Introduction
Breast cancer (BC) is one of the most commonly
diagnosed cancers and the leading cause of cancer death among women
worldwide (1). Improved diagnostic
capabilities has led to an increased rate of BC identified
at an early stage (2).
However, despite early diagnosis and treatment, the rate of
recurrence and metastasis following radical treatment remains
disappointingly high, and survival varies considerably between
patients with closely matching tumour characteristics.
Inflammation and angiogenesis are the main drivers
of cancer. These processes are tightly interconnected in the sense
that many pro-inflammatory proteins possess proangiogenic
properties and vice versa. The formation of new blood vessels in
malignant tumours ensures the supply of nutrients and oxygen, hence
promoting the spread of tumour cells (3). Microvessel density is a pivotal risk
factor for metastasis and a predictor of poor BC prognosis
(4).
The vascular endothelial growth factor A (VEGFA, or
VEGF) is the best-known proangiogenic molecule. VEGFA mediates the
growth of new blood vessels by binding to the endothelial cell
surface receptors. It promotes endothelial cell proliferation,
migration and the formation of tubular structures (5). Another mechanism of tumour
neovascularization is the so-called inflammatory angiogenesis. Such
pro-inflammatory cytokines as interleukin-1β (IL-1β), interlukin-1α
(IL-1α) and interleukin-6 (IL-6) through a variety of signalling
pathways promote endothelial cell migration and proliferation,
contributing to tumour angiogenesis that facilitates the survival
of cancer cells (6,7). Apart from stimulating angiogenesis,
VEGFA, IL-1β1, IL-1α and IL-6 are also involved in inflammatory
processes. These proteins may prevent apoptosis and promote cancer
cell proliferation, differentiation, migration and metastasis.
The association between the above-mentioned proteins
and BC prognosis was demonstrated by several authors. The elevated
serum level of VEGFA in metastatic BC patients is linked to worse
progression-free survival (PFS) and overall survival (OS) (8). Another cytokine, IL-6, induces
epithelial-mesenchymal phenotype and therapeutic resistance in BC
cells (9). Higher circulating levels
of IL6 were observed in more advanced stages of the disease
(10). It was also found that high
tumour co-expression of the VEGF and IL-6 family cytokines
significantly lowers the human epidermal growth factor receptor 2
(HER2) negative BC survival (11).
Moreover, higher expression of pro-inflammatory IL-1β cytokine is
correlated with higher BC stage and significantly worse survival
(12). In addition, IL-1α acts as a
pro-inflammatory molecule itself and also promotes the activity of
IL-1β, resulting in an increased growth of BC cells and tumour
progression (13).
Common polymorphisms in proinflammatory and
proangiogenic cytokine genes may influence their coded protein
production and play a role in the course of BC. The polymorphisms
with proved functional activity are VEGFA (rs699947,
rs833061, rs25648, rs1005230), IL-1β (rs1143634, rs16944),
IL-1α (rs1800587) and IL-6 (rs1800795, rs1800797)
(14–21). Candidate gene studies as well as
moderate-sized genome-wide association studies (GWAS) highlight the
important role of these polymorphisms in BC risk and aggressiveness
(22–32), although substantial heterogeneity
across studies exists. The currently available results are
inconsistent in terms of different ethnicity and cancer stage;
therefore, it is necessary to further investigate the role of
VEGFA, IL-1β, IL-1α and IL-6 gene polymorphisms in
breast carcinogenesis and cancer progression.
This paper describes a cohort study that aimed to
examine the contribution of VEGFA, IL-1β, IL-1α and
IL-6 gene polymorphisms to the clinicopathologic features
and survival in a homogeneous group of Eastern European
(specifically, Lithuanian) early-stage BC patients.
Materials and methods
Patients
The study consisted of 202 adult Lithuanian women
with primary I–II stage BC. All patients were treated in the
Hospital of Lithuanian University of Health Sciences Kaunas
Clinics. The exclusion criteria were other malignancies,
significant comorbidities and/or incomplete medical documentation.
Surgery and adjuvant therapy were chosen by the clinicians, based
on pathomorphologic characteristics and validated prognostic
factors. The patients were followed until 30 April, 2019 (censoring
date).
Candidate polymorphisms
The genes and polymorphisms known to modulate
inflammation and angiogenesis were selected. The selection criteria
included: i) functional single nucleotide polymorphisms (SNPs) in
the VEGFA, IL-1β, IL-1α and IL-6 genes predicting
alterations in the protein level; ii) SNP relevant to outcomes in
other settings; and iii) SNP with a minor allele frequency greater
than 15% in the study population. We selected nine SNPs: The
VEGFA gene rs699947, rs833061, rs25648, and rs1005230; the
IL-1β gene rs1143634 and rs16944; the IL-1α gene
rs1800587; and the IL-6 gene rs1800795 and rs1800797.
Assay methods
Peripheral blood samples from the study population
were collected in 2009–2017. For genomic DNA extraction from
peripheral blood leukocytes, a commercially available DNA
extraction kit (i.e., Thermo Fisher Scientific Baltics, Lithuania)
was used. The DNA was stored at −20°C prior to usage.
Genotyping of the selected polymorphisms was
performed at the Dr. K. Janusauskas Laboratory of Genetics of the
Institute of Biology Systems and Genetic Research of Lithuanian
University of Health Sciences Kaunas Clinics. The SNPs of the
target genes were estimated by using TaqMan SNP Genotyping Assays
(C_8311602_10, C_1647381_10, C_791476_10, C_8311612_10,
C_1839697_20, C_1839695_20, C_9546517_10, C_1839943_10,
C_1839943_10; Applied Biosystems; Thermo Fisher Scientific, Inc.).
The polymerase chain reaction was performed in a reaction volume of
25 µl containing template DNA (2 ng), 2X TaqMan Universal Master
Mix II, no UNG (Applied Biosystems; Thermo Fisher Scientific,
Inc.)-12,5 µl, 20X TaqMan SNP Genotyping Assay stock (initial stock
of 40X or 80X was diluted to get 20X working stock)-1,25 µl. The
final volume of 25 µl was adjusted by adding nuclease free
ddH2O. Finally, the 2 µl of DNA was added from each
sample. For negative control, nuclease free ddH2O was
used instead of patient DNA, while for positive control, the DNA of
the known genotype was used. Each sample genotyping was repeated
twice for accuracy.
The Applied Biosystems 7900HT Real-Time Polymerase
Chain Reaction System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used for SNP detection. The cycling program
started from heating up to 95°C for 10 min followed by 40 cycles
(at 95°C for 15 sec and at 60°C for 1 min). Finally, allelic
discrimination was done by using the SDS 2.3 software provided by
Applied Biosystems; Thermo Fisher Scientific, Inc.
Study design
A prospective cohort study was conducted at the
Hospital of Lithuanian University of Health Sciences. For the case
selection, information on primarily BC patients was retrieved from
the hospital's Pathology Department. The patients who fulfilled the
inclusion criteria and signed the informed consent document
(approved by the Kaunas Regional Ethics Committee for Biomedical
research; Protocol number BE-2-10) were enrolled in the study and
their peripheral blood samples were obtained. The characteristics
of clinical and pathological features and the course of the disease
were obtained for all study subjects. The date of histological BC
verification was time zero in the survival analysis. The endpoints
of interest were disease-free survival (DFS), metastasis-free
survival (MFS) and OS. We checked for associations of SNPs with the
known BC prognostic factors and survival endpoints. The guidelines
for the reporting of tumour marker prognostic studies were applied
while conducting the study (33,34).
Statistical analysis
The allele frequency distributions of the
investigated SNPs were compared with the European population data
from the 1000 Genomes project phase 3 database (35). For each SNP a Hardy-Weinberg
equilibrium was assessed by using Pearson's chi square and Fisher's
exact tests. The Haploview v4.1 software was used to check for the
linkage disequilibrium between SNPs (36). The VEGFA and IL-6
haplotypes were inferred from the tested SNPs by Bayesian methods
as implemented in the Phase software (v2.1; Department of
Statistics, University of Washington, Seattle, WA, USA) (37,38). The
SNPs were analysed under genotype, allelic and haplotype (for
IL-6 and VEGFA SNPs) models. The associations of
polymorphisms with clinicopathologic variables were evaluated by
Pearson's Chi-square or Fisher's exact test. The
Bonferroni-corrected α level was used in the association analysis
for multiple comparisons. Moreover, the Cox proportional hazards
model was used to estimate the prognostic factors for DFS, MFS and
OS. In addition, multivariate analysis was used to determine the
interdependency of genotypes and other known prognostic factors,
such as age, tumour differentiation grade, tumour size, lymph node
status, oestrogen receptor status, progesterone receptor status and
human epidermal growth factor receptor 2 (HER2) status. Hazard
ratios (HR) and their 95% confidence intervals (CI) were recorded
for each tested marker. Finally, the Kaplan-Meier analysis with the
log-rank test was applied to compare the survival of the patients
with different genotypes. Statistical significance was set at 5%
(P<0.05). Statistical analysis was performed using SPSS for
Windows v20.0 (Released 2011; IBM Corp).
