Introduction
Bisphenol A (BPA) is known endocrine-disrupting
chemical (1,2). Previous studies have investigated
exposure to BPA as a leading cause of the development of endocrine
diseases, including breast cancer (1–5). The use
of BPA has been identified in a variety of consumer products,
including water bottles, food/beverage containers and linings,
thermal receipts, medical equipment and paper cups (6,7). It has
been reported that the use of BPA-containing products introduces
BPA into the food chains of industrialized countries (8). It has been demonstrated that 95% of BPA
may be absorbed by the intestinal tract, when BPA enters the human
body (9). It has been indicated that
BPA binds to glucuronic acid in the liver subsequent to being
metabolized through the phase II pathway (9,10).
It has been demonstrated that fetal BPA exposure
increases the induction of mammary gland ductal hyperplasia and
carcinoma in situ (3). In a
previous study, it was identified that when pregnant mice were
treated with various doses (0–250 µg/kg) of BPA, a change in the
morphology of the mammary gland was observed, possibly due to BPA,
in addition to mammary tumorigenesis indicated in the offspring
during adulthood (1,11–13). It
has been demonstrated that perinatal exposure to BPA changes the
long-term progesterone response and increases the adult mice
mammary cell number, resulting in the development of breast cancer
(4). Previous studies have
determined specific molecular alterations in the presence of BPA in
high-risk breast tissues. They used non-malignant random
periareolar fine-needle aspirates from unafflicted and
contralateral breast tissue of the primary breast lesion and
determined that the BPA response profile was significantly
associated with breast tumors that were characterized by an
advanced histological grade, large tumor size and poor patient
survival rate (14–16). Another study identified that BPA
increased antioxidant proteins, including B-cell lymphoma 2
apoptosis regulator and superoxidase dismutase 1, and activated
epidermal growth factor receptor pathways, inducing inflammatory
breast cancer proliferation (17).
Forkhead box protein A1 (FOXA1) has been identified as a
hormone-response mediator in breast cancer (18). It has been identified that BPA may
downregulate FOXA1, inducing mesenchymal transition and promoting
the in vitro motility of estrogen receptor (ER)-negative
breast cancer cells (19).
In rodents, perinatal exposure to low doses of BPA
has been demonstrated to be associated with weight gain and the
metabolic syndrome (MetSyn) when fed on a normal diet and more
severe metabolic diseases when fed on a high-fat diet (20). Urinary BPA has been indicated to be
associated with general and central obesity (21), and may be further associated with
Type 2 diabetes mellitus (22,23).
The aforementioned trends are generating the
so-called ‘diabesity phenotype’ (24). A previous study indicated that, owing
to the pregnant mice's exposure to BPA, the male offspring
presented symptoms of diabesity (25). Another study indicated that BPA may
disrupt the functions of pancreatic β-cells, adipocytes and
hepatocytes, which are all targets of BPA, indicating an
association with metabolic diseases, including diabesity (26). Williams (27) indicated that exposure to
xenoestrogens, including BPA, may result in obesity, Type 2
diabetes, breast cancer, uterine overgrowth and prostate cancer.
Obesity has also been reported to be associated with breast cancer
(28). It has been identified that
central obesity may be associated with breast cancer in
premenopausal women (28). It has
been reported that the presence of high blood pressure, insulin
resistance and visceral obesity may constitute as a MetSyn, thus
suggesting an association with aggressive breast cancer (29). Visceral adiposity and MetSyn have
been indicated to be associated with increased tumor aggressiveness
and distant metastasis of breast cancer (30,31).
It has been suggested that BPA may exacerbate the
risk of obesity, which may lead to an increase in breast cancer.
Therefore, we hypothesized that BPA-associated pathways and
candidate genes are developmentally active in breast cells, and
that these molecular markers are associated with breast cancer
susceptibility and aggressiveness. To test the aforementioned
hypothesis, microarray datasets from an identical platform were
used to determine gene sets that are associated with the
development and aggressiveness of various types of breast cancer. A
total of three different statistical comparison procedures were
applied to test the hypothesis. Common gene sets were identified,
which were significantly expressed by comparing BPA-exposed cells
with breast tissue datasets and identified potential pathways that
associate BPA exposure with breast cancer. In addition, the common
gene sets that were associated with BPA-regulated high-grade breast
cancer in different patient datasets were assessed. The probes of
BPA-associated significant gene sets were further pooled to
identify significant canonical pathways in various types of breast
cancer.
Materials and methods
Microarray datasets
The six selected datasets, GSE50705 (32), GSE32158 (33), GDS4114 (34), GDS3853 (35), GSE5460 (36) and GSE6532 (37), used an identical gene expression
platform obtained using a Human Genome U133 Plus 2.0 software array
(GPL570 platform; Affymetrix; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and were organized into three types. The first
type consisted of two datasets for breast cancer MCF-7 and MCF-10
cell lines treated with BPA. ER-positive MCF-7 cells were exposed
to 1, 0.5 and 0.25 µM BPA for 48 h and were monitored on arrays
obtained from GSE50705 (32). The
normal breast epithelial MCF-10F cell line was exposed to 10 and 1
µM BPA for 2 weeks and cells were monitored on arrays obtained from
GSE32158 (33). The second type
consisted of two breast tissue datasets, GDS4114 and GDS3853,
associated with the microarray expression profiles. GDS4114
consisted of six samples of stroma-surrounding invasive type of
breast tumor and six matched samples of normal stroma. Host tissue
stroma interacting with tumor cells has been identified to be
associated with tumor progression and metastasis (34). GDS3853 consisted of 14 samples of
nine ductal carcinoma in situ and five healthy tissue
samples (35). The third type
consisted of two high-grade tissue datasets, GSE5460 and GSE6532.
The GSE5460 microarray profile consisted of 129 patients with a
tumor, including 76 ER-positive patients and 53 ER-negative
patients, which were divided into 70 high-grade and 59 low-grade
tumor samples (36). GSE6532
consisted of 16 high-grade and 54 low-grade ER-positive tumor
samples from patients undergoing tamoxifen treatment (37).
The present study was approved by the Institutional
Review Board of Kaohsiung Medical University (Kaohsiung, Taiwan).
All microarray profiles were obtained from the NCBI Gene Expression
Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) from original
submitter-supplied records, including series, samples and
platforms, and curated datasets.
Bioinformatics and statistical
methods
The normalization method of the Microarray Suite
software (version 5.0; Affymetrix; Thermo Fisher Scientific, Inc.)
was implemented, according to the manufacturer's protocols. The
BRB-Array Tools software Beta 2 (version 4.3.0; linus.nci.nih.gov/BRB-ArrayTools.html) was utilized in
the Bioconductor software suite, based on the R programming
language (38,39). The array tool program obtained the
curated Biocarta pathways from the Cancer Genome Anatomy Project
(cgap.nci.nih.gov/Pathways). For the gene
set analysis, the least squares/Kolmogorov-Smirnov permutation test
(40), Efron-Tibshirani's Gene Set
Analysis maxmean test and the 1000-permutation statistics
enrichment method were employed to identify significant gene sets.
