAP4 levels. Notably, the previously mentioned
study reported that a combined biomarker panel of AFP and CKAP4
presents improved diagnostic accuracy for HCC compared with
stand-alone analysis of either AFP or CKAP4, even in diagnosing
early HCC. Additionally, a good association has been observed
between CKAP4 mRNA and protein expression in tissues from patients
with HCC (19). These findings
indicate that both CKAP4 mRNA in tumor tissue and CKAP4 protein in
serum are potential biomarkers for HCC detection that complement
AFP evaluation.
The findings of the present study also confirmed the
prognostic value of CKAP4 mRNA in patients with HCC. Univariate Cox
regression analysis revealed that a high CKAP4 mRNA level was one
of the three risk factors for OS and DFS in patients with HCC. In
the present study, patients with HCC who had a high CKAP4 mRNA
level were observed to have unfavorable OS and DFS rates (based on
Kaplan-Meier survival curve analyses) and a shorter survival time
or shorter recurrence time after curative surgical resection. These
results are consistent with previous findings in numerous tumors
(14–16) but differ from a report by Li et
al (19), which demonstrated
that patients with HCC and high CKAP4 mRNA (tissue microarray
analysis) and protein (western blot analysis) expression had
favorable OS and DFS rates. The present study considers that CKAP4
is associated with poor prognosis in HCC patients, as this
conclusion is supported directly by the results of survival
analysis, but also indirectly by the significant associations found
between CKAP4 mRNA levels and typical prognosis-related
clinicopathological factors (Table
I), including histological differentiation grade, TNM stage and
AFP level.
CKAP4 promotes tumor growth in numerous human
cancers via different mechanisms. Kimura et al (14) demonstrated that CKAP4 expressed at
the plasma membrane stimulated pancreatic and lung cancer cell
proliferation by activating the phosphoinositide 3-kinase-AKT
pathway; upregulation of CKAP4 was also strongly correlated with
tumor growth and poor prognosis in patients. Shinno et al
(16) reported that CKAP4 was
upregulated in ~40% of patients with ESCC and as a DKK1 receptor in
cells activated the AKT pathway and promoted ESCC cell
proliferation when DKK1 was highly expressed. DKK1 has also been
reported to be upregulated in HCC tissues and associated with
advanced tumor stage, metastasis, recurrence, OS and DFS rates
(28,29). However, a previous study revealed
that CKAP4 inhibited HCC growth and metastasis by suppressing EGFR
signaling, which indicates that the effect of CKAP4 in HCC is the
opposite of that observed in the other aforementioned cancer types
(20). This is also contradictory to
previous findings that CKAP4 mRNA is upregulated in tissues from
patients with HCC, and significantly associated with tumor size,
intrahepatic metastasis and portal venous invasion (19). More rigorous experiments are required
to elucidate the effect and mechanism of CKAP4 in HCC growth and
progression.
The present study has several limitations. Firstly,
the HCC clinical data from the GDC TCGA-LIHC database were
incomplete; thus, the sample size was insufficient for multivariate
Cox regression analysis, and to subsequently conclude that CKAP4
mRNA is an independent risk factor for HCC prognosis. Secondly,
although the upregulation of CKAP4 mRNA was confirmed in each of
the four independent datasets, only one available dataset included
clinical data that could be used to evaluate the diagnostic and
prognostic significance of CKAP4 mRNA in HCC. Thus, cohorts with
more patients with HCC are required to validate the results of the
present study. Also, there were no data demonstrating CKAP4 protein
expression in tumor and corresponding adjacent tissues for
simultaneous analysis, although a strong correlation between CKAP4
mRNA and protein expression has been observed previously (19). Future large-scale studies without the
aforementioned limitations are required to verify the findings of
the present study.