Results
Sample characteristics
A total of 202 Lithuanian women with early-stage BC
were included in the current analysis. The frequency distributions
of clinical and tumour biological factors are shown in Table I. For all study participants, primary
treatments included surgery (100%), chemotherapy (77%), hormone
therapy (71%), trastuzumab (19%) and radiation therapy (97%).
 | Table I.Frequency data for clinical and
tumour biological factors. |
Table I.
Frequency data for clinical and
tumour biological factors.
| Factor | Patients, % |
|---|
| Age at diagnosis,
years |
|
| <50
years | 65 |
| ≥50
years | 35 |
| Tumour size,
cm |
|
|
<2 | 64 |
|
2–5 | 36 |
| Lymph node
status |
|
|
Positive | 55 |
|
Negative | 45 |
| Grade |
|
| G1 and
G2 | 78 |
| G3 | 22 |
| ER status |
|
| ER
positive | 68 |
| ER
negative | 32 |
| PR status |
|
| PR
positive | 60 |
| PR
negative | 40 |
| HER2 status |
|
| HER2
positive | 19 |
| HER2
negative | 81 |
All the patients were genotyped for a panel of nine
SNPs: The VEGFA gene rs699947, rs833061, rs25648 and
rs1005230; the IL-1β gene rs1143634 and rs16944; the
IL-1α gene rs1800587; the IL-6 gene rs1800795 and
rs1800797. The genotypes were found to be in Hardy-Weinberg
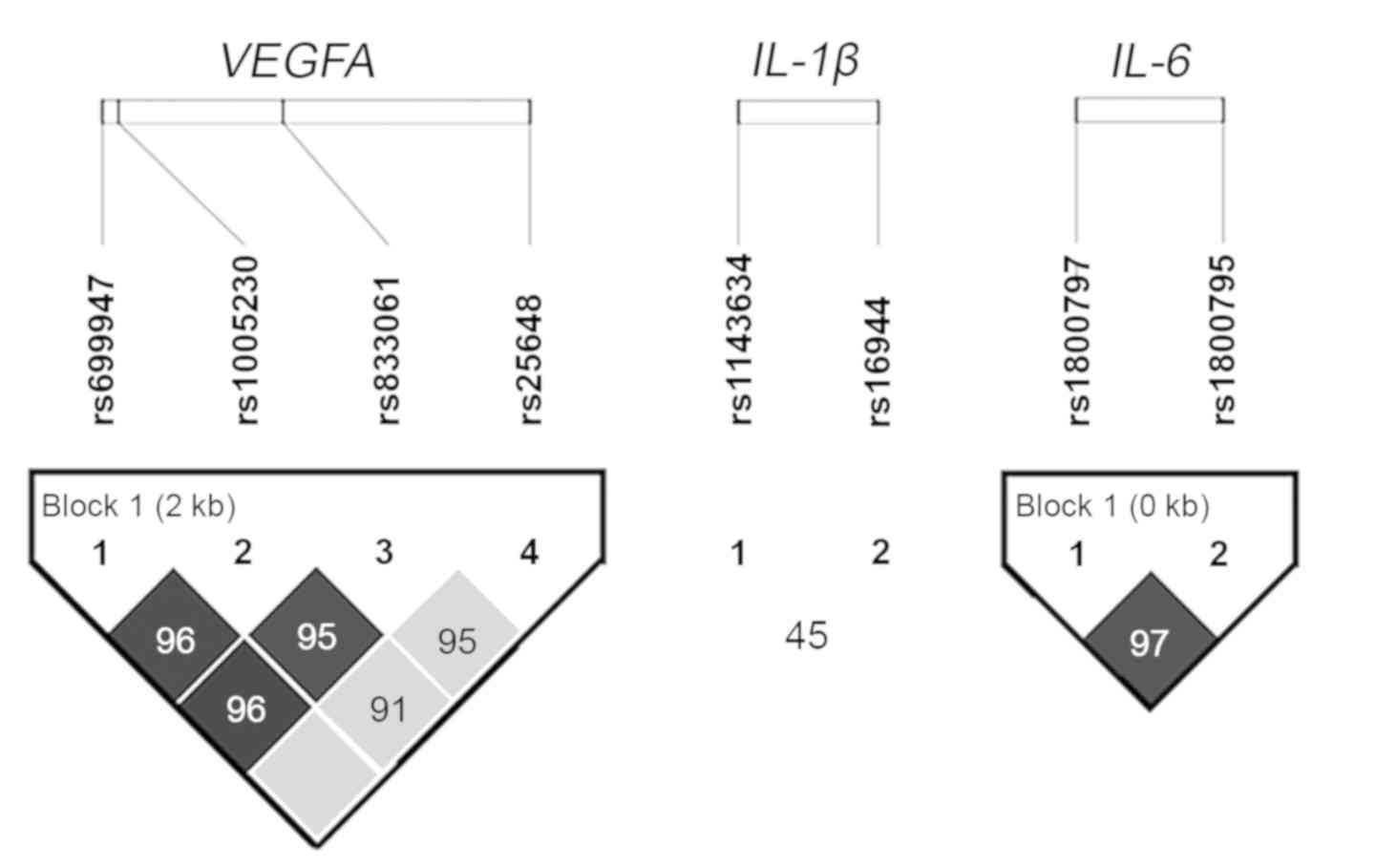
equilibrium in all the nine SNPs. A strong linkage disequilibrium
between four VEGFA and two IL-6 polymorphisms was
confirmed (Fig. 1). Our cohort had
similar allele distribution to that of the 1000 Genomes project
phase 3 for European population. The genotype and allele frequency
data is presented in Table II.
 | Table II.Allele and genotype frequencies of
analysed single nucleotide polymorphisms in the study population
and in European population data from the 1000 Genomes Project Phase
3 database. |
Table II.
Allele and genotype frequencies of
analysed single nucleotide polymorphisms in the study population
and in European population data from the 1000 Genomes Project Phase
3 database.
| Gene | Polymorphism | Study allele and
genotype frequencies (1000 Genomes Project Phase 3 database allele
frequencies) |
|---|
| VEGFA | rs699947 | A, 0.53 (0.50) | C, 0.47 (0.50) | AA, 0.28 | CA, 0.50 | CC, 0.22 |
|
| rs833061 | T, 0.47 (0.50) | C, 0.53 (0.50) | TT, 0.21 | TC, 0.51 | CC, 0.28 |
|
| rs25648 | C, 0.81 (0.83) | T, 0.19 (0.17) | TT, 0.03 | CT, 0.32 | CC, 0.65 |
|
| rs1005230 | T, 0.52 (0.50) | C, 0.48 (0.50) | TT, 0.28 | CT, 0.49 | CC, 0.23 |
| IL-6 | rs1800795 | C, 0.47 (0.42) | G, 0.53 (0.58) | CC, 0.24 | GC, 0.47 | GG, 0.29 |
|
| rs1800797 | A, 0.46 (0.41) | G, 0.54 (0.59) | AA, 0.23 | GA, 0.47 | GG, 0.30 |
| IL-1β | rs1143634 | G, 0.73 (0.75) | A, 0.27 (0.25) | GG, 0.52 | GA, 0.42 | AA, 0.06 |
|
| rs16944 | G, 0.66 (0.65) | A, 0.34 (0.35) | GG, 0.43 | GA, 0.47 | AA, 0.10 |
| IL-1α | rs1800587 | C, 0.69 (0.71) | T, 0.31 (0.29) | CC, 0.48 | CT, 0.43 | TT, 0.09 |
Inferential analysis
The data on associations between the analysed
polymorphisms and clinicopathologic tumour features is shown in
Tables SI–SVI. In the single-locus analysis, the
genotype model revealed a significant link between the IL-6
rs1800797 genotypes and the oestrogen receptor (P=0.005) and the
progesterone receptor (P=0.007) status (Bonferoni adjusted;
significant p value <0.008). The allelic model showed that the A
allele of this SNP is associated with positive oestrogen receptors
(OR 2.23; 95% CI: 1.19–4.08; P=0.014). Patients carrying the
IL-6 rs 1800797 A allele were also predisposed to higher
rates of HER2 negative BC (OR 2.21; 95% CI: 1.07–4.57; P=0.042).
