Introduction
Bladder cancer (BCa) is one of the major causes of
cancer related morbidity in the US and worldwide (1). In 2019, around 80,270 new cases of BCa
were expected, and 17,670 estimated BCa patient deaths in the US
(2). Most cases of BCa are
non-muscle invasive and transurethral resection is commonly
performed but with a high recurrence rate (3). However, nearly 25% of newly diagnosed
BCa patients have muscle invasive bladder cancer (MIBC), and
approximately half of the patients with MIBC recurrence eventually
die from BCa because of the lack of treatment options (4,5). Radical
cystectomy followed with pelvic lymphadenectomy is the gold
standard treatment for MIBC (6).
Bladder preserving trimodal therapies are also evolving as an
effective alternate (7), however,
combined with current platinum-based chemotherapy (such as
MVAC-methotrexate, vinblastine, adriamycin, and cisplatin), 25–30%
patients still require salvage cystectomy (8). The high morbidity of definitive therapy
for BCa along with poor prognosis of advanced BCa warrants
identification of novel targets and subsequent therapeutic
interventions to achieve complete remission of BCa.
To overcome the disadvantages of traditional
chemotherapy and radiotherapy, the research paradigm has shifted
towards elimination of cancer cells specifically by targeting
specific molecular targets. To achieve this for use in clinical
practice, the current preferred approaches are search for novel and
targeted small-molecule agents (9)
and monoclonal antibodies (mAbs) (10). mAbs are usually large molecular
weight proteins (~150 kDa), whereas small molecule cancer drugs can
transfer through the plasma membranes owing to their much smaller
in size (≤500 Da) (11). The
cost-effectiveness and their amenable nature to oral administration
make them a better choice than mAbs, which are mostly administered
intravenously (12).
Small molecules are being extensively used to target
oncogenic pathways that are aberrantly activated in most cancers.
These molecules function by targeting the kinases that include
receptors as well as their downstream regulators thus inhibiting
cancer cell survival and proliferation (13,14). A
better understanding of oncogenic mechanisms, as well as
identification of specific genes/proteins may help design novel
strategies to not only improve the efficacy of current drugs but
can also be a major aid in the identification of novel agents.
Advancement of systems biology can help us analyze and identify
interactions between important gene networks that can provide us
significant insight into biological pathways (15). The genetic processes are very
complex, and these interactions change significantly when a cancer
cell is treated with a small molecule.
Based on the structure-activity relationship studies
focused on the Withaferin A (a dietary compound which exhibits
anti-cancer effect against many cancer types) (16–22)
analogs designed in our laboratory, we have identified a novel
small molecule, ASR488, designed by protecting-OH group at
4-position of Withaferin A by thiophene-2-carbonyl functionality.
ASR-488 demonstrated cell growth arrest in BCa cells and, more
importantly, is non-toxic to normal BCa cells. In the current
study, differential gene network analysis was performed to detect
the changes in gene expression in ASR488 treated MIBC cells. Also,
functional annotation and network analyses were performed to
identify differential gene expression (DEGs). By analyzing the
biological functions and networks of ASR488-treated MIBC cells, our
findings will help to gain a better understanding of the effect of
small molecules and to explore the candidate BCa treatments.
Materials and methods
Synthesis of ASR488
ASR488 was synthesized starting from Withaferin A
according to a synthetic strategy recently developed in our
laboratory (manuscript under preparation). Briefly, to a mixture of
Withaferin A and trimethylamine in methylene chloride at 0°C was
added 2-thiophenecarbonyl chloride and the resulting reaction
mixture was stirred overnight at room temperature. The reaction
mixture was quenched with saturated NaHCO3 solution,
extracted with methylene chloride, and purified by column
chromatography. The compound was characterized by NMR and MS and
its purity (≥98%) was determined by HPLC.
Cell culture and viability assay
BCa cell lines TCCSUP (ATCC® HTB5™), and
HT1376 (ATCC® CRL-1472™) were purchased from ATCC. Cell
lines were maintained in Eagle's Minimum Essential Medium at 37°C
and 5% CO2 (16). The
anti-proliferative effect of ASR488 was determined by the MTT
(3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide)
assay. TCCSUP and HT1376 cells were treated with varying
concentrations of ASR488 (0.2–12.5 µM) for 24, 48 and 72 h.
Detection of apoptosis by flow
cytometry and immunoblotting
Annexin V-fluorescein isothiocyanate (FITC) against
propidium iodide (PI) assay (FITC Annexin V Apoptosis Detection Kit
I, BD Pharminogen) was used for detecting apoptosis as described
previously (17). Total protein
extracts from TCCSUP cells were prepared with the Mammalian Protein
Extraction Reagent (Thermo Scientific) according to the
manufacturer's instructions. Western blotting was performed using
specific antibodies against Cleaved PARP (cat. no. 5625), BAX (cat.
no. 5023), , Bcl-2 (cat. no. 4223), p65 (cat. no. 8242) (Cell
signaling Technology), and β-actin (Santa Cruz Biotechnologies).
The positive bands were detected using enhanced
chemiluminescence.