The cut-off level for determining significant gene sets was
0.05.
In the ER-positive MCF-7 cell line microarray, a
total of 42 common gene sets were identified when exposed to 0.25,
0.5 and 1 µM BPA, with each treatment containing, out of the 271
investigated significant gene sets, 60, 55 and 66 significant gene
sets, respectively (Table I). In the
normal breast epithelial cell MCF-10F microarray, a total of 72
common gene sets were identified when exposed to 1 and 10 µM BPA,
with each condition containing, out of the 279 investigated
significant gene sets, 140 and 105 gene sets, respectively.
However, two breast tissue profiles, GDS4114 and GDS3853, contained
83 and 77 of the investigated significant gene sets, respectively
(Table I). In the primary and
untreated breast cancer array GSE5460, 31 overlapping gene sets,
from the 50 and 53 significant gene sets, were obtained from the
axillary grade (high vs. low) and ER status (positive vs. negative)
assessments, respectively. GSE6532 consisted of 73 significant gene
sets from ER-positive groups, comparing the high vs. low grade
under tamoxifen treatment (Table
I).
 | Table I.Significant gene set pathways from
six microarray datasets. |
Table I.
Significant gene set pathways from
six microarray datasets.
| Microarray
dataset | Exposure | No. of significant
gene sets/total number of gene sets investigateda | Overlapping gene
sets |
|---|
| GSE50705 | 0.25 vs. 0 µM
BPA | 60/271 | 42 |
|
| 0.5 vs. 0 µM
BPA | 55/271 |
|
|
| 1 vs. 0 µM BPA | 66/271 |
|
| GSE32158 | 1 vs. 0 µM BPA | 140/279 | 72 |
|
| 10 vs. 0 µM
BPA | 105/279 |
|
| GDS4114 | Breast cancer vs.
healthy control | 83/295 | – |
| GDS3853 | Breast cancer vs.
healthy control | 77/295 | – |
| GSE5460 | ER-positive vs.
ER-negative | 53/295 | 31 |
|
| High grade vs. low
grade | 50/295 |
|
| GSE6532 | High grade vs. low
grade | 73/297 | – |
Two class comparisons were sequentially performed
with a two-sample t-test and a gene set expression comparison
analysis to identify individual candidate genes. The following fold
changes were separately assessed under the supervised methods: BPA
exposure vs. the control (0 µM); tumor vs. normal tissue;
ER-positive vs. ER-negative and high-grade (grade III) vs.
low-grade (grade I) (36). The
selection criteria was a fold-change cut-off value ≥2 which
indicated upregulation, and ≤0.5 indicating downregulation between
the two classes, as previously described (41). This fold change was considered
significant at P<0.01 as previously described (42). Furthermore, Entrez IDs of the genes
without current gene symbols were also excluded.
Ingenuity Pathway Analysis (IPA)
The candidate genes originated from the significant
Biocarta pathways in the gene set analysis and the overlap of
individual candidate genes from the comparisons of breast cancer
datasets of BPA-treated cells. The probes of BPA-associated
significant gene sets were further pooled to present a cellular
pathway in breast cancer. IPA (2017 release; www.ingenuity.com) was performed to determine the
significant canonical pathway. The pathway was determined and
ranked on the basis of the ratio of the number of genes matched to
the curated pathways in the IPA knowledge database and the P-value
was determined by Fisher's exact test. The molecular network
indicated that relevant associations were connected to biological
interactions. The score for the aforementioned network is the
negative logarithm of the P-value calculated with the right-tailed
Fisher's exact test with a hypergeometric distribution.
Results
Gene sets associated with
BPA-associated breast cancer from individual datasets
The gene sets, which were associated with BPA
exposure were assessed in the ER-positive breast cancer cell line
MCF-7, using the BRB-Array Tool and analyzed on the basis of a
signaling network database of the Biocarta pathways. It was
indicated that seven gene sets significantly overlapped between the
MCF-7 cells exposed to BPA and the stroma of breast cancer profile
GDS4114. In addition, there were 18 overlapping gene sets between
the MCF-7 cells exposed to BPA and the breast cancer profile
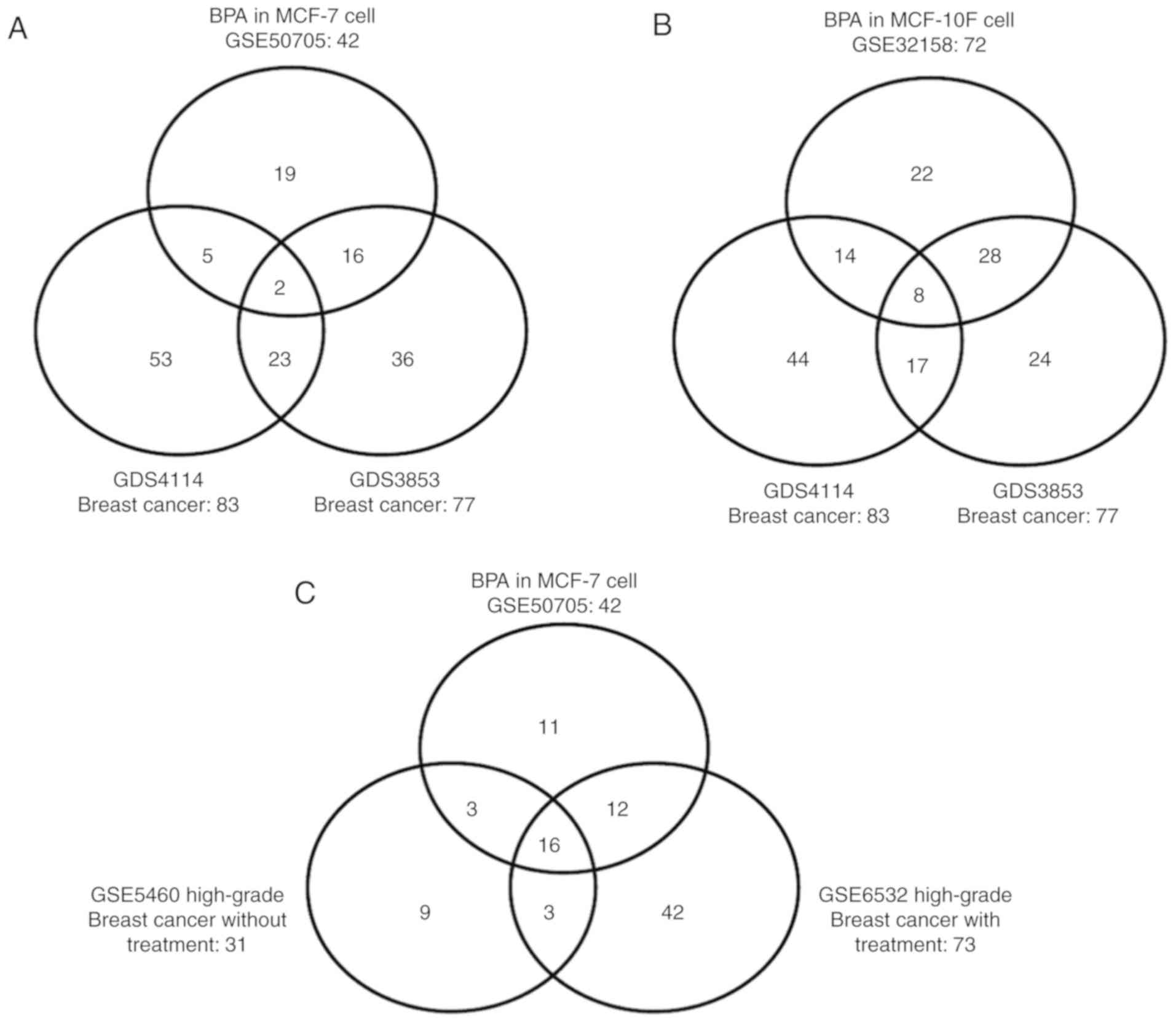
GDS3853 (Fig. 1A). Furthermore, two
consistent gene sets overlapped among all three datasets: The
visceral obesity pathway, involved in visceral fat deposits and
MetSyn, and the cell cycle pathway, involved in cyclins and cell
cycle regulation (Table II).