Additionally, another IL-6 polymorphism, rs1800795, in both
genotype and allelic models was linked to oestrogen receptor
positive BC. Specifically, 72% of patients carrying the IL-6
rs1800795 C allele had oestrogen receptor positive disease,
compared to 58% of non-carriers (OR 1.92; 95% CI: 1.04–3.57;
P=0.04).
Linkage disequilibrium analysis showed a high degree
of disequilibrium between the two IL-6 SNPs
(r2=0.89), meaning that the associations were not
independent. As such, haplotype analysis was performed to further
explore the relationship between IL-6 variations and other
prognostic factors. Phasing revealed three possible IL-6
(rs1800797, rs1800795) haplotypes: GG (51.7%), AC (45.5%) and GC
(2.8%). It was further demonstrated that the AC haplotype was
positively associated with oestrogen receptor positive BC (HR 2.13;
95% CI 1.14–3.98; P=0.018; Table
SII).
By analysing the associations between the
IL-1α SNP and clinicopathologic factors (Table SIII), we found a link between the
IL-1α rs1800587 SNP C allele and larger primary tumour size
(comparing tumours sized <2 vs. 2–5 cm) (OR 4.91; 95% CI:
1.09–22.01; P=0.022).
Other analysed polymorphisms of IL-1β and
VEGFA revealed no associations with the evaluated prognostic
factors in neither genotype nor allelic models (Tables SIV–SVI). Due to the strong linkage
disequilibrium, four VEGFA polymorphisms were also analysed
in the haplotype model. Three main haplotypes were identified: CCTC
(45.5%), ATCC (33.4%) and ATCT (17.6%), as well as several rare
variants. None of the VEGFA haplotypes was associated with
clinicopathologic prognostic variables.
Survival analysis
In the mean follow-up time of 67 months (range
28–202), progression of the disease was observed for 33 patients.
Of those who progressed, 28 had distant metastases. Twenty-two
patients with progressive disease died, all due to cancer. The
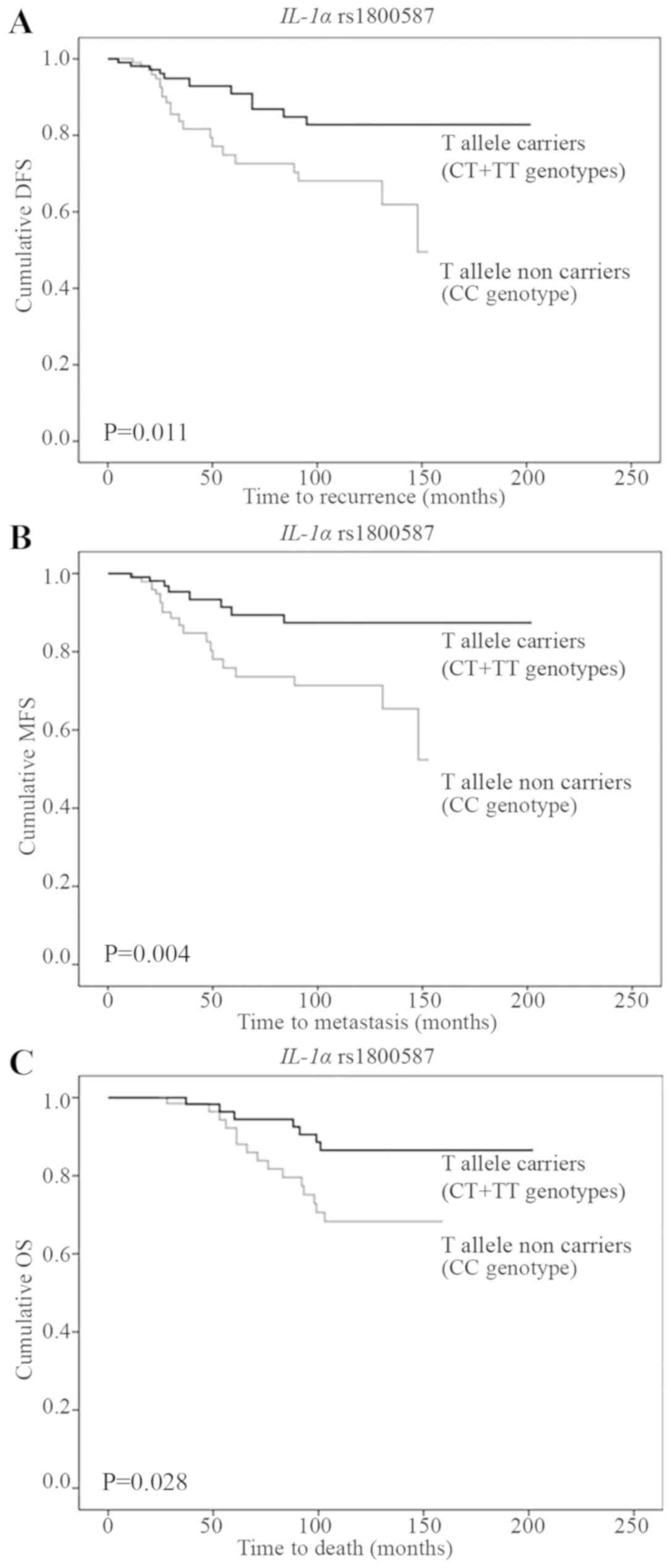
Kaplan-Meier survival analysis showed that the IL-1α
rs1800587 SNP is associated with early-stage breast cancer DFS, MFS
and OS. In particular, patients homozygous for the C allele (CC)
had worse survival rates than patients homozygous and heterozygous
for the A allele (CT and TT) (Fig.
2A-C).
The univariate Cox regression analysis (presented in
Tables III–V) revealed that L-1α rs1800587 CC
genotype carriers had 2.48 times higher risk of disease recurrence
(95% CI: 1.19–5.11; P=0.014), 3.12 times higher risk of metastasis
(95% CI: 1.37–7.10; P=0.007) and 2.63 times higher risk of death
(95% CI: 1.07–6.46; P=0.035) than the carriers of CT and TT
genotypes (Table III).
 | Table III.Cox's univariate model for
IL-1β and IL-1α SNPs. |
Table III.
Cox's univariate model for
IL-1β and IL-1α SNPs.