In view of the controversial role of CKAP4 in HCC
growth and progression, the present study extracted data from the
GEO and TCGA public databases to systematically investigate the
relationship between CKAP4 mRNA and HCC. The findings of the
present study confirmed that significantly higher CKAP4 mRNA levels
are present in tumor tissues compared with adjacent liver tissues
in patients with HCC, and that this upregulation is closely
associated with poor clinicopathological conditions and outcome;
this indicated the potential value of CKAP4 in HCC diagnosis and
prognosis. To the best of our knowledge, this is the first study to
investigate the association between CKAP4 mRNA expression in tumor
tissues and the clinical characteristics of HCC based on data in
public databases.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
ZYC and KHZ designed the present study. TW, XG, SHC,
YTH and YQW conducted the study and analyzed the data. ZYC wrote
the manuscript and KHZ revised the manuscript. All the authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Global Burden of Disease Liver Cancer
Collaboration, ; Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu
MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, et al: The
burden of primary liver cancer and underlying etiologies from 1990
to 2015 at the global, regional, and national level: Results from
the global burden of disease study 2015. JAMA Oncol. 3:1683–1691.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shimada K, Sano T, Sakamoto Y and Kosuge
T: A long-term follow-up and management study of hepatocellular
carcinoma patients surviving for 10 years or longer after curative
hepatectomy. Cancer. 104:1939–1947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Poon RT: Prevention of recurrence after
resection of hepatocellular carcinoma: A daunting challenge.
Hepatology. 54:757–759. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tralhao JG, Dagher I, Lino T, Roudie J and
Franco D: Treatment of tumour recurrence after resection of
hepatocellular carcinoma. Analysis of 97 consecutive patients. Eur
J Surg Oncol. 33:746–751. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gupta N, Manevich Y, Kazi AS, Tao JQ,
Fisher AB and Bates SR: Identification and characterization of p63
(CKAP4/ERGIC-63/CLIMP-63), a surfactant protein A binding protein,
on type II pneumocytes. Am J Physiol Lung Cell Mol Physiol.
291:L436–L446. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schweizer A, Ericsson M, Bachi T,
Griffiths G and Hauri HP: Characterization of a novel 63 kDa
membrane protein. Implications for the organization of the
ER-to-Golgi pathway. J Cell Sci. 104:671–683. 1993.PubMed/NCBI
|
|
9
|
Nikonov AV, Hauri HP, Lauring B and
Kreibich G: Climp-63-mediated binding of microtubules to the ER
affects the lateral mobility of translocon complexes. J Cell Sci.
120:2248–2258. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shibata Y, Shemesh T, Prinz WA, Palazzo
AF, Kozlov MM and Rapoport TA: Mechanisms determining the
morphology of the peripheral ER. Cell. 143:774–788. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bates SR, Kazi AS, Tao JQ, Yu KJ, Gonder
DS, Feinstein SI and Fisher AB: Role of P63 (CKAP4) in binding of
surfactant protein-A to type II pneumocytes. Am J Physiol Lung Cell
Mol Physiol. 295:L658–L669. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Conrads TP, Tocci GM, Hood BL, Zhang CO,
Guo L, Koch KR, Michejda CJ, Veenstra TD and Keay SK: CKAP4/p63 is
a receptor for the frizzled-8 protein-related antiproliferative
factor from interstitial cystitis patients. J Biol Chem.
281:37836–37843. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shahjee HM, Koch KR, Guo L, Zhang CO and
Keay SK: Antiproliferative factor decreases Akt phosphorylation and
alters gene expression via CKAP4 in T24 bladder carcinoma cells. J
Exp Clin Cancer Res. 29:1602010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kimura H, Fumoto K, Shojima K, Nojima S,
Osugi Y, Tomihara H, Eguchi H, Shintani Y, Endo H, Inoue M, et al:
CKAP4 is a Dickkopf1 receptor and is involved in tumor progression.