 | Table II.Significant pathways that link BPA
exposure to breast cancer. |
Table II.
Significant pathways that link BPA
exposure to breast cancer.
| A, MCF-7
(GSE50705), GDS4114 and GDS3853 |
|---|
|
|---|
| No. | Biocarta
pathway | Pathway
description | No. of probes |
P-valuea |
|---|
| 1 |
h_cellcyclePathway | Cyclins and cell
cycle regulation | 29 |
1.00×10-5–5.00×10-3 |
| 2 |
h_vobesityPathway | Visceral fat
deposits and the metabolic syndrome | 7 |
1.00×10-5–1.29×10-2 |
|
| B, MCF-10F
(GSE32158), GDS4114 and GDS3853 |
|
| No. | Biocarta
pathway | Pathway
description | No. of
probes |
P-valuea |
|
| 1 | h_tsp1Pathway | TSP-1 induced
apoptosis in microvascular endothelial cells | 16 |
1.00×10-5–6.51×10-3 |
| 2 | h_ctcfPathway | CTCF: First
multivalent nuclear factor | 47 |
1.00×10-5–2.44×10-2 |
| 3 |
h_extrinsicPathway | Extrinsic
prothrombin activation pathway | 9 |
2.00×10-5–2.42×10-3 |
| 4 | h_reckPathway | Inhibition of
matrix metalloproteinases | 14 |
<1.00×10-3–2.33×10-2 |
| 5 |
h_vobesityPathway | Visceral fat
deposits and the metabolic syndrome | 11 | 1.00×10-5
−1.29×10-2 |
| 6 |
h_longevityPathway | IGF-1 receptor and
longevity | 19 |
1.00×10-5–2.89×10-3 |
| 7 | h_achPathway | Role of nicotinic
acetylcholine receptors in the regulation of apoptosis | 13 |
<1.00×10-3–4.24×10-2 |
| 8 |
h_cellcyclePathway | Cyclins and cell
cycle regulation | 33 |
1.00×10-5–5.00×10-3 |
|
| C, MCF7
(GSE50705), GDS6532 and GDS5460 with high grade |
|
| No. | Biocarta
pathway | Pathway
description | No. of
probes |
P-valuea |
|
| 1 |
h_cellcyclePathway | Cyclins and cell
cycle regulation | 29 |
1.00×10-5–1.10×10-4 |
| 2 | h_EfpPathway | EFP controls the
cell cycle and breast tumor growth | 28 |
1.00×10-5–4.75×10-3 |
| 3 | h_g1Pathway | Cell cycle: G1/S
checkpoint | 49 |
1.00×10-5–2.29×10-3 |
| 4 | h_g2Pathway | Cell cycle: G2/M
checkpoint | 40 |
1.00×10-5–1.80×10-2 |
| 5 | h_mcmPathway | CDK regulation of
DNA replication | 31 |
1.00×10-5–1.00×10-5 |
| 6 | h_rbPathway | Rb tumor
suppressor/checkpoint signaling in response to DNA damage | 24 |
1.00×10-5–1.80×10-2 |
| 7 | h_cdc25Pathway | CDC25 and CHK1
regulatory pathway in response to DNA damage | 18 |
1.00×10-5–1.00×10-3 |
| 8 | h_ptc1Pathway | Sonic Hedgehog
receptor PTC1 regulates cell cycle | 16 |
1.00×10-5–7.76×10-3 |
| 9 | h_ranMSpathway | Role of Ran in
mitotic spindle regulation | 20 |
1.00×10-5–1.52×10-3 |
| 10 | h_p27Pathway | Regulation of p27
phosphorylation during cell cycle progression | 22 |
5.00×10-5–5.73×10-3 |
| 11 |
h_atrbrcaPathway | Role of BRCA1,
BRCA2 and ATR in cancer susceptibility | 45 |
5.30×10-4–1.75×10-2 |
| 12 |
h_srcRPTPPathway | Activation of Src
by protein tyrosine phosphatase α | 17 |
1.00×10-5–6.45×10-3 |
| 13 |
h_skp2e2fPathway | E2F1 destruction
pathway | 18 |
4.60×10-4–8.06×10-3 |
| 14 |
h_akap95Pathway | Role of AKAP95 in
mitosis and chromosome dynamics | 18 |
<1.00×10-3–3.75×10-2 |
| 15 | h_plk3Pathway | Regulation of cell
cycle progression by PLK3 | 15 |
2.00×10-5–3.90×10-2 |
| 16 | h_fbw7Pathway | Cyclin E
destruction pathway | 19 |
7.97×10-3–4.81×10-2 |
It was identified that 22 gene sets significantly
overlapped between the normal breast epithelial cell line, MCF-10F,
exposed to BPA and the stroma of breast cancer profile GDS4114, in
addition to 36 overlapping gene sets between the MCF-10F cells
exposed to BPA and the breast cancer profile GDS3853 (Fig. 1B). Furthermore, the following 8
consistent gene sets overlapped among all three datasets: The
thrombospondin 1 (TSP-1) pathway, where TSP-1-induced apoptosis in
microvascular endothelial cells; the CCCTC-binding factor pathway
(first multivalent nuclear factor); the extrinsic pathway
(extrinsic prothrombin activation pathway); the reversion inducing
cysteine-rich protein with Kazal motifs pathway (inhibition of
matrix metalloproteinases); the longevity pathway (insulin-like
growth factor 1 receptor and longevity); the acetylcholine pathway
(role of nicotinic acetylcholine receptors in the regulation of
apoptosis); the visceral obesity pathway (visceral fat deposits and
MetSyn), and the cell cycle pathway (cyclins and cell cycle
regulation) (Table II).