RNA isolation, cDNA library
construction, and DNA sequencing
TCCSUP cells treated with vehicle (DMSO) or ASR488
were subjected to RNA isolation using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Nano Photometer®
spectrophotometer (Implen, Inc.) was used to measure the RNA
concentration and purity of the samples. A cDNA library was then
constructed using an NEB Next® Ultra™ RNA Library Prep
kit for Illumina® (New England Biolabs, Inc.) by
Novogene Bioinformatics Technologies Co. Ltd., following the
manufacturer's protocols. Subsequently, polymerase chain reaction
(PCR) was, Universal PCR primers, and Index (X) Primer for
double-stranded cDNA amplification. The PCR products generated for
PCR performed using Phusion High-Fidelity DNA polymerase were
purified using the AMPure XP system, and Agilent Bioanalyzer 2100
system was used to analyze the library quality. The cDNA library
was sequenced using an Illumina Hiseq 2000/2500 platform and 100
bp/50 bp single-end reads were generated.
Data analysis
Bioinformatics analysis was performed using a
combination of programs including STAR, HTseq, Cufflink, and our
wrapped scripts. Tophat program was used to parse the alignments
and DESeq2/edgeR was utilized to ascertain differential
expressions. To determine GO and KEGG enrichment Cluster Profiler
was used.
Clustering
Fragments Per Kilobase of transcript per Million
mapped reads (FPKM) expression level was used to cluster different
samples to identify the correlation between differences. To analyze
the difference between DEGs and the correlation, hierarchical
clustering distance method was used with default parameters in R
with the function of the heatmap, SOM (Self-organization mapping),
and k means using silhouette coefficient.
GO and KEGG enrichment analysis of
differentially expressed genes
Cluster profiler R package with corrected gene
length bias was used for analyzing GO enrichment of DEGs. P<0.05
were considered significantly enriched by DEGs. Cluster profiler R
package was also used to examine the statistical enrichment of
differential expression genes in KEGG pathways for understanding
functions as well as molecular level information of the dataset
generated by the RNASeq.
Differential expression analysis
For DESeq2 with biological replicates
DESeq2 R package (2_1.6.3) was used to analyze
differential expression between the ASR488 treated and control
groups (two biological replicates per condition). The P-values
obtained from the analysis were adjusted using the Benjamini and
Hochberg's approach so that the false discovery rate (FDR) can be
controlled. The genes having adjusted P-value <0.05 were
assigned as differentially expressed.