|
|
|
|
| PFS | MFS | OS |
|---|
|
|
|
|
|
|
|
|
|---|
| Reference SNP
ID | Model |
Genotype/allele/haplotype | Patients n | Univariate hazard
ratio (95% CI) | P-value | Univariate hazard
ratio (CI) | P-value | Multivariate hazard
ratio (95% CI) | P-value |
|---|
| IL-1β
rs1143634 | Genotype | AA | 12 | 1 | 0.043 | 1 | 0.062 | 1 | 0.271 |
|
|
| GA | 85 | 0.443
(0.094–2.100) | 0.305 | 0.945
(0.116–7.688) | 0.360 | 0.773
(0.093–6.421) | 0.811 |
|
|
| GG | 105 | 1.243
(0.292–5.291) | 0.768 | 2.555
(0.343–19.048) | 0.958 | 1.667
(0.220–12.630) | 0.621 |
|
| Allelic | A allele
carriers | 97 | 1 |
| 1 |
| 1 |
|
|
|
| A allele non
carriers | 105 | 2.490
(0.944–5.246) | 0.056 | 2.686
(0.932–6.102) | 0.058 | 2.087
(0.851–5.122) | 0.108 |
|
|
| G allele
carriers | 190 | 1 |
| 1 |
| 1 |
|
|
|
| G allele non
carriers | 12 | 1.171
(0.279–4.913) | 0.829 | 0.563
(0.076–4.148) | 0.573 | 0.799
(0.107–5.939) | 0.826 |
| IL-1-β
rs16944 | Genotype | AA | 20 | 1 | 0.335 | 1 | 0.347 | 1 | 0.715 |
|
|
| GA | 95 | 1.657
(0.387–7.096) | 0.496 | 1.386
(0.320–6.005) | 0.663 | 1.183
(0.264–5.287) | 0.826 |
|
|
| GG | 87 | 0.974
(0.216–4.396) | 0.973 | 0.763
(0.165–3.533) | 0.729 | 0.814
(0.173–3.834) | 0.794 |
|
| Allelic | A allele
carriers | 115 | 1 |
| 1 |
| 1 |
|
|
|
| A allele non
carriers | 87 | 0.623
(0.302–1.294) | 0.201 | 0.573
(0.259–1.267) | 0.169 | 0.706
(0.296–1.684) | 0.432 |
|
|
| G allele
carriers | 182 | 1 |
| 1 |
| 1 |
|
|
|
| G allele non
carriers | 20 | 0.754
(0.180–3.155) | 0.70 | 0.926
(0.220–3.905) | 0.917 | 0.999
(0.233–4.277) | 0.99 |
| IL-1α
rs1800587 | Genotype | TT | 18 | 1 | 0.049 | 1 | 0.025 | 1 | 0.108 |
|
|
| CT | 87 | 0.805
(0.174–3.729) | 0.782 | 1.292
(0.159–10.506) | 0.811 | 1.015
(0.122–8.428) | 0.989 |
|
|
| CC | 97 | 2.067
(0.486–8.794) | 0.326 | 3.891
(0.521–29.034) | 0.185 | 2.664
(0.352–20.187) | 0.343 |
|
| Allelic | T allele
carriers | 105 | 1 |
| 1 |
| 1 |
|
|
|
| T allele non
carriers | 97 | 2.480
(1.199–5.114) | 0.014a | 3.122
(1.372–7.104) | 0.007a | 2.632
(1.072–6.461) | 0.035a |
|
|
| C allele
carriers | 184 | 1 |
| 1 |
| 1 |
|
|
|
| C allele non
carriers | 18 | 0.703
(0.168–2.940) | 0.630 | 0.392
(0.053–2.888) | 0.358 | 0.551
(0.074–4.096) | 0.560 |
 | Table V.Cox's univariate model for
VEGFA SNPs. |
Table V.
Cox's univariate model for
VEGFA SNPs.
|
|
|
|
| PFS | MFS | OS |
|---|
|
|
|
|
|
|
|
|
|---|
| Reference SNP
ID | Model |
Genotype/allele/haplotype | Patients, n | Univariate hazard
ratio (95% CI) | P-value | Univariate hazard
ratio (95% CI) | P-value | Univariate hazard
ratio (95% CI) | P-value |
|---|
| VEGFA
rs699947 | Genotype | AA | 56 | 1 | 0.373 | 1 | 0.538 | 1 | 0.162 |
|
|
| CA | 101 | 1.483
(0.616–3.565) | 0.387 | 1.104
(0.447–2.754) | 0.317 | 1.141 |
|
|
|
|
|
|
|
|
|
| (0.383–3.414) | 0.813 |
|
|
| CC | 45 | 2.03
(0.76–5.47) | 0.160 | 1.689
(0.613–4.622) | 0.845 | 2.524 |
|
|
|
|
|
|
|
|
|
| (0.822–7.723) | 0.105 |
|
| Allelic | A allele
carriers | 157 | 1 |
| 1 |
| 1 |
|
|
|
| A allele non
carriers | 45 | 1.568
(0.727–3.383) | 0.251 | 1.585
(0.697–3.604) | 0.272 | 2.322 |
|
|
|
|
|
|
|
|
|
| (0.973–5.543) | 0.058 |
|
|
| C allele
carriers | 146 | 1 |
| 1 |
| 1 |
|
|
|
| C allele non
carriers | 56 | 0.613
(0.266–1.415) | 0.252 | 0.791
(0.336–1.864) | 0.592 | 0.651 |
|
|
|
|
|
|
|
|
|
| (0.240–1.766) | 0.399 |
| VEGFA
rs833061 | Genotype | CC | 56 | 1 | 0.295 | 1 | 0.430 | 1 | 0.115 |
|
|
| TC | 103 | 1.445
(0.600–3.499) | 0.410 | 1.081
(0.429–2.703) | 0.875 | 1.124 |
|
|
|
|
|
|
|
|
|
|
(0.377–3.344) | 0.842 |
|
|
| TT | 43 | 2.189
(0.813–5.892) | 0.121 | 1.801
(0.652–4.974) | 0.257 | 2.694 |
|
|
|
|
|
|
|
|
|
| (0.880–8.250) | 0.083 |
|
| Allelic | C allele
carriers | 159 | 1 |
| 1 |
| 1 |
|
|
|
| C allele non
carriers | 43 | 1.709
(0.792–3.687) | 0.172 | 1.718
(0.755–3.905) | 0.197 | 2.511 |
|
|
|
|
|
|
|
|
|
| (0.952–5.994) | 0.058 |
|
|
| T allele
carriers | 146 | 1 |
| 1 |
| 1 |
|
|
|
| T allele non
carriers | 56 | 0.609
(0.264–1.405) | 0.245 | 0.786
(0.334–1.853) | 0.583 | 0.649 |
|
|
|
|
|
|
|
|
|
| (0.239–1.760) | 0.395 |
| VEGFA
rs1005230 | Genotype | TT | 57 | 1 | 0.438 | 1 | 0.657 | 1 | 0.268 |
|
|
| CT | 99 | 1.553
(0.646–3.755) | 0.327 | 1.153
(0.465–2.891) | 0.763 | 1.204 |
|
|
|
|
|
|
|
|
|
| (0.402–3.593) | 0.740 |
|
|
| CC | 46 | 1.894
(0.702–5.083) | 0.207 | 1.584
(0.571–4.355) | 0.381 | 2.281 |
|
|
|
|
|
|
|
|
|
| (0.753–6.974) | 0.149 |
|
| Allelic | C allele
carriers | 57 | 1 |
| 1 |
| 1 |
|
|
|
| C allele non