It was subsequently determined whether BPA-affected
gene sets were associated with aggressiveness in breast cancer. A
total of 16 gene sets, including the cyclins and cell cycle
regulation pathway, significantly overlapped among all three
datasets of MCF-7 cells exposed to BPA, and the high-grade breast
cancer profiles GSE5460 and GSE6532 (Fig. 1C; Table
II). In addition, the following 12 gene sets were identified in
BPA-associated aggressive tumors treated with tamoxifen: Breast
cancer-associated 1 (BRCA1), DNA repair associated-dependent
ubiquitin-ligase activity (h_bard1Pathway); apoptotic DNA
fragmentation and tissue homeostasis (h_DNAfragmentPathway); ataxia
telangiectasia mutated (ATM) serine/threonine kinase signaling
pathway (h_atmPathway); progesterone initiation of oocyte
maturation (h_mPRPathway); stathmin and breast cancer resistance to
antimicrotubular agents (h_stathminPathway); Fas cell-surface death
receptor signaling pathway (h_fasPathway); p53 signaling pathway
(h_p53Pathway); erythrocyte differentiation pathway
(h_erythPathway); signal transduction through interleukin 1
receptor (IL1R) (h_il1rPathway); the function of small leucine-rich
proteoglycan in bone (h_slrp2Pathway); nuclear factor-κB activation
by non-typeable Haemophilus influenzae (h_nthiPathway) and
the visceral fat deposits and MetSyn pathway (h_vobesityPathway).
Therefore, the visceral fat deposits and MetSyn pathway
(h_vobesityPathway), and the cyclins and cell cycle regulation
(cell cycle pathway) were involved in BPA-associated breast cancer
development and aggressiveness (Table
II).
Pooled analysis of the gene sets in
the development and aggressiveness of BPA-associated breast
cancer
The probes of BPA-associated significant gene sets
were further pooled to present a cellular pathway using IPA. The
most significant cellular pathway of ER-positive breast cancer was
the cell cycle G1/S checkpoint regulation (with GDS4114,
P=3.63×10−55 and with GDS3853, P=2.40×10−28),
whereas other statistically significant cellular pathways were
associated with cyclin and cell cycle regulation (with GDS4114,
P=7.76×10−52 and with GDS3853, P=1.74×10−35),
cell cycle G2/M damage check point regulation (with
GDS3853, P=2.14×10−37), aryl hydrocarbon receptor
signaling (with GDS4114, P=3.63×10−40 and with GDS3853,
P=4.68×10−24) and hereditary breast cancer signaling
(with GDS3853, P=1.00×10−30) (Fig. 2A). The most significant cellular
pathway from gene sets of normal breast cells and breast cancer was
the ‘molecular mechanisms of cancer’ (with GDS4114,
P=2.34×10−61 and with GDS3853, P=2.14×10−82)
(Fig. 2B) from the cellular
canonical pathways analysis. It was subsequently determined whether
BPA-regulated cellular pathways were associated with high-grade
breast cancer. The most significant cellular pathway for the
ER-positive and high-grade breast cancer was the ‘molecular
mechanisms of cancer’ (with GSE5460, P=3.16×10−63 and
with GSE6532, P=4.68×10−84) (Fig. 2C). Numerous cell cycle pathways,
including cyclin and cell cycle regulation, G2/M damage
and G1/S checkpoint regulation, for the ER-positive and
high-grade breast cancer were also observed (Fig. 2C).
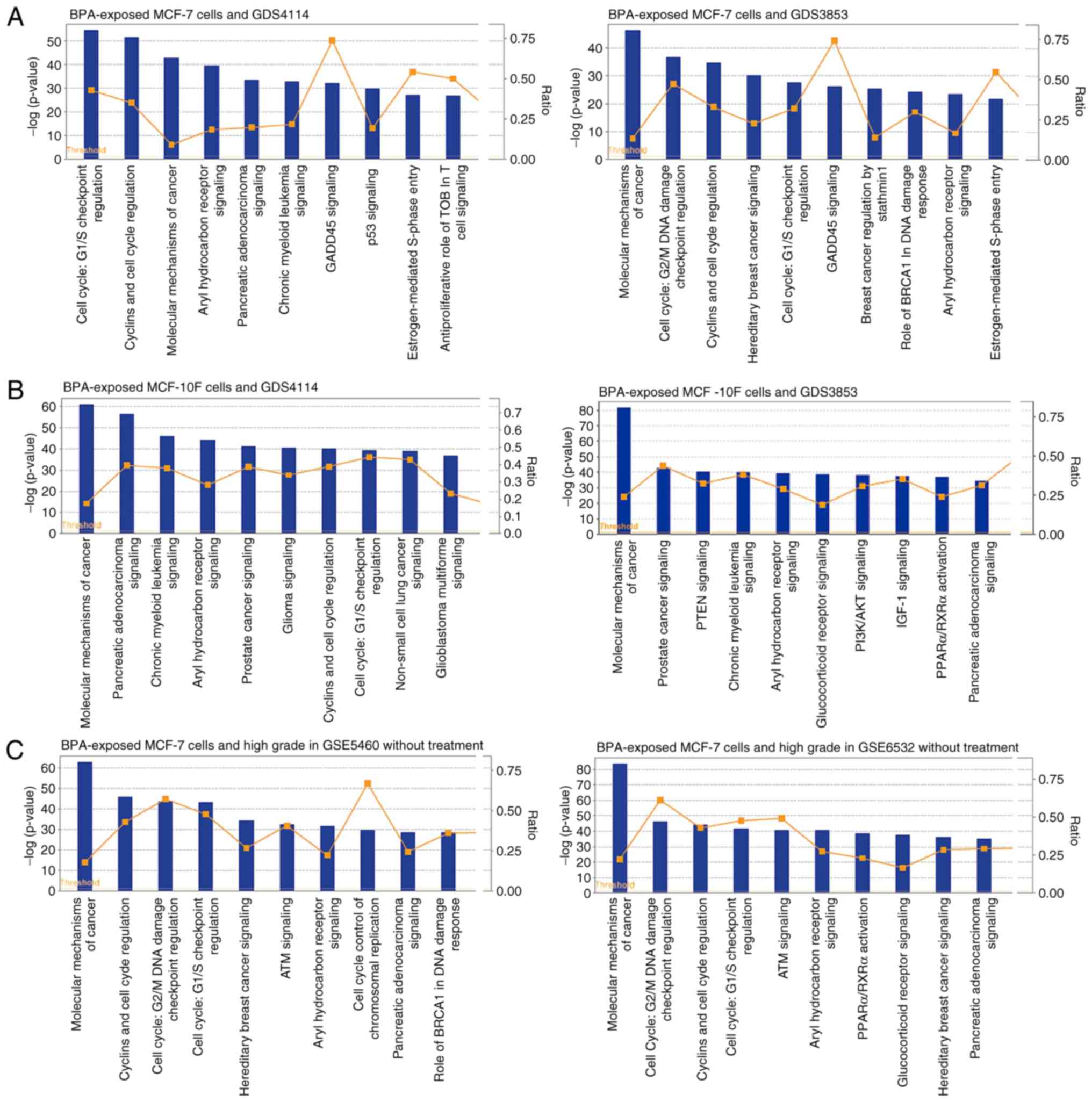 | Figure 2.Cellular canonical pathways
associated with the development and high grade of breast cancer.