For edge R without biological replicates
EdgeR program package (3.16.5) was used to adjust
the read counts for each sequenced library before differential gene
expression analysis, through one scaling normalized factor. The
P-values were adjusted using the Benjamini and Hochberg method. A
corrected P-value of 0.05 and absolute fold-change of 1 were set as
the thresholds for significantly differential expression. The Venn
diagrams were prepared using the function Venn diagram in R based
on the gene list for different groups.
Statistical analysis
The data are presented as the mean ± SD. The
significance of the differences between the groups was determined
using the unpaired Student's t-test or multiple comparisons between
groups were performed using a one-way ANOVA with a post hoc
Dunnett's test. P<0.05 was considered to indicate a
statistically significant difference. All of the statistical
analyses were performed using Prism 6 software (GraphPad Software
Inc.).
Results
ASR488 treatment inhibits MIBC cell
growth
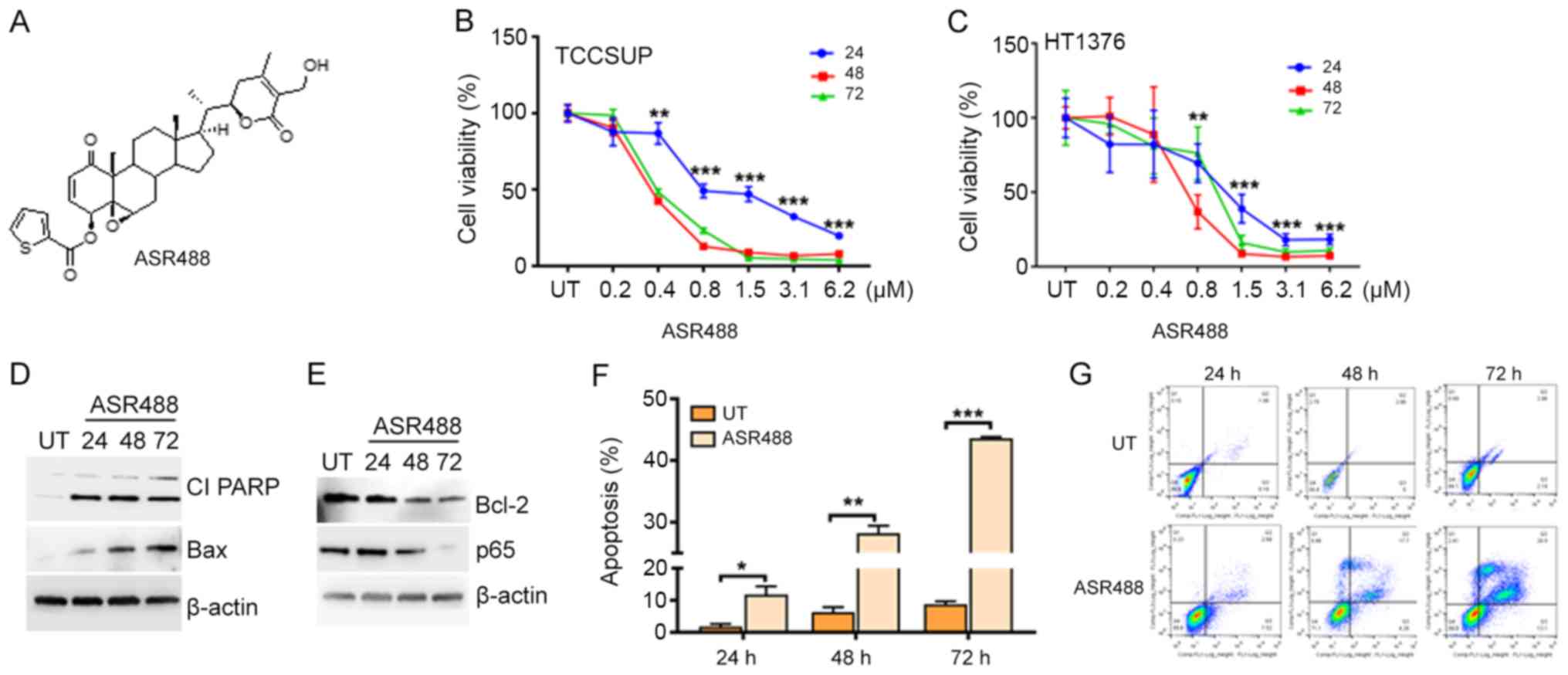
To determine the therapeutic potency of ASR488
(Fig. 1A) on MIBC, we examined the
effect of ASR488 treatment on cell viability of TCCSUP and HT1376
cells using the MTT assay. Significant reductions in cell viability
were observed in both TCCSUP (IC50 at 800, 480 and 450
nM at 24, 48 and 72 h, respectively) and HT1376 (IC50 at
1.28 µM, 750 and 850 nM at 24, 48 and 72 h, respectively) cell
lines (Fig. 1B and C). Induction of
apoptosis could be interpreted by observation of increased
expression of BAX and Cleaved PARP (Fig.
1D). ASR488 treatment inhibited survival signaling such as
downregulation of p65 and Bcl-2 expression in ASR488 treated MIBC
cell lines (Fig. 1E). Annexin V-FITC
staining further corroborated the results by showing significant
increases in apoptosis in both cell lines (TCCSUP: 30.5%, P=0.0382
and HT1376: 23.2%, P=0.0131) after 24 h treatment of ASR488
(Fig. 1F and G). Overall, these
results suggest that ASR488 effectively initiated apoptotic
signaling, which resulted in significant growth inhibition of MIBC
cells.
Identification of differentially
expressed genes in ASR488-treated TCCSUP cells
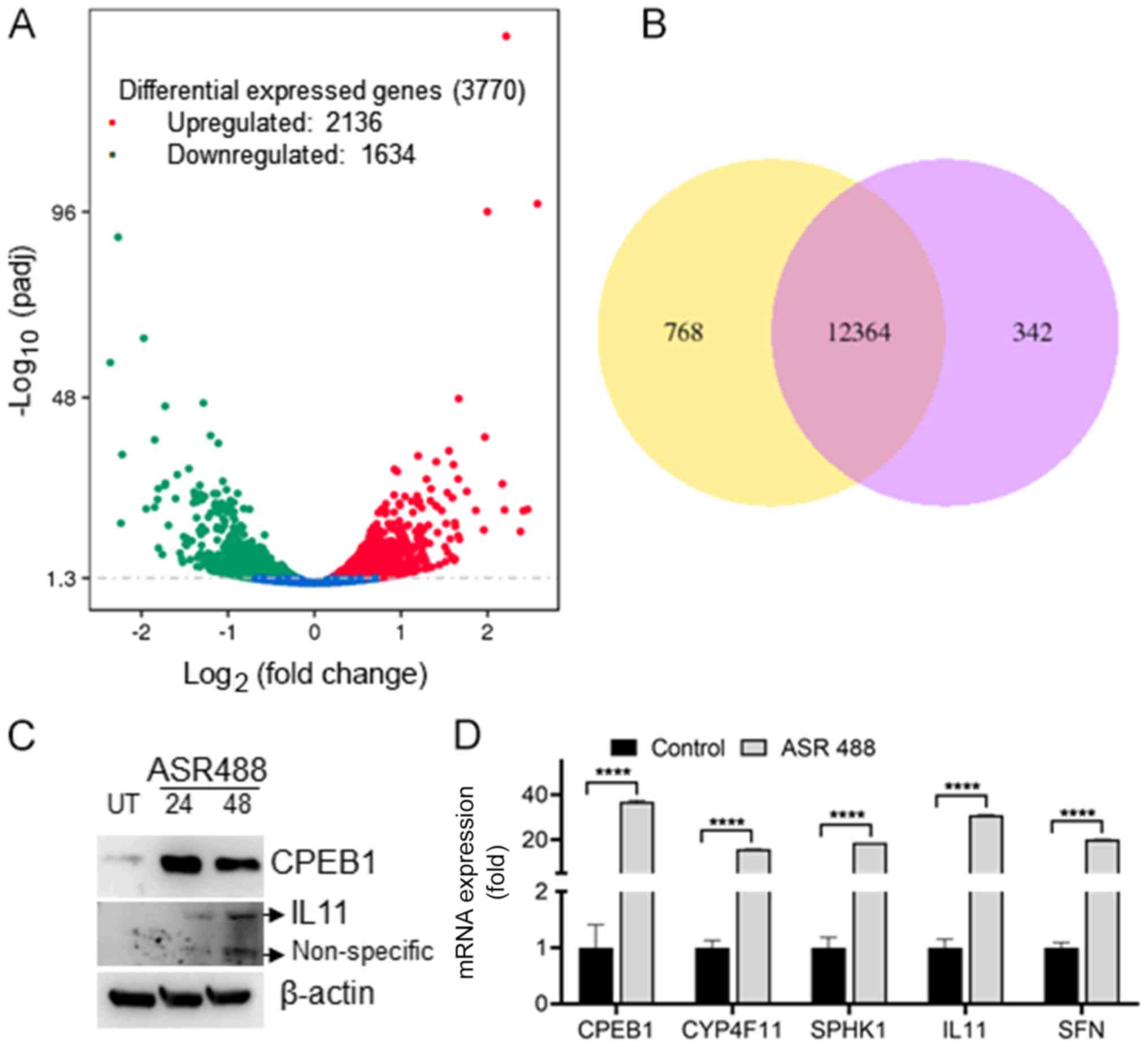
To identify whether there is a significant change in
expression of key regulatory genes in ASR488 treated cells, we
performed and analyzed RNASeq data for DEGs in ASR488-treated and
vehicle-treated TCCSUP cells. A volcano map demonstrated the
overall distribution of DEGs between control and ASR488-treated BCa
cells. The genes presented as red dots are the upregulated genes,
whereas the downregulated genes are represented as green dots, and