carriers | 145 | 0.604
(0.262–1.393) | 0.237 | 0.779
(0.331–1.835) | 0.568 | 0.647 |
|
|
|
|
|
|
|
|
|
| (0.238–1.753) | 0.392 |
|
|
| T allele
carriers | 156 | 1 |
| 1 |
| 1 |
|
|
|
| T allele non
carriers | 46 | 2.034
(0.857–4.844) | 0.110 | 1.440
(0.634–3.269) | 0.384 | 2.031 |
|
|
|
|
|
|
|
|
|
| (0.851–4.844) | 0.110 |
| VEGFA
rs25648 | Genotype | CC | 131 | 1 | 0.425 | 1 | 0.727 | 1 | 0.602 |
|
|
| CT | 65 | 0.604
(0.271–1.345) | 0.217 | 0.796 | 0.589 | 0.706 |
|
|
|
|
|
|
|
| (0.348–1.820) |
| (0.274–1.819) | 0.471 |
|
|
| TT | 6 | 1.373
(0.185–10.181) | 0.756 | 1.706 | 0.603 | 1.861 | 0.548 |
|
|
|
|
|
|
| (0.228–12.794) |
| (0.246–14.087) |
|
|
| Allelic | T allele
carriers | 71 | 1 |
| 1 |
| 1 |
|
|
|
| T allele non
carriers | 131 | 1.553
(0.721–3.343) | 0.261 | 1.181 | 0.681 | 1.291 | 0.577 |
|
|
|
|
|
|
| (0.534–2.612) |
| (0.526–3.168) |
|
|
|
| C allele
carriers | 196 | 1 |
| 1 |
| 1 |
|
|
|
| C allele non
carriers | 6 | 1.593
(0.217–11.703) | 0.647 | 1.835 | 0.552 | 2.083 | 0.474 |
|
|
|
|
|
|
| (0.248–13.556) |
| (0.280–15.494) |
|
By analysing the SNPs of the IL-6 gene, we
found that the rs1800797 GG genotype was a negative prognostic
factor for DFS and MFS. Furthermore, IL-6 rs1800797 GG and
IL-6 rs1800795 GG genotype carriers displayed a shorter OS
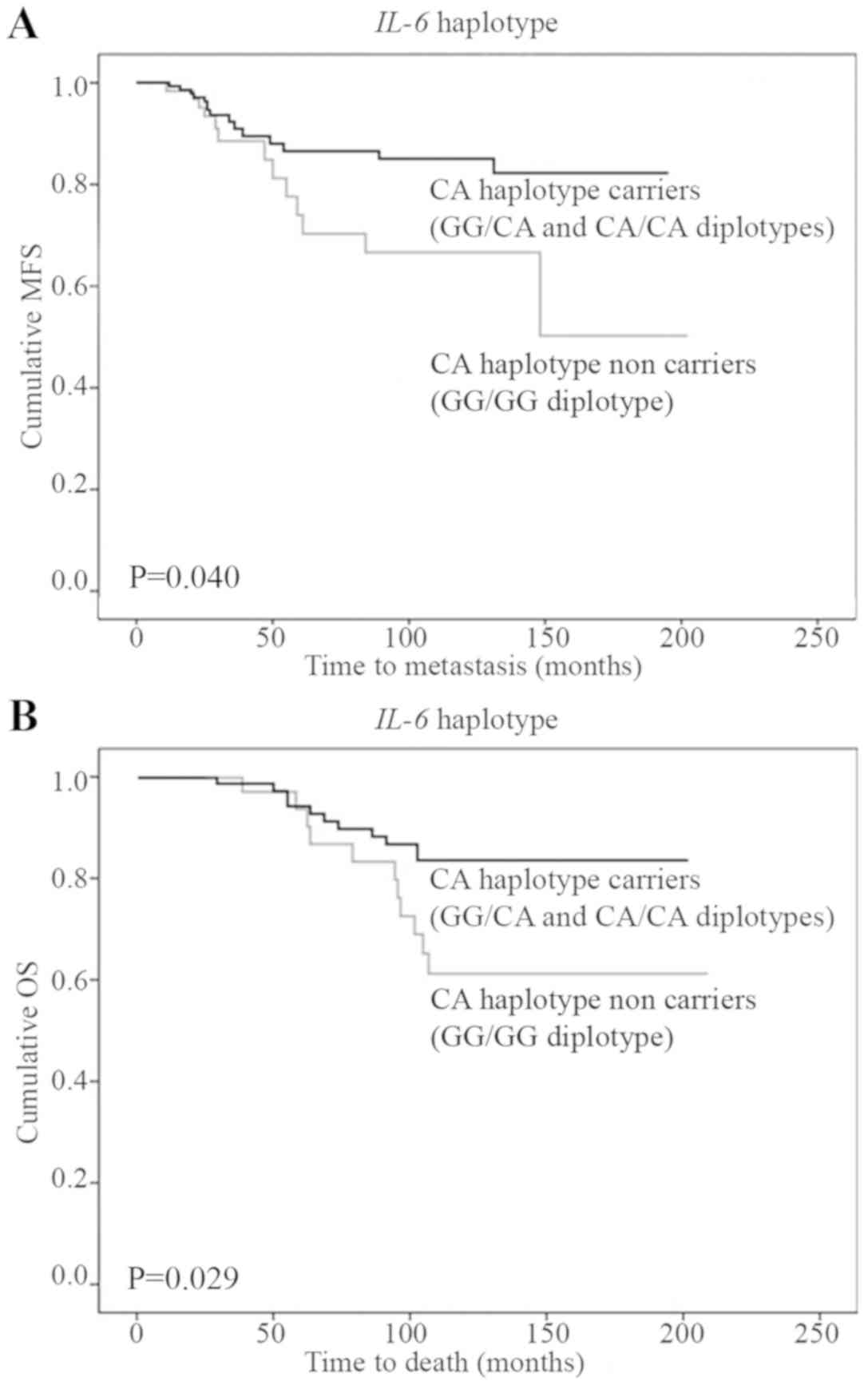
(Table IV). In a haplotype model,
the patients who inherited the GG/GG diplotype (AC haplotype
non-carriers) had a higher risk of disease recurrence, metastasis
and death (Table IV; Fig. 3A and B).
 | Table IV.Cox's univariate model for
IL-6 SNPs. |
Table IV.
Cox's univariate model for
IL-6 SNPs.
|
|
|
|
| PFS | MFS | OS |
|---|
|
|
|
|
|
|
|
|
|---|
| Reference SNP
ID | Model |
Genotype/allele/haplotype | Patients, n | Univariate hazard
ratio (95% CI) | P-value | Univariate hazard
ratio (95% CI) | P-value | Univariate hazard
ratio (95% CI) | P-value |
|---|
| IL-6
rs1800795 | Genotype | CC | 48 | 1 | 0.239 | 1 | 0.160 | 1 | 0.101 |
|
|
| GC | 95 | 1.441
(0.518–4.010) | 0.484 | 0.983
(0.336–2.881) | 0.972 | 1.216
(0.321–4.560) | 0.779 |
|
|
| GG | 49 | 2.257
(0.811–6.277) | 0.119 | 2.053
(0.736–5.761) | 0.174 | 2.842
(0.798–10.193) | 0.109 |
|
| Allelic | C allele
carriers | 143 | 1 |
| 1 |
| 1 |
|
|
|
| C allele non
carriers | 49 | 1.750
(0.874–3.502) | 0.114 | 2.074
(0.983–4.375) | 0.055 | 2.484
(1.075–5.737) | 0.033a |
|
|
| G allele
carriers | 144 | 1 |
| 1 |
| 1 |
|
|
|
| G allele non
carriers | 48 | 0.568
(0.219–1.473) | 0.245 | 0.716
(0.272–1.888) | 0.499 | 0.552
(0.163–1.866) | 0.339 |
| IL-6
rs1800797 | Genotype | AA | 46 | 1 | 0.170 | 1 | 0.136 | 1 | 0.117 |
|
|
| GA | 95 | 1.724
(0.621–4.787) | 0.296 | 1.173
(0.406–3.441) | 0.771 | 1.536
(0.418–5.783) | 0.528 |
|
|
| GG | 61 | 2.596
(0.934–7.215) | 0.067 | 2.361
(0.841–6.627) | 0.104 | 3.174
(0.858–11.355) | 0.077 |
|
| Allelic | A allele
carriers | 141 | 1 |
| 1 |
| 1 |
|
|
|
| A allele non
carriers | 61 | 2.370
(1.032–5.464) | 0.044a | 2.154
(1.009–4.476) | 0.047a | 2.373
(1.033–5.461) | 0.044a |
|
|
| G allele
carriers | 156 | 1 |
| 1 |
| 1 |
|
|
|
| G allele non
carriers | 46 | 0.483
(0.186–1.251) | 0.134 | 0.610
(0.232–1.606) | 0.317 | 0.458
(0.135–1.548) | 0.209 |
| IL-6
ts1800797- rs1800795 | Diplotype | AC/AC | 48 | 1 | 0.339 | 1 | 0.292 | 1 | 0.250 |
|
|
| GG/AC | 80 | 1.666
(0.599–4.634) | 0.328 | 1.125
(0.383–3.305) | 0.830 | 1.141
(0.382–5.434) | 0.589 |
|
|
| GG/GG | 63 | 2.146
(0.772–5.967) | 0.143 | 1.953
(0.695–5.485) | 0.204 | 2.555
(0.711–9.161) | 0.150 |
|
| Haplotype | AC haplotype
carriers | 136 | 1 |
| 1 |
| 1 |
|
|
|
| AC haplotype non
carriers | 60 | 1.809
(0.905–3.617) | 0.094 | 2.142
(1.021–4.514) | 0.045a | 2.455
(1.062–5.661) | 0.035a |
|
|
| GG haplotype
carriers | 149 | 1 |
| 1 |
| 1 |
|
|
|
| GG haplotype non
carriers | 48 | 1.818
(0.701–4.713) | 0.219 | 1.438
(0.546–3.791) | 0.462 | 1.894
(0.560–6.401) | 0.304 |
In a multivariate Cox regression model including age
at diagnosis, tumour size, lymph node status, tumour
differentiation grade, oestrogen receptor, progesterone receptor
and HER2 status, the IL-1α 1800587 CC genotype remained a
significant predictor of poor DFS, MFS and OS. Furthermore, the
IL-6 GG/GG diplotype (non-carrying the AC haplotype) was an
independent negative prognostic factor for MFS and OS (Table VI). Finally, other polymorphisms and
VEGFA haplotypes were not associated with any of the
survival endpoints.
 | Table VI.Cox's multivariate model. |
Table VI.