(A) The results of BPA-exposed MCF-7 cells (GSE50705), GDS4114 and
GDS3853. (B) The results of BPA-exposed MCF-10F cells (GSE32158),
GDS4114 and GDS3853. (C) The results of BPA-exposed MCF-7 cells
(GSE50705), GSE5460 (high grade without treatment) and GSE6532
(high grade with treatment). The left y-axis indicates the
negative logarithm of the P-value calculated using a right-tailed
Fisher's exact test with the threshold set at P<0.05. The right
y-axis indicates the ratios of the number of genes in the
pathway that meet the cut-off criteria to the total number of genes
in that pathway. BPA, bisphenol A; GADD45, growth arrest and DNA
damage-inducible 45; TOB, transducer of ERBB2; BRCA, breast
cancer-associated; IGF-1, insulin-like growth factor 1; PTEN,
phosphatase and tensin homolog; AKT, protein kinase B; PPARα,
peroxisome-proliferator-activated receptor α; ATM, ataxia
telangiectasia mutated. |
Biological interaction analysis of the
gene sets
Biological interaction maps of genes that are
commonly expressed in human breast cells and breast tissue
following BPA exposure were generated using IPA. Simplified
versions of two network maps for the visceral obesity pathway and
cell cycle pathway are presented in Fig.
3. A multiple interaction network of the visceral obesity
pathway was involved in visceral fat deposits and MetSyn, revealing
adiponectin, retinoid X receptor α and nuclear receptor subfamily 3
group C member 1 (Fig. 3A). The
network interaction map for the cell cycle pathway identified a
number of genes with multiple interactions that were associated
with cyclins and cell cycle regulation, including cyclin-dependent
kinases 1, 2 and 4, cyclin-dependent kinase inhibitors 1A, 2B and
2C, cyclin B1, E2F transcription factor 1 and retinoblastoma 1
(Fig. 3B).
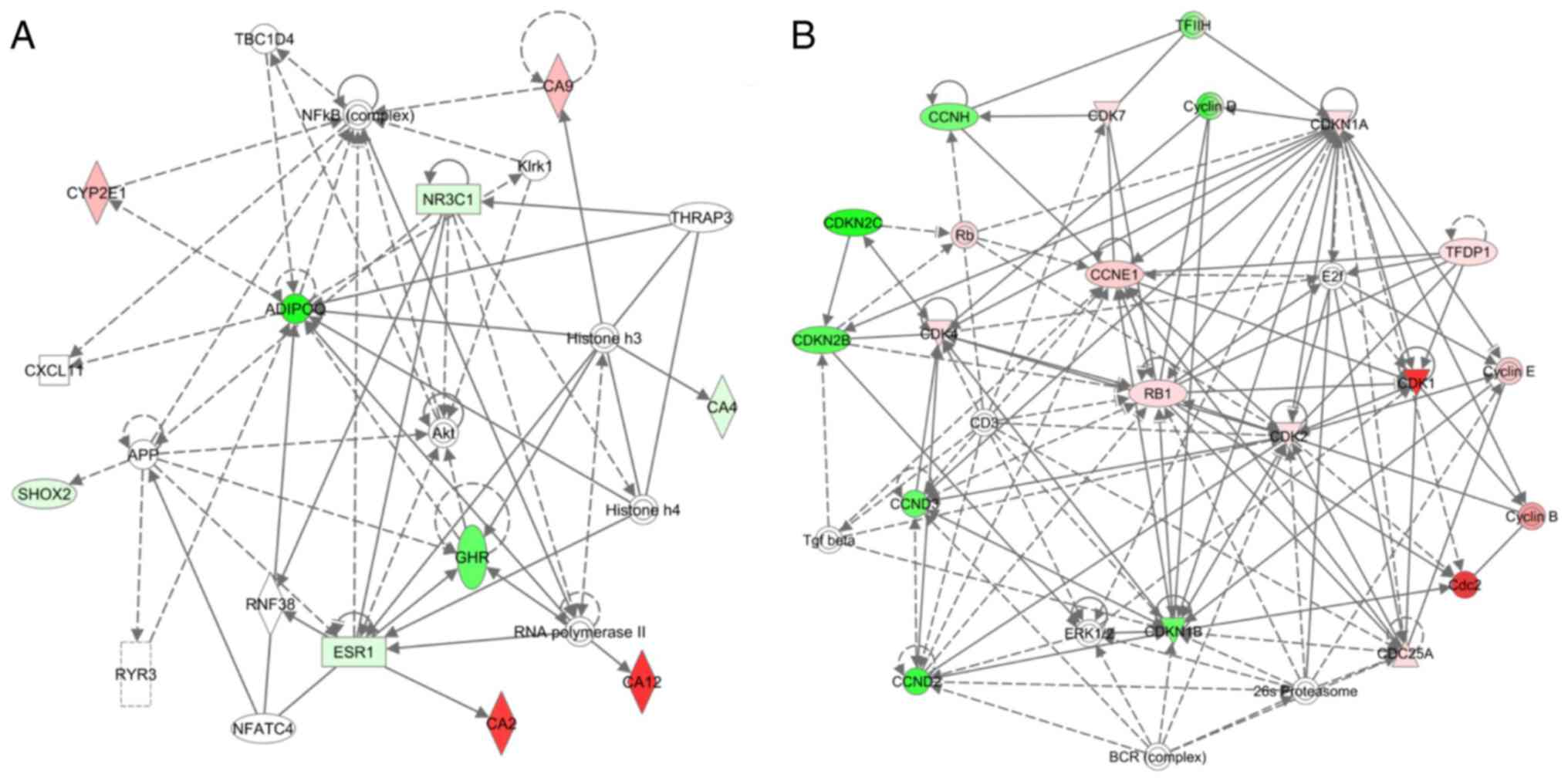 | Figure 3.Interaction network analysis of the
(A) visceral obesity pathway and (B) the cell cycle pathway, using
the path designer of IPA. A simplified version of IPA pathway maps
generated for genes that were commonly expressed in microarrays of
human breast cells and breast tissue treated with BPA. Red and
green nodes represent upregulated or downregulated genes. White
nodes represent the genes in a pathway not available in the
microarray for testing. Symbols represent the following: Concentric
circle, complex/group; square, cytokine; rhombus, enzyme; dashed
rectangle, ion channel; inverted triangle, kinase; rectangle,
ligand-dependent nuclear receptor; circle, other; triangle,
phosphatase; horizontal oval, transcription regulator; vertical
oval, transmembrane receptor. BPA, bisphenol A; IPA, Ingenuity
Pathway Analysis. |
Discussion
The aim of the present study was to identify the
potential pathways of the progression from normal breast epithelial
cells to the development and aggressiveness of breast cancer
following exposure to BPA. The two BPA-treated datasets and the two
breast tissue datasets all displayed an overlap of two consistent
gene sets: The visceral obesity pathway, involved in visceral fat
deposits and MetSyn, and the cell cycle pathway, involved in
cyclins and cell cycle regulation. Visceral adipose tissue has been
recognized as an important metabolic tissue, secreting factors that
may alter metabolism and immunity (43). Visceral obesity, insulin resistance
and pro-inflammatory state were defined as MetSyn. It has been
identified that MetSyn was associated with the pathogenesis of
breast cancer and colorectal cancer (43). It has been demonstrated that cyclin
D1 serves as a cell cycle regulatory switch and is important in the
regulation of proliferation during mammary oncogenesis (44). Cyclin-dependent kinase 4 (CDK4) and
CDK6 are proteins in the cell cycle regulatory pathway (45). When there is an imbalance in the
cyclin D and cyclin-dependent kinase (CDK) pathway in cancer cells,
the cells may progress towards a more proliferative phenotype. The
present study demonstrated that oral CDK inhibitors may improve the
outcomes in patients with ER-positive breast cancer (45). It has been reported that the R-point
is important in cell cycle control and it switches on the
G1-to-S transition process (46). It has been demonstrated that if the
cells pass the R-point, they can subsequently evade apoptosis,
proliferation and metastasis (46).