blue dots represent the genes that remained unchanged (Fig. 2A). Our current study obtained 3770
genes that were differentially expressed in TCCSUP cells upon
ASR488 treatment, of which 2,136 genes were upregulated, and 1634
were downregulated (Fig. 2A). Lists
of the ten most upregulated and downregulated genes in
ASR488-treated MIBC cells are given in Tables I and II. Specifically, expression levels of
CPEB1, ACTG2, SFN, HSPA6, CYP4F11, TAGLN, LINC00707, IL11,
MAP1A, SPHK1, and GNGT2 were upregulated in treated
TCCSUP cells, whereas expression levels of SFRP4, DDX60, GBP4,
BBOX1, RSAD2, OASL, FOS, IFIT2, CMPK2, STEAP4, and
IFI44L were the downregulated. The top five upregulated
genes were confirmed by reverse transcription-quantitative PCR
analysis: CPEB1 (36-fold), IL11 (30-fold), SFN
(20.12-fold) and CYP4F11 (15.8-fold) (Fig. 2D, primer details: Table SI), while no significant change was
observed in downregulated genes. The top two upregulated genes
CPEB1 and IL11 expressions were confirmed by immunoblotting
(Fig. 2C). To identify significant
DEGs during ASR488 treatment, the expression quantity of each gene
in untreated and ASR488-treated TCCSUP cells was also compared
pairwise and filtered with [log2(fold-change)]>1 and
q value <0.005. 13,474 DEGs were detected in both datasets
(Fig. 2B). Among these, 12,364 genes
showed significantly differential expression in both groups.
Three-hundred-forty-two genes in the ASR488 treated cells and 768
genes in the control cells showed significantly differential
expression (Fig. 2B). To visualize
the similarities between the two groups and also to determine if
the expression profile of ASR488-treated TCCSUP cells and control
cells are different, the genes that were differentially expressed
in pairwise comparison were clustered. The dendrogram showed that
the gene profile from vehicle-treated BCa cells was distant from
that of ASR488-treated TCCSUP cells (Fig. S1). These results confirm that
treating metastatic BCa cells with ASR488 leads to differential
expression of key genes.
 | Table I.List of top 10 upregulated genes in
ASR488-treated TCCSUP cells. |
Table I.
List of top 10 upregulated genes in
ASR488-treated TCCSUP cells.
| Gene symbol | Log2
(fold-change) | P-value |
|---|
| CPEB1 | 6.3347 |
4.17×10−3 |
| CYP4F11 | 8.3568 |
1.09×10−8 |
| SPHK1 | 4.6389 |
1.32×10−41 |
| IL11 | 2.9265 |
8.14×10−10 |
| SFN | 3.7239 |
1.54×10−9 |
| ACTG2 | 3.0293 |
8.70×10−12 |
| HSPA6 | 3.5985 |
6.27×10−9 |
| TAGLN | 2.3661 |
4.54×10−8 |
| MAP1A | 2.1205 |
8.78×10−7 |
| GNGT2 | 2.5802 |
3.62×10−6 |
 | Table II.List of top 10 downregulated genes in
ASR488-treated TCCSUP cells. |
Table II.