Cox's multivariate model.
|
|
| PFS | MFS | OS |
|---|
|
|
|
|
|
|
|---|
| Variable | Patients, n | Multivariate hazard
ratio (95% CI) | P-value | Multivariate hazard
ratio (95% CI) | P-value | Multivariate hazard
ratio (95% CI) | P-value |
|---|
| Age at diagnosis,
years |
|
|
|
|
|
|
|
|
≥50 | 71 | 1 |
| 1 |
| 1 |
|
|
<50 | 131 | 7.523 | 0.049a | 6.081 | 0.082 | 9.237 | 0.091 |
|
|
| (0.990–56.690) |
| (0.786–46.619) |
| (0.983–57.232) |
|
| Tumor size, cm |
|
|
|
|
|
|
|
|
<2 | 129 | 1 |
| 1 |
| 1 |
|
|
2–5 | 73 | 1.102 | 0.795 | 1.018 | 0.631 | 1.021 | 0.669 |
|
|
| (0.583–2.399) |
| (0.522–1.824) |
| (0.529–1.919) |
|
| Lymph node
status |
|
|
|
|
|
|
|
|
Negative | 111 | 1 |
| 1 |
| 1 |
|
|
Positive | 91 | 2.331 | 0.030a | 2.413
(1.051–5.587) | 0.039a | 2.151
(0.832–5.560) | 0.115 |
|
|
| (1.086–4.989) |
|
|
|
|
|
| Grade |
|
|
|
|
|
|
|
| G1 and
G2 | 158 | 1 |
| 1 |
| 1 |
|
| G3 | 44 | 2.281 | 0.079 | 2.145 | 0.123 | 1.904 | 0.247 |
|
|
| (0.916–5.719) |
| (0.815–5.628) |
| (0.636–5.645) |
| Estrogen receptor
status |
|
|
|
|
|
|
|
|
Positive | 137 | 1 |
| 1 |
| 1 |
|
|
Negative | 65 | 1.182 | 0.721 | 1.304 | 0.594 | 1.580 | 0.395 |
|
|
| (0.482–2.939) |
| (0.496–3.455) |
| (0.545–4.555) |
|
| Progesterone
receptor status |
|
|
|
|
|
|
|
|
Positive | 121 | 1 |
| 1 |
| 1 |
|
|
Negative | 82 | 3.088 | 0.020a | 3.284 | 0.025a | 4.670 | 0.018a |
|
|
| (1.193–7.993) |
| (1.164–9.228) |
| (1.315–16.435) |
|
| Human epidermal
growth factor |
|
|
|
|
|
|
|
| receptor 2
status |
|
|
|
|
|
|
|
|
Negative | 164 | 1 |
| 1 |
| 1 |
|
|
Positive | 38 | 2.393 | 0.104 | 1.904 | 0.246 | 1.718 |
|
|
|
| (0.841–6.817) |
| (0.641–5.621) |
| (0.518–5.735) | 0.376 |
| IL-6 haplotype |
|
|
|
|
|
|
|
| AC
haplotype carriers (GG/AC+AC/AC) | 128 | – |
| 1 |
| 1 |
|
| AC
haplotype non carriers (GG/GG) | 63 | – | – | 2.750
(1.184–7.690) | 0.048 | 2.631
(1.042–7.670) | 0.049 |
| IL-1α
rs1800587 |
| 1 |
| 1 |
| 1 |
|
| T
allele carriers (CT+TT genotypes) | 105 | 2.704
(1.280–5.683) | 0.009a | 3.442
(1.484–7.994) | 0.004a | 3.181
(1.252–8.134) | 0.016a |
| T
allele non carriers (CC genotype) | 97 | – | – | 2.750
(1.184–7.690) | 0.048a | 2.631
(1.042–7.670) | 0.049 a |
Discussion
In the present study, we investigated the
associations between nine functional SNPs in four cytokine genes
(i.e., VEGFA, IL-1α, IL-1β and IL-6) and the
clinicopathologic profiles and survival rates in a group of
Lithuanian women with early-stage BC. We found an association
between the IL-6 SNPs and the oestrogen receptor positive,
progesterone receptor positive and HER2 negative status, and a link
between IL-1α SNP and larger primary tumour size. Furthermore, we
confirmed a negative prognostic value of the IL-6
rs1800797-rs1800795 GG/GG diplotype on MFS and OS and of the
IL-1α rs1800587 CC genotype on DFS, MFS and OS in a highly
homogeneous group of patients.
Several authors have demonstrated that carrying the
IL-6 rs1800795 G or IL-6 rs1800795 G alleles is
associated with higher IL-6 protein production (19–21).
Specifically, IL-6 is a cytokine which is considered a prognostic
marker as well as a potential therapeutic target for BC patients.
This cytokine acts through several pathways, regulating the
proliferation, apoptosis and metabolism of BC cells. However, the
most important role of IL-6 in breast carcinogenesis is its
potential to induce breast metastasis by enhancing angiogenesis and
tumour cell migration (39).
According to previous studies, IL-6 rs1800797 and rs1800795
polymorphisms appear to be biologically important; however, the
data on their clinical importance is still heterogeneous.
The association of IL-6 polymorphisms with BC
survival was analysed in several studies (Table VII). Snoussi et al (32) and DeMichele et al (27) analysed patients with non-metastatic
stage I–III BC. In both studies the IL-6 rs1800795 GG
genotype was associated with decreased DFS and OS. In DeMichele
et al (27) the presence of
at least one copy of the haplotype
rs1800797G-rs1800796G-373(10A/11T)-rs1800795G was associated with
worse DFS. Snoussi et al (32) also found a significant link between
IL-6 rs1800797 GG polymorphism and decreased DFS and OS.
However, in DeMichele et al (28), where only locally advanced stage III
patients with more than 10 positive lymph nodes were studied, the
results were not replicated. Specifically, there was no impact of
IL-6 polymorphisms on DFS and OS in the study population. In the
same study, only an unplanned sub-analysis showed an association of
the rs1800795 GG and rs1800797 GG genotypes with lower DFS but not
OS in the oestrogen receptor positive patient subgroup. Our study
is the first to confirm the prognostic value of IL-6 polymorphisms
(namely, the IL-6 GG/GG diplotype) on MFS and OS in
early-stage BC. Therefore, IL-6 GG/GG may potentially be
used for the selection of patients who need intensified adjuvant BC
treatment.
 | Table VII.Studies evaluating the assocaitions
of IL-6 rs1800795, IL-6 rs1800797 and IL-1α rs1800587 single
nucleotide polymorphisms with breast cancer survival. |
Table VII.
Studies evaluating the assocaitions
of IL-6 rs1800795, IL-6 rs1800797 and IL-1α rs1800587 single
nucleotide polymorphisms with breast cancer survival.
| Author, year | Polymorphism | Study design | Number (inclusion
criteria) | Ethnicity | Finding | (Refs.) |
|---|
| Abana et al,
2017 | IL-6
rs1800795 | Case/control | 157/158 (stage IV
vs. stage I–III) | Non-Hispanic
white | GG genotype with
distant metastasis | (26) |
| DeMichele et
al, 2003 | IL-6
rs1800795 | Cohort | 80 (stage
I–III) | Various | GG genotype with
worse DFS and OS | (27) |
| DeMichele et
al, 2009 | IL-6
rs1800795, rs1800797 | Cohort | 346
(non-metastatic, ≥10 positive nodes) | Various | GG genotypes of
both SNPs with worse DFS in estrogen receptor positive
subgroup | (28) |
| Markkula et
al, 2014 | IL-6
rs1800795 | Cohort | 634 (stage
I–III) | No data | C allele in
estrogen receptor negative cases with early events | (29) |
| Snoussi et
al, 2005 | IL-6
rs1800795, rs1800797 | Cohort | 305 (stage
I–III) | North African | GG genotypes in
both SNPs with worse DFS, but not OS | (32) |
|
| IL-1α
rs1800587 |
|
|
| TT genotype with
worse DFS and OS |
|
| Grimm et al,
2009 | IL-1α
rs1800587 | Cohort | 262 (stage
I–III) | Caucasian | C allele with OS in
the univariate analysis only | (31) |
| Escala-Garcia et
al, 2019 | GWAS | Cohort | 96.661 (stage
I–IV) | Various | No association of
IL-1α and IL-6 SNPs with breast cancer-specific mortality | (23) |
| Khan et al,
2017 | GWAS | Meta-analysis | 3.136 (stage I–IV
estrogen receptor positive) | Various | No association of
IL-1α and IL-6 SNPs with breast cancer-specific mortality | (24) |
| Present study | IL-6
rs1800795, rs1800797 | Cohort | 202 (early stage
I–II) |
Eastern-European | GG/GG diplotype
with worse MFS (Lithuanian) and OS | – |
|
| IL-1α
rs1800587 |
|
|
| CC genotype with
worse DFS, MFS and OS |
|
The above-mentioned studies by Snoussi et al
(32) and DeMichele et al
(27,28) did not analyse MFS as a survival
endpoint. To the best of our knowledge, the only study that
evaluated the association between IL-6 polymorphisms and BC
metastasis was a case-control study conducted by Abana et al
(26). The authors demonstrated the
significant link between the IL-6 rs1800795 GG genotype and
the development of BC metastases with an OR of 1.52. However, due
to case-control design, we have no data on MFS differences. In our
study IL-6 GG/GG predicted MFS and OS, but not DFS. Taking
into account the critical role of IL-6 in the development of BC
metastasis and our study results, we propose the hypothesis that
IL-6 polymorphisms may predict the development of metastasis rather
than loco-regional relapse.