The results of the present study suggest that BPA-affected gene
sets cause breast carcinogenesis, through the visceral fat deposits
and MetSyn pathway, and the cell cycle pathway. In addition, the
aforementioned processes lead to high-grade aggressiveness in
breast cancer. The visceral fat deposits and MetSyn pathway, and
the cell cycle pathway may be involved when BPA affects high-grade
tumor aggressiveness.
Previous studies have indicated that BPA causes
obesity and metabolic disruption in animals and humans (21–23). The
National Health and Nutrition Examination Surveys reported that
urinary BPA concentrations are positively associated with general
and central obesity (21). It has
been suggested that exposure to BPA may lead to insulin resistance
and the development of Type 2 diabetes mellitus, which may occur
through the overactivation of pancreatic β-cells (22,23). A
previous study conducted in a Chinese population identified an
association among urinary BPA concentrations and obesity, insulin
resistance, and disturbances in body weight regulation (47). The aforementioned results are
consistent with the results of the present study in which exposure
to BPA initiated the visceral obesity pathway, involved in visceral
fat deposits and MetSyn. A murine study identified that BPA
injections into the fat pads enhanced epithelial tumor
proliferation rather than subcutaneous proliferation (48). Furthermore, a number of cohort
studies have indicated that abdominal obesity and MetSyn increased
the risk of breast cancer (49–51). It
has been identified that numerous inflammatory cells in the
visceral adipose tissue, including macrophages and T cells,
generate a pro-tumorigenic environment (43).
A proteomic analysis of mammary fat identified
numerous signaling molecules involved in a variety of biological
processes, including signal transduction and cell communication,
energy and protein metabolism, proliferation and apoptosis, all of
which served an important function in the breast tumor
microenvironment (52). The
underlying molecular mechanisms of breast carcinogenesis by
visceral adiposity and MetSyn have been indicated to include
microenvironmental alterations in cell signaling pathways and
adipokine secretion (43). A gene
set analysis of BPA, combined with 17β-estradiol and progesterone
exposure in eight non-malignant breast epithelial cell culture
samples, elucidated six biological pathways implicated in
tumorigenesis. Exposure to BPA led to the overexpression of genes
facilitating cell cycle progression and created a high-risk
microenvironment for breast tumor development and aggressiveness
(14). The results of the present
study support those of the aforementioned previous studies. In the
present study, it was identified that the cell cycle pathway
involved in cyclins and cell cycle regulation is associated with
BPA-regulated breast carcinogenesis.
It has been reported that breast cancer consists of
multiple different subtypes and biological processes, and therefore
distinct gene set pathways are associated with prognosis and
chemotherapy sensitivity in these disease subsets (53). In the individual dataset analysis for
the aforementioned biological pathways, it was indicated that two
pathways overlapped among the predictive gene sets in the
BPA-treated cells and breast cancer datasets. This result indicated
that different cellular pathways following BPA exposure were
associated with the progression of normal breast cells to breast
cancer cells. From the pooled analysis, the gene sets involved in
the pathways of G2/M DNA damage, growth arrest and DNA
damage-inducible 45, p53 and T-cell signalling, and
estrogen-mediated S phase entry were associated with BPA-regulated
carcinogenesis of ER-positive breast cancer. A previous review of
numerous multiple oncogenic signaling pathways following BPA
exposure in ER-positive breast cancer cells identified vascular
endothelial growth factor signaling, the DNA repair pathway,
extracellular-signal-regulated kinase 1/2 activation and signal
transducer and activator of transcription 3 signaling (54). Furthermore, DNA damage, involving
cell cycle G2/M and BRCA1, ATM and hereditary breast
cancer signaling gene sets were associated with BPA-regulated
high-grade aggressiveness. The result is consistent with a
functional study that indicated that gene expression alteration,
due to BPA, was significantly associated with an advanced
histological grade and large tumor size in breast tumors (14).
In addition to the breast cancer cell line MCF-7,
the normal breast epithelial cell line MCF-10F was also selected to
mimic the cells of healthy subjects exposed to BPA. In the present
study, the gene set analysis of MCF-10F exposed to BPA compared
with breast cancer datasets elucidated two important biological
pathways, the visceral fat deposits and MetSyn pathway, and cyclins
and cell cycle regulation, which are implicated in tumorigenesis
associated to BPA. The stimulation of immortalized breast cells
suggests a possible function for these pathways in modulating the
carcinogenesis and aggressiveness of breast cancer. The microarray
association results indicate that exposure to BPA initiates
processes that are a part of the breast cancer developmental cycle.
The combination of the aforementioned results with clinical and
epidemiologic evidence associates BPA exposure with subsequent
breast cancer.
A number of limitations are noted in the present
study. First, the tumor datasets were from different patient
cohorts with different inclusion and exclusion criteria, along with
the breast cell line datasets being treated with different doses of
BPA. However, the six datasets selected used an identical gene
expression platform, GPL570, to decrease any bias resulting from
quantifying different initial gene sets and inconsistent
associative intensity values for a candidate gene. Secondly,
different methodologies may have been applied in patient archiving
and specimen preparation, and different pre-analytical variations
may have occurred. This variability may decrease the
reproducibility of the results of the present study. Thirdly, owing
to the limitation of the selected platform, the sample size of the
datasets and study subjects for patients with breast cancer were
limited in each of the array datasets. Patients included in one
high-grade dataset were treated with tamoxifen regimens, whereas
patients in the other high-grade dataset received no treatment.
Therefore, the probability of coincidental discoveries cannot be
excluded due to the limited sample size and low statistical
power.