List of top 10 downregulated genes in
ASR488-treated TCCSUP cells.
| Gene symbol | Log2
(fold-change) | P-value |
|---|
| DDX60 | −2.5069 |
1.59×10−8 |
| GBP4 | −2.2704 |
2.98×10−8 |
| BBOX1 | −2.2384 |
7.26×10−6 |
| RSAD2 | −2.2235 |
9.71×10−8 |
| OASL | −1.9737 |
8.15×10−7 |
| FOS | −1.9465 |
9.91×10−5 |
| IFIT2 | −1.8473 |
1.66×10−6 |
| CMPK2 | −1.8470 |
8.78×10−7 |
| STEAP4 | −1.8141 |
2.70×10−5 |
| IFI44L | −1.9791 |
1.04×10−5 |
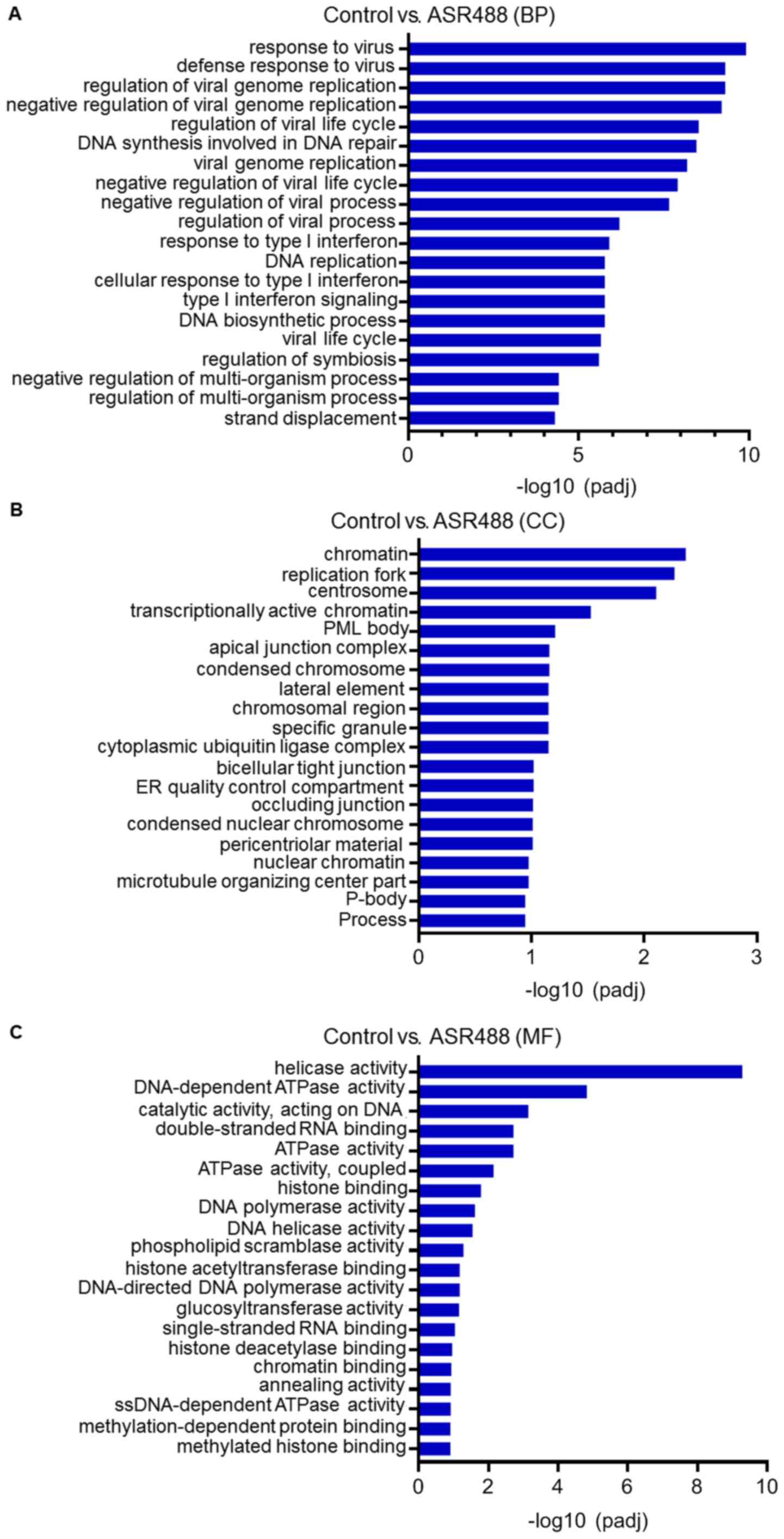
Functional enrichment of DEGs via
GO
To further visualize the relationship between genes
in ASR488-treated TCCSUP in context of their expression, distinct
clusters of genes were extracted and submitted to gene set
enrichment analysis. The GO terms as well as pathways that were
significantly over-represented among genes were identified from the
clusters. The top GO terms (from BP (Biological Process), CC
(Cellular Component), and MF (Molecular Function) categories)
enriched by the upregulated and downregulated DEGs were identified
(Figs. 3 and 4). The results revealed that the
downregulated genes were involved in viral defense response, DNA
synthesis and repair (BP category, Fig.