It is important to mention that, in contrast with
the findings of the above-mentioned studies, Markkula et al
(29), who analysed a Swedish cohort
of patients with any primary stage non-metastatic BC, demonstrated
that the carriers of the C allele in rs1800795 SNP had a higher
risk of early BC events. The authors, however, conducted no DFS,
MFS or OS analyses. Furthermore, none of the GWAS studies showed a
statistically significant association between IL-6
polymorphisms and BC survival (23,24). The
contrasting findings in IL-6 polymorphism studies may be due
to sampling errors or differences in patient ethnicity.
Nevertheless, most of the experimental data and the possible
biological pathway support our findings.
IL-6 also acts as a regulator of oestrogen synthesis
and may modulate the tumour cell growth related to the hormone
receptor status (40).
Hormone-sensitive cells exhibit a higher response to IL-6, while
ER-negative cells are suppressed by IL-6 (41). However, there is no clear mechanism
of association between IL-6 SNPs and the oestrogen receptor
positive, progesterone receptor positive and HER2 negative status.
Therefore, further studies are needed.
As far as IL-1α SNP is concerned, Um et
al (18) demonstrated that the
IL-1α rs1800587 CC genotype is associated with higher
transcriptional activity of the IL−1α gene. Overexpression
of the IL-1α promotes tumour invasiveness and metastasis by
inducing the expression of angiogenic genes and growth factors
(12). A large meta-analysis
performed by Xia et al (30)
showed that the IL-1α rs1800587 C allele and CC genotype are
associated with increased cancer risk in general. The studies which
investigated the associations between IL-1α rs1800587
polymorphism and BC survival are presented in Table VII. Grimm et al (31) analysed 262 Caucasian BC patients and
found that the IL-1α rs1800758 C allele in the univariate
model is associated with OS; however, the multivariate model failed
to repeat the association. In our study, the multivariate survival
analysis confirmed the statistically significant impact of the CC
genotype on DFS, MFS and OS. Contrasting data is provided by
Snoussi et al (32), who
analysed North African BC patients and found that the rs1800587
homozygous TT genotype showed a significant association with
reduced DFS and OS rate. However, the allele and genotype frequency
of rs1800587 SNP in African population differs substantially from
that in the population with European ancestry. This difference may
be the cause of the observed inconsistency between these studies.
We demonstrated that in an Eastern-European population the
IL-1α rs1800758 C allele is associated with more aggressive
local disease (i.e., larger tumour size) and for the first time we
proposed that the IL-1α rs1800758 CC genotype is an
independent negative prognostic biomarker for early-stage BC. As
the allele frequencies of the IL-1α rs1800758 SNP in our
study correspond to those of the 1000 Genomes project phase 3
database for European population, these findings could potentially
be replicated for European population, but larger confirmatory
studies are warranted.
Although experimental data suggests that
VEGFA and IL-1β play a role in BC, our findings,
along with the results from several other studies (22–24), do
not suggest that several common polymorphisms in these genes are
associated with BC clinical and morphological variables and BC
survival rates.
Potential limitations of our study include a limited
sample size, lack of access to tumour and/or tumour stromal tissue
(which would have allowed for the assessment of the presence of
SNPs in those tissues), the risk of other confounders, possible
gene-gene and gene-environment interactions, and non-random
sampling. We also acknowledge that VEGFA, IL-1α, IL-1β and
IL-6 measurements were not available in the current study.
However, our study supports the relevance of functional IL-6
and IL-1α germline polymorphisms to BC prognosis. Further
investigations, preferable on larger cohorts with different ethnic
origins, are needed to confirm the results of the current
study.
In conclusion, we found an association between the
IL−1α rs1800587 C allele and larger primary tumour size. The
IL-6 rs1800797 A allele, GA genotype, IL-6 rs1800795
C allele and IL-6 (rs1800797-re1800795) AC diplotype were
linked to hormonal receptor positive BC. Additionally, IL-6
rs1800797 A was associated with HER2 negative status. The
multivariate IL-1α rs1800587 CC genotype was confirmed as an
independent negative prognostic factor for DFS, MFS and OS, and the
IL6 GG/GG diplotype for MFS and OS in early-stage BC patients. Our
findings confirm the hypothesis that functional SNPs in
angiogenesis- and inflammation-associated genes are associated with
early-stage BC prognosis in Lithuanian population.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was partially supported by the
Lithuanian University of Health Sciences Science Foundation.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
EK, RU and EJu designed the research study. EK, RU,
RI, LP, VR, EJa and EJu performed the research. EK and RU analyzed
the data. RU and RI contributed essential reagents or tools. EK
wrote the manuscript.
Ethics approval and consent to
participate
The present study was approved by Kaunas Regional
Ethics Committee for Biomedical Research (protocol no. BE-2-10).
Informed consent for participation in the study and use of tissue
was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BC
|
breast cancer
|
|
SNP
|
single nucleotide polymorphism
|
|
VEGFA
|
vascular endothelial growth factor
A
|
|
IL-6
|
interleukin-6
|
|
IL-1α
|
interleukin-1α
|
|
IL-1β
|
interleukin-1β
|
|
A
|
adenine
|
|
G
|
guanine
|
|
T
|
thymine
|
|
C
|
cytosine
|
|
DFS
|
disease-free survival
|
|
MFS
|
metastasis-free survival
|
|
OS
|
overall survival
|
|
HR
|
hazard ratio
|
|
OR
|
odds ratio
|
|
CI
|
confidence interval
|
|
HER2
|
human epidermal growth factor
receptor 2
|
|
GWAS
|
genome-wide association study
|
References
|
1
|
IARC: World Cancer Report 2014. Stewart BW
and Wild CP: IARC; Lyon: 2014
|
|
2
|
Cancer Trends Progress Report. National
Cancer Institute, NIH, DHHS; Bethesda, MD: 2019, https://progressreport.cancer.gov
|
|
3
|
Zhao Y and Adjei AA: Targeting
angiogenesis in cancer therapy: Moving beyond vascular endothelial
growth factor. Oncologist. 20:660–673. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bielenberg DR and Zetter BR: The
contribution of angiogenesis to the process of metastasis. Cancer
J. 21:267–273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cartland SP, Genner SW, Zahoor A and
Kayurma MM: Comparative evaluation of TRAIL, FGF-2 and
VEGF-A-induced angiogenesis in vitro and in vivo. Int J Mol Sci.
17:E20252016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Palma M, Biziato D and Petrova TV:
Microenvironmental regulation of tumour angiogenesis. Nat Rev
Cancer. 17:457–474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marech I, Leporini C, Ammendola M,
Porcelli M, Gadaleta CD, Russo E, De Sarro G and Ranieri G:
Classical and non-classical proangiogenic factors as a target of
antiangiogenic yherapy in tumor microenvironment. Cancer Lett.