Furthermore, the selection criteria for the searched
microarray datasets from the NCBI's GEO database were BPA, Homo
sapiens and gene expression array until July 6, 2017. It was
indicated that five databases were breast-associated cell lines, as
well as BPA-treated, two of which, GSE50705 and GSE32158, were
included in the present study. The other three datasets were MCF-7
cells treated with 0.4 µM BPA for 48 h, MCF-10F cells treated with
10 and 1 µM BPA, and MCF-10F cells treated with 10 and 1 µM for 2
weeks. The datasets selected had a lower BPA concentration compared
with the other three datasets, including GSE50705, MCF-7-treated
with 0.25, 0.5 and 1 µM BPA for 48 h. However, doses of BPA were
not biologically relevant in humans. Further functional studies are
required to validate the predictive and prognostic value of the
proposed pathways and markers of breast cancer. The results of the
present study suggest that normal cell exposure to BPA may lead to
breast cancer development and aggressiveness, through the
regulation of key BPA pathways, particularly for the visceral
obesity pathway and the cell cycle pathway.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Science Council of Taiwan (grant no. NSC 102-2632-B-037-001-MY3)
and partially from the Kaohsiung Medical University ‘Aim for the
Top Universities Grant’ (grant nos. KMU-TP103A16, KMU-TP104A01 and
KMU-TP105A05).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TNW conceived and designed the study and drafted the
manuscript. PJY, YTT and YST edited the manuscript and analyzed the
datasets. PLK, CCC, SSL, THH, MFH and EMT were involved in the
conception of the study. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BPA
|
bisphenol A
|
|
GEO
|
Gene Expression Omnibus
|
|
IPA
|
Ingenuity Pathway Analysis
|
|
MetSyn
|
the metabolic syndrome
|
|
ER
|
estrogen receptor
|
References
|
1
|
Weber Lozada K and Keri RA: Bisphenol A
increases mammary cancer risk in two distinct mouse models of
breast cancer. Biol Reprod. 85:490–497. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Katchy A, Pinto C, Jonsson P, Nguyen-Vu T,
Pandelova M, Riu A, Schramm KW, Samarov D, Gustafsson JÅ, Bondesson
M and Williams C: Coexposure to phytoestrogens and bisphenol a
mimics estrogenic effects in an additive manner. Toxicol Sci.
138:21–35. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Murray TJ, Maffini MV, Ucci AA,
Sonnenschein C and Soto AM: Induction of mammary gland ductal
hyperplasias and carcinoma in situ following fetal bisphenol A
exposure. Reprod Toxicol. 23:383–390. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ayyanan A, Laribi O, Schuepbach-Mallepell
S, Schrick C, Gutierrez M, Tanos T, Lefebvre G, Rougemont J,
Yalcin-Ozuysal O and Brisken C: Perinatal exposure to bisphenol a
increases adult mammary gland progesterone response and cell
number. Mol Endocrinol. 25:1915–1923. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jenkins S, Wang J, Eltoum I, Desmond R and
Lamartiniere CA: Chronic oral exposure to bisphenol A results in a
nonmonotonic dose response in mammary carcinogenesis and metastasis
in MMTV-erbB2 mice. Environ Health Perspect. 119:1604–1609. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cooper JE, Kendig EL and Belcher SM:
Assessment of bisphenol A released from reusable plastic, aluminium
and stainless steel water bottles. Chemosphere. 85:943–947. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Geens T, Goeyens L, Kannan K, Neels H and
Covaci A: Levels of bisphenol-A in thermal paper receipts from
Belgium and estimation of human exposure. Sci Total Environ
435-436. 30–33. 2012. View Article : Google Scholar
|
|
8
|
Sutton P, Wallinga D, Perron J, Gottlieb
M, Sayre L and Woodruff T: Reproductive health and the
industrialized food system: A point of intervention for health
policy. Health Aff (Millwood). 30:888–897. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dekant W and Völkel W: Human exposure to
bisphenol A by biomonitoring: Methods, results and assessment of
environmental exposures. Toxicol Appl Pharmacol. 228:114–134. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Völkel W, Colnot T, Csanády GA, Filser JG
and Dekant W: Metabolism and kinetics of bisphenol a in humans at
low doses following oral administration. Chem Res Toxicol.
15:1281–1287. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Acevedo N, Davis B, Schaeberle CM,
Sonnenschein C and Soto AM: Perinatally administered bisphenol a as
a potential mammary gland carcinogen in rats. Environ Health
Perspect. 121:1040–1046. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vandenberg LN, Schaeberle CM, Rubin BS,
Sonnenschein C and Soto AM: The male mammary gland: A target for
the xenoestrogen bisphenol A. Reprod Toxicol. 37:15–23. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dhimolea E, Wadia PR, Murray TJ, Settles
ML, Treitman JD, Sonnenschein C, Shioda T and Soto AM: Prenatal
exposure to BPA alters the epigenome of the rat mammary gland and
increases the propensity to neoplastic development. PLoS One.
9:e998002014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dairkee SH, Seok J, Champion S, Sayeed A,
Mindrinos M, Xiao W, Davis RW and Goodson WH: Bisphenol A induces a
profile of tumor aggressiveness in high-risk cells from breast
cancer patients. Cancer Res. 68:2076–2080. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dairkee SH, Luciani-Torres MG, Moore DH
and Goodson WH III: Bisphenol-A-induced inactivation of the p53
axis underlying deregulation of proliferation kinetics, and cell
death in non-malignant human breast epithelial cells.
Carcinogenesis. 34:703–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goodson WH III, Luciani MG, Sayeed SA,
Jaffee IM, Moore DH II and Dairkee SH: Activation of the mTOR
pathway by low levels of xenoestrogens in breast epithelial cells
from high-risk women. Carcinogenesis. 32:1724–1733. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sauer SJ, Tarpley M, Shah I, Save AV,
Lyerly HK, Patierno SR, Williams KP and Devi GR: Bisphenol A
activates EGFR and ERK promoting proliferation, tumor spheroid
formation and resistance to EGFR pathway inhibition in estrogen
receptor-negative inflammatory breast cancer cells. Carcinogenesis.
38:252–260. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Robinson JL and Carroll JS: FoxA1 is a key
mediator of hormonal response in breast and prostate cancer. Front
Endocrinol (Lausanne). 3:682012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang XL, Wang HS, Liu N and Ge LC:
Bisphenol A stimulates the epithelial mesenchymal transition of
estrogen negative breast cancer cells via FOXA1 signals. Arch
Biochem Biophys. 585:10–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei J, Lin Y, Li Y, Ying C, Chen J, Song
L, Zhou Z, Lv Z, Xia W, Chen X and Xu S: Perinatal exposure to
bisphenol A at reference dose predisposes offspring to metabolic
syndrome in adult rats on a high-fat diet. Endocrinology.
152:3049–3061. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carwile JL and Michels KB: Urinary
bisphenol A and obesity: NHANES 2003-2006. Environ Res.