3A), transcriptionally active chromatin, endoplasmic reticulum
quality (CC Category, Fig. 3B),
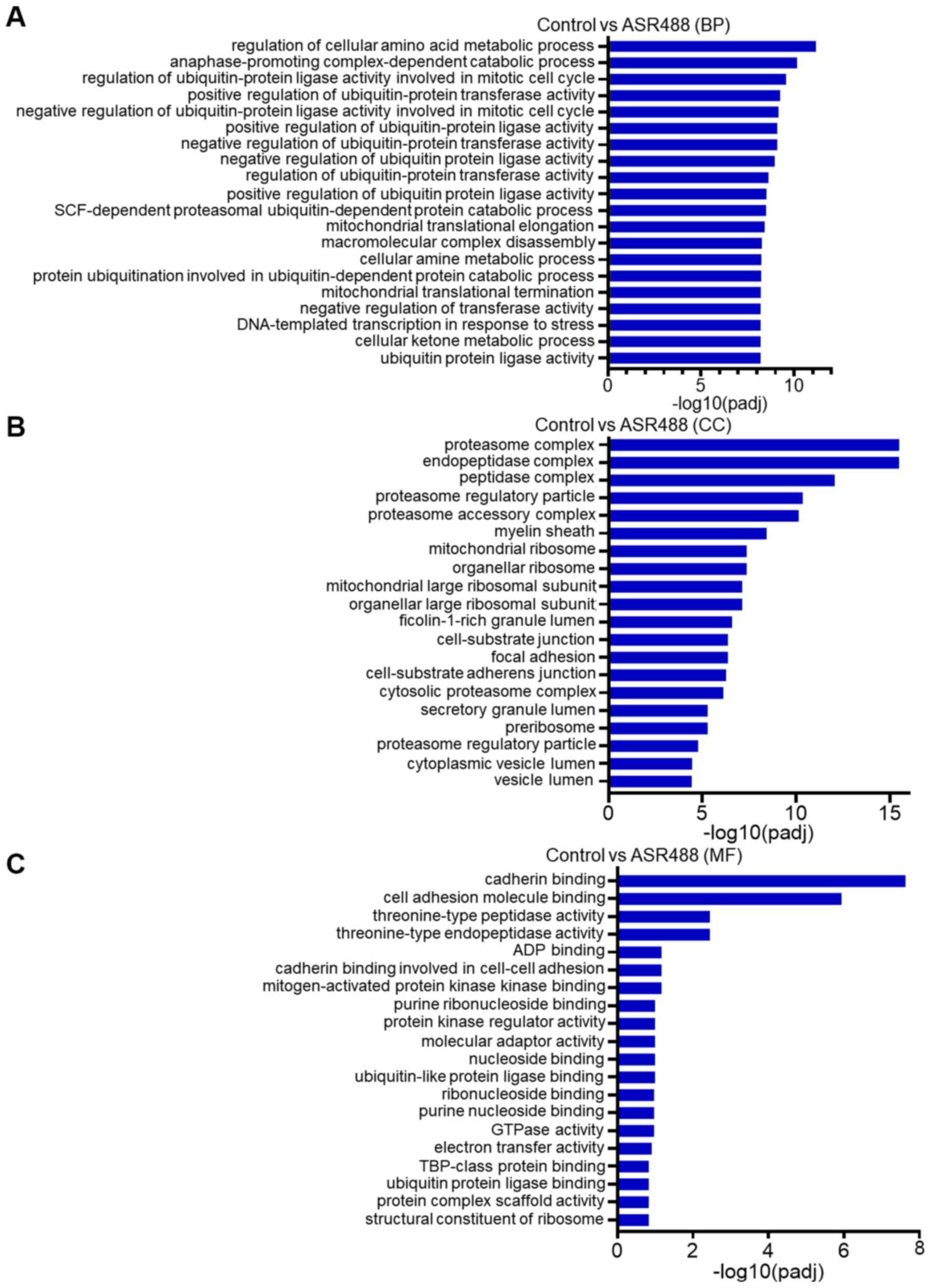
kinase activity, and DNA polymerase activity (MF category, Fig. 3C). On the other hand, the upregulated
genes were mainly associated with regulation of cellular metabolic
processes and regulation of ubiquitin-protein ligase activity in
the BP category (Fig. 4A). In the CC
category, these upregulated genes were involved in the regulation
of the proteasome complex, endopeptidase complex, and myelin sheath
(Fig. 4B). Whereas, in the MF
category, these were mainly involved in cadherin binding, cell
molecular adhesion binding, and threonine-type endopeptidase
activity (Fig. 4C).