380:216–226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Banys-Paluchoothtwski M, Witzel I,
Riethdorf S, Pantel K, Rack B, Janni W, Fasching PA, Aktas B,
Kasimir-Bauer S, Hartkopf A, et al: The clinical relevance of serum
vascular endothelial growth factor (VEGF) in correlation to
circulating tumor cells and other serum biomarkers in patients with
metastatic breast cancer. Breast Cancer Res Treat. 172:93–104.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gyamfi J, Eom M, Koo JS and Choi J:
Multifaceted roles of interleukin-6 in adipocyte-breast cancer cell
interaction. Transl Oncol. 11:275–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Knüpfer H and Preiss R: Significance of
interleukin-6 (IL-6) in breast cancer (Review). Breast Cancer Res
Treat. 102:129–135. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tawara K, Scott H, Emathinger J, Ide A,
Fox R, Greiner D, LaJoie D, Hedeen D, Nandakumar M, Oler AJ, et al:
Co-expression of VEGF and IL-6 family cytokines is associated with
decreased survival in HER2 negative breast cancer patients:
Subtype-specific IL-6 family cytokine-mediated VEGF secretion.
Transl Oncol. 12:245–255. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Litmanovich A, Khazim K and Cohen I: The
role of interleukin-1 in the pathogenesis of cancer and its
potential as a therapeutic target in clinical practice. Oncol Ther.
6:109–127. 2018. View Article : Google Scholar
|
|
13
|
Balasubramanian SP, Azmy IA, Higham SE,
Wilson AG, Cross SS, Cox A, Brown NJ and Reed MW: Interleukin gene
polymorphisms and breast cancer: A case control study and
systematic literature review. BMC Cancer. 6:1882006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koukourakis MI, Papazoglou D,
Giatromanolaki A, Bougioukas G, Maltezos E and Siviridis E: VEGF
gene sequence variation defines VEGF gene expression status and
angiogenic activity in non-small cell lung cancer. Lung Cancer.
46:293–298. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Watson CJ, Webb NJ, Bottomley MJ and
Brenchley PE: Identification of polymorphisms within the vascular
endothelial growth factor (VEGF) gene: Correlation with variation
in VEGF protein production. Cytokine. 12:1232–1235. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ray D, Mishra M, Ralph S, Read I, Davies R
and Brenchley P: Association of the VEGF gene with proliferative
diabetic retinopathy but not proteinuria in diabetes. Diabetes.
53:861–864. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hall SK, Perregaux DG, Gabel CA, Woodworth
T, Durham LK, Huizinga TW, Breedveld FC and Seymour AB: Correlation
of polymorphic variation in the promoter region of the
interleukin-1 beta gene with secretion of interleukin-1 beta
protein. Arthritis Rheum. 50:1976–1983. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Um JY, Rim HK, Kim SJ, Kim HL and Hong SH:
Functional polymorphism of IL-1 alpha and its potential role in
obesity in humans and mice. PLoS One. 6:e295242011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rana BK, Flatt SW, Health DD, Pakiz B,
Quintana EL, Natarajan L and Rock CL: The IL-6 gene promoter SNP
and plasma IL-6 in response to diet intervention. Nutrients.
9:E5522017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Boeta-Lopez K, Duran J, Elizondo D,
Gonzales E, Rentfro A, Schwarzbach AE and Nair S: Association of
interleukin-6 polymorphisms with obesity or metabolic traits in
young Mexican-Americans. Obes Sci Pract. 4:85–96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sampaio AM, Balseiro SC, Silva MR, Alarcão
A, d'Aguiar MJ, Ferreira T and Carvalho L: Association between IL-4
and IL-6 expression variants and gastric cancer among portuguese
population. GE Port J Gastroenterol. 22:143–152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Langsenlehner U, Hofmann G, Renner W,
Gerger A, Krenn-Pilko S, Thurner EM, Krippl P and Langsenlehner T:
Association of vascular endothelial growth factor--a gene
polymorphisms and haplotypes with breast cancer metastases. Acta
Oncol. 54:368–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Escala-Garcia M, Guo Q, Dörk T, Canisius
S, Keeman R, Dennis J, Beesley J, Lecarpentier J, Bolla MK, Wang Q,
et al: Genome-wide association study of germline variants and
breast cancer-specific mortality. Br J Cancer. 120:647–657. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Khan S, Fagerholm R, Kadalayil L, Tapper
W, Aittomäki K, Liu J, Blomqvist C, Eccles D and Nevanlinna H:
Meta-analysis of three genome-wide association studies identifies
two loci that predict survival and treatment outcome in breast
cancer. Oncotarget. 9:4249–4257. 2017.PubMed/NCBI
|
|
25
|
Peng X, Shi J, Sun W, Ruan X, Guo Y, Zhao
L, Wang J and Li B: Genetic polymorphisms of IL-6 promoter in
cancer susceptibility and prognosis: A meta-analysis. Oncotarget.
9:12351–12364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Abana CO, Bingham BS, Cho JH, Graves AJ,
Koyama T, Pilarski RT, Chakravarthy AB and Xia F: IL-6 variant is
associated with metastasis in breast cancer patients. PLoS One.
12:e01817252017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
DeMichele A, Martin AM, Mick R, Gor P,
Wray L, Klein-Cabral M, Athanasiadis G, Colligan T, Stadtmauer E
and Weber B: Interleukin-6 −174G->C polymorphism is associated
with improved outcome in high-risk breast cancer. Cancer Res.
63:8051–8056. 2003.PubMed/NCBI
|
|
28
|
DeMichele A, Gray R, Horn M, Chen J,
Aplenc R, Vaughan WP and Tallman MS: Host genetic variants in the
interleukin-6 promoter predict poor outcome in patients with
estrogen receptor-positive, node-positive breast cancer. Cancer
Res. 69:4184–4191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Markkula A, Simonsson M, Ingvar C, Rose C
and Jernström H: IL6 genotype, tumour ER-status, and treatment
predicted disease-free survival in a prospective breast cancer
cohort. BMC Cancer. 14:7592014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xia H, Chen Y, Meng J and Liang C: Effect
of polymorphisms on IL1A to cancer susceptibility: Evidence based
on 34,016 subjects. Artif Cells Nanomed Biotechnol. 47:3138–3152.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grimm C, Kantelhardt E, Heinze G,
Polterauer S, Zeillinger R, Kölbl H, Reinthaller A and Hefler L:
The prognostic value of four interleukin-1 gene polymorphisms in
Caucasian women with breast cancer: A multicenter study. BMC
Cancer. 9:782009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Snoussi K, Strosberg AD, Bouaouina N, Ben
Ahmed S and Chouchane L: Genetic variation in pro-inflammatory
cytokines (interleukin-1beta, interleukin-1alpha and interleukin-6)
associated with the aggressive forms, survival, and relapse
prediction of breast carcinoma. Eur Cytokine Netw. 16:253–260.
2005.PubMed/NCBI
|
|
33
|
Hayes DF, Ethier S and Mippman ME: New
guidelines for reporting of tumor marker studies in breast cancer
research and treatment: REMARK. Breast Cancer Res Treat.
100:237–238. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McShane LM, Altman DG, Sauerbrei W, Taube
SE, Gion M and Clark GM; Statistics Subcommittee of the NCI-EORTC
Working Group on Cancer Diagnostics, : Reporting recommendations
for tumor marker prognostic studies (REMARK). J Natl Cancer Inst.
97:1180–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
1000 Genomes Project Consortium, ; Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini
JL, McCarthy S, McVean GA and Abecasis GR: A global reference for
human genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stephens M, Smith NJ and Donnelly P: A new
statistical method for haplotype reconstruction from population
data. Am J Hum Genet. 68:978–989. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stephens M and Scheet P: Accounting for
decay of linkage disequilibrium in haplotype inference and
missing-data imputation. Am J Hum Genet. 76:449–462. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumari N, Dwarakanath BS, Das A and Bhatt
AN: Role of interleukin-6 in cancer progression and therapeutic
resistance. Tumour Biol. 37:11553–11572. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dethlefsen C, Højfeldt G and Hojman P: The
role of intratumoral and systemic IL-6 in breast cancer. Breast
Cancer Res Treat. 138:657–664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Masjedi A, Hashemi V, Hojjat-Farsangi M,
Ghalamfarsa G, Azizi G, Yousefi M and Jadidi-Niaragh F: The
significant role of IL-6 and its signaling pathway in the
immunopathogenesis and treatment of breast cancer. Biomed
Pharmacother. 108:1415–1424. 2018. View Article : Google Scholar : PubMed/NCBI
|