J Clin Invest. 126:2689–2705. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kikuchi A, Fumoto K and Kimura H: The
Dickkopf1-cytoskeleton-associated protein 4 axis creates a novel
signalling pathway and may represent a molecular target for cancer
therapy. Br J Pharmacol. 174:4651–4665. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shinno N, Kimura H, Sada R, Takiguchi S,
Mori M, Fumoto K, Doki Y and Kikuchi A: Activation of the
Dickkopf1-CKAP4 pathway is associated with poor prognosis of
esophageal cancer and anti-CKAP4 antibody may be a new therapeutic
drug. Oncogene. 37:3471–3484. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Johnston HE, Carter MJ, Larrayoz M, Clarke
J, Garbis SD, Oscier D, Strefford JC, Steele AJ, Walewska R and
Cragg MS: Proteomics profiling of CLL versus healthy B-cells
identifies putative therapeutic targets and a subtype-independent
signature of spliceosome dysregulation. Mol Cell Proteomics.
17:776–791. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kimura H, Yamamoto H, Harada T, Fumoto K,
Osugi Y, Sada R, Maehara N, Hikita H, Mori S, Eguchi H, et al:
CKAP4, a DKK1 receptor, is a biomarker in exosomes derived from
pancreatic cancer and a molecular target for therapy. Clin Cancer
Res. 25:1936–1947. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li SX, Tang GS, Zhou DX, Pan YF, Tan YX,
Zhang J, Zhang B, Ding ZW, Liu LJ, Jiang TY, et al: Prognostic
significance of cytoskeleton-associated membrane protein 4 and its
palmitoyl acyltransferase DHHC2 in hepatocellular carcinoma.
Cancer. 120:1520–1531. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li SX, Liu LJ, Dong LW, Shi HG, Pan YF,
Tan YX, Zhang J, Zhang B, Ding ZW, Jiang TY, et al: CKAP4 inhibited
growth and metastasis of hepatocellular carcinoma through
regulating EGFR signaling. Tumour Biol. 35:7999–8005. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Yu W, He M, Huang Y, Wang M and
Zhu J: Serum cytoskeleton-associated protein 4 as a biomarker for
the diagnosis of hepatocellular carcinoma. Onco Targets Ther.
12:359–364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao Y, Ding W, Zhang J, Gao Q, Yang H, Cao
W, Wang Z, Fang L and Du R: Significant down-regulation of urea
cycle generates clinically relevant proteomic signature in
hepatocellular carcinoma patients with macrovascular invasion. J
Proteome Res. 18:2032–2044. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramalho FS, Ramalho LN, Della Porta L and
Zucoloto S: Comparative immunohistochemical expression of p63 in
human cholangiocarcinoma and hepatocellular carcinoma. J
Gastroenterol Hepatol. 21:1276–1280. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grinchuk OV, Yenamandra SP, Iyer R, Singh
M, Lee HK, Lim KH, Chow PK and Kuznetsov VA: Tumor-adjacent tissue
co-expression profile analysis reveals pro-oncogenic ribosomal gene
signature for prognosis of resectable hepatocellular carcinoma. Mol
Oncol. 12:89–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mah WC, Thurnherr T, Chow PK, Chung AY,
Ooi LL, Toh HC, The BT, Saunthararajah Y and Lee CG: Methylation
profiles reveal distinct subgroup of hepatocellular carcinoma
patients with poor prognosis. PLoS One. 9:e1041582014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li MH, Dong LW, Li SX, Tang GS, Pan YF,
Zhang J, Wang H, Zhou HB, Tan YX, Hu HP and Wang HY: Expression of
cytoskeleton-associated protein 4 is related to lymphatic
metastasis and indicates prognosis of intrahepatic
cholangiocarcinoma patients after surgery resection. Cancer Lett.
337:248–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Desert R, Mebarki S, Desille M, Sicard M,
Lavergne E, Renaud S, Bergeat D, Sulpice L, Perret C, Turlin B, et
al: ‘Fibrous nests’ in human hepatocellular carcinoma express a
Wnt-induced gene signature associated with poor clinical outcome.
Int J Biochem Cell Biol. 81:195–207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang Y, Yang X, Zhao F, Shen Q, Wang Z,
Lv X, Hu B, Yu B, Fan J and Qin W: Overexpression of Dickkopf-1
predicts poor prognosis for patients with hepatocellular carcinoma
after orthotopic liver transplantation by promoting cancer
metastasis and recurrence. Med Oncol. 31:9662014. View Article : Google Scholar : PubMed/NCBI
|