111:825–830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Silver MK, O'Neill MS, Sowers MR and Park
SK: Urinary bisphenol A and type-2 diabetes in U.S. adults: Data
from NHANES 2003-2008. PLoS One. 6:e268682011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shankar A and Teppala S: Relationship
between urinary bisphenol A levels and diabetes mellitus. J Clin
Endocrinol Metab. 96:3822–3826. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barouki R, Gluckman PD, Grandjean P,
Hanson M and Heindel JJ: Developmental origins of non-communicable
disease: Implications for research and public health. Environ
Health. 11:422012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
García-Arevalo M, Alonso-Magdalena P,
Rebelo Dos Santos J, Quesada I, Carneiro EM and Nadal A: Exposure
to bisphenol-A during pregnancy partially mimics the effects of a
high-fat diet altering glucose homeostasis and gene expression in
adult male mice. PLoS One. 9:e1002142014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Porreca I, Ulloa-Severino L, Almeida P,
Cuomo D, Nardone A, Falco G, Mallardo M and Ambrosino C: Molecular
targets of developmental exposure to bisphenol A in diabesity: A
focus on endoderm-derived organs. Obes Rev. 18:99–108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Williams GP: The role of oestrogen in the
pathogenesis of obesity, type 2 diabetes, breast cancer and
prostate disease. Eur J Cancer Prev. 19:256–271. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Harvie M, Hooper L and Howell AH: Central
obesity and breast cancer risk: A systematic review. Obes Rev.
4:157–173. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Healy LA, Ryan AM, Carroll P, Ennis D,
Crowley V, Boyle T, Kennedy MJ, Connolly E and Reynolds JV:
Metabolic syndrome, central obesity and insulin resistance are
associated with adverse pathological features in postmenopausal
breast cancer. Clin Oncol (R Coll Radiol). 22:281–288. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Capasso I, Esposito E, de Laurentiis M,
Maurea N, Cavalcanti E, Botti G, Petrillo A, Montella M, D'Aiuto M,
Coppola C, et al: Metabolic syndrome-breast cancer link varies by
intrinsic molecular subtype. Diabetol Metab Syndr. 6:1052014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berrino F, Villarini A, Traina A, Bonanni
B, Panico S, Mano MP, Mercandino A, Galasso R, Barbero M, Simeoni
M, et al: Metabolic syndrome and breast cancer prognosis. Breast
Cancer Res Treat. 147:159–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shioda T, Rosenthal NF, Coser KR, Suto M,
Phatak M, Medvedovic M, Carey VJ and Isselbacher KJ: Expressomal
approach for comprehensive analysis and visualization of ligand
sensitivities of xenoestrogen responsive genes. Proc Natl Acad Sci
USA. 110:16508–16513. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
NCBI, . Bisphenol A Regulates the
Expression of DNA Repair Genes in Human Breast Epithelial Cells
(expression data). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32158Septemper
16–2011
|
|
34
|
Planche A, Bacac M, Provero P, Fusco C,
Delorenzi M, Stehle JC and Stamenkovic I: Identification of
prognostic molecular features in the reactive stroma of human
breast and prostate cancer. PLoS One. 6:e186402011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kretschmer C, Sterner-Kock A, Siedentopf
F, Schoenegg W, Schlag PM and Kemmner W: Identification of early
molecular markers for breast cancer. Mol Cancer. 10:152011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X
and Richardson AL: Predicting features of breast cancer with gene
expression patterns. Breast Cancer Res Treat. 108:191–201. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Loi S, Haibe-Kains B, Desmedt C, Lallemand
F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, et
al: Definition of clinically distinct molecular subtypes in
estrogen receptor-positive breast carcinomas through genomic grade.
J Clin Oncol. 25:1239–1246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu X, Zhao Y and Simon R: Gene set
expression comparison kit for BRB-ArrayTools. Bioinformatics.
24:137–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Simon R, Lam A, Li MC, Ngan M, Menenzes S
and Zhao Y: Analysis of gene expression data using BRB-ArrayTools.
Cancer Inform. 3:11–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Takashima S, Usui S, Kurokawa K, Kitano T,
Kato T, Murai H, Furusho H, Oda H, Maruyama M, Nagata Y, et al:
Altered gene expression in T-cell receptor signalling in peripheral
blood leucocytes in acute coronary syndrome predicts secondary
coronary events. Open Heart. 3:e0004002016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Korkor MT, Meng FB, Xing SY, Zhang MC, Guo
JR, Zhu XX and Yang P: Microarray analysis of differential gene
expression profile in peripheral blood cells of patients with human
essential hypertension. Int J Med Sci. 8:168–179. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y, Li J, Fang C, Shi L, Tan J, Xiong Y,
Bin Fan and Li C: Genome-wide differential expression of genes and
small RNAs in testis of two different porcine breeds and at two
different ages. Sci Rep. 6:268522016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Doyle SL, Donohoe CL, Lysaght J and
Reynolds JV: Visceral obesity, metabolic syndrome, insulin
resistance and cancer. Proc Nutr Soc. 71:181–189. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Caldon CE, Daly RJ, Sutherland RL and
Musgrove EA: Cell cycle control in breast cancer cells. J Cell
Biochem. 97:261–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Murphy CG and Dickler MN: The role of
CDK4/6 inhibition in breast cancer. Oncologist. 20:483–490. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tenga MJ and Lazar IM: Proteomic snapshot
of breast cancer cell cycle: G1/S transition point. Proteomics.
13:48–60. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang T, Li M, Chen B, Xu M, Xu Y, Huang Y,
Lu J, Chen Y, Wang W, Li X, et al: Urinary bisphenol A (BPA)
concentration associates with obesity and insulin resistance. J
Clin Endocrinol Metab. 97:E223–E227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Elliott BE, Tam SP, Dexter D and Chen ZQ:
Capacity of adipose tissue to promote growth and metastasis of a
murine mammary carcinoma: Effect of estrogen and progesterone. Int
J Cancer. 51:416–424. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Calle EE, Rodriguez C, Walker-Thurmond K
and Thun MJ: Overweight, obesity, and mortality from cancer in a
prospectively studied cohort of U.S. Adults. N Engl J Med.
348:1625–1638. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Reeves GK, Pirie K, Beral V, Green J,
Spencer E and Bull D; Million Women Study Collaboration, : Cancer
incidence and mortality in relation to body mass index in the
Million women study: Cohort study. BMJ. 335:11342007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
John EM, Sangaramoorthy M, Hines LM, Stern
MC, Baumgartner KB, Giuliano AR, Wolff RK and Slattery ML: Overall
and abdominal adiposity and premenopausal breast cancer risk among
hispanic women: The breast cancer health disparities study. Cancer
Epidemiol Biomarkers Prev. 24:138–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Celis JE, Moreira JM, Cabezón T, Gromov P,
Friis E, Rank F and Gromova I: Identification of extracellular and
intracellular signaling components of the mammary adipose tissue
and its interstitial fluid in high risk breast cancer patients:
Toward dissecting the molecular circuitry of epithelial-adipocyte
stromal cell interactions. Mol Cell Proteomics. 4:492–522. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Iwamoto T, Bianchini G, Booser D, Qi Y,
Coutant C, Shiang CY, Santarpia L, Matsuoka J, Hortobagyi GN,
Symmans WF, et al: Gene pathways associated with prognosis and
chemotherapy sensitivity in molecular subtypes of breast cancer. J
Natl Cancer Inst. 103:264–272. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gao H, Yang BJ, Li N, Feng LM, Shi XY,
Zhao WH and Liu SJ: Bisphenol A and hormone-associated cancers:
Current progress and perspectives. Medicine (Baltimore).
94:e2112015. View Article : Google Scholar : PubMed/NCBI
|