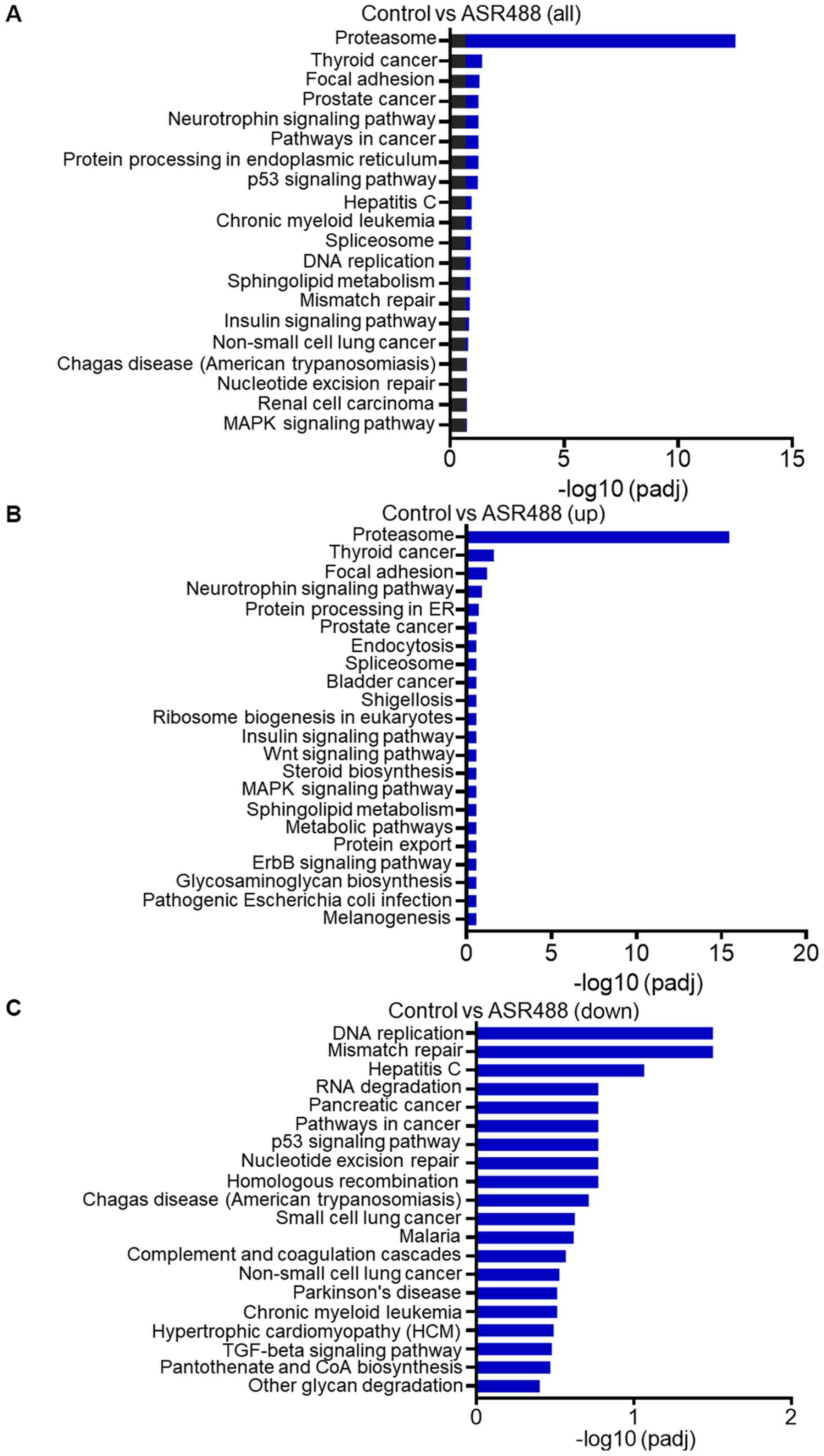
KEGG pathway analysis of DEGs
To analyze the functional status of DEGs i.e., which
DEGs are activated and suppressed in different classes of pathways,
the information we got from gene expression analysis of
ASR488-treated TCCSUP cells was mapped to the KEGG pathway. Pathway
analysis and functional annotation for up- and down-regulated genes
were performed. The analysis revealed that 156 up-regulated genes
(padj<0.001, log2 FC>2) and 82 down-regulated
genes (padj<0.001, log2 FC<-2) were mapped to 238
KEGG pathways. The top 20 enriched pathways are displayed in
Fig. 5. The results indicate that
the DEGs are highly clustered in several signaling pathways, such
as focal adhesion, neurotrophin-signaling, and p53 signaling, as
well as in protein processing in the endoplasmic reticulum and BCa
(Fig. 5B). More interestingly, the
down-regulated pathways in ASR488-treated BCa cells were enriched
in DEGs involved in DNA replication, mismatch repair, RNA
degradation, nucleotide excision repair, TGFβ signaling, and
pathways in cancer (Fig. 5C).
Downregulation of the DNA replication, mismatch repair, and
pathways in cancer make the ASR488 treated TCCSUP cells less
proliferative and invasive, finally contributing to the decreased
tumorigenic capacity of the cells.
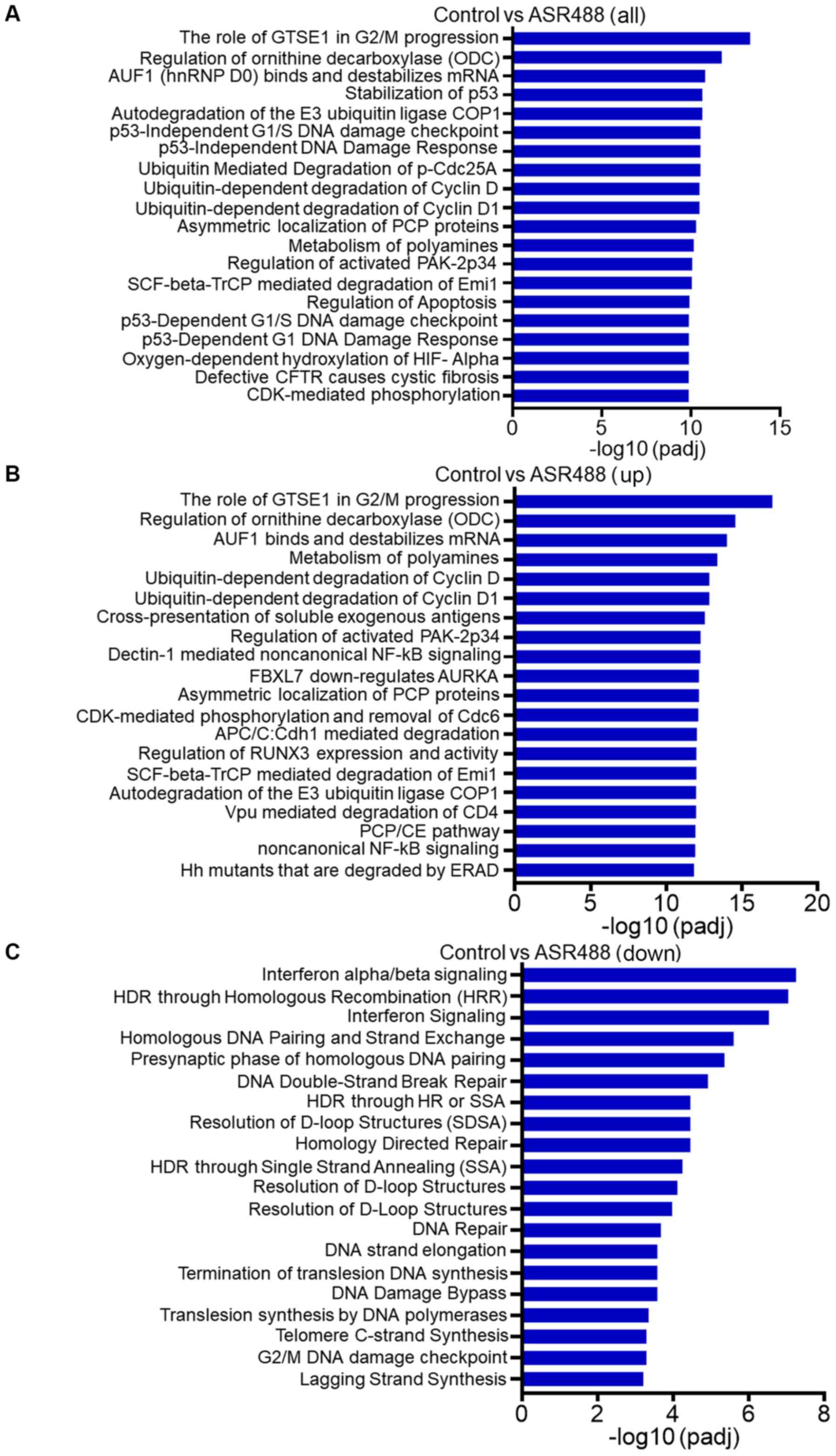
Reactome pathway analysis of DEGs
To further analyze gene sets (pre-defined groups of
genes that are functionally related), a reactome enrichment
analysis was performed (Fig. 6A). It
is well established that consistent perturbations over such gene
sets frequently cause mechanistic changes. The results from our
study demonstrate that the significantly enriched reactome pathways
of upregulated DEGs were related to ornithine decarboxylase
regulation, regulation of tumor suppressor RUNX3 expression, and
non-canonical NFκB signaling (Fig.
6B). Interestingly, the reactome data indicated that gene sets
related to ubiquitin-dependent degradation of cyclin D1 were
significantly upregulated, which indicated arrest of the cell cycle
in the treatment group and supported the growth inhibitory effect
of ASR488 treatment in TCCSUP cells (Fig. 6B). The data also demonstrate
significant downregulation of gene networks involved in telomere C
strand synthesis and DNA damage checkpoints (Fig. 6C).
Discussion
Among the limited options available to patients with
BCa, programmed cell death protein 1 (PD-1) pathway inhibitors are
a major category of inhibitors (18,19).
However, only a small group of patients respond, and options after
disease progression are a significant unmet need. Over 20 small
molecule drugs are being successfully used in cancer treatment
after being approved for clinical use. Nevertheless, these are not
without limitations. Non-specific binding to multiple molecular
targets such as cell surface receptors increase the the risk of
toxicity (20). It is thus important
to screen the promising small molecules for their effect on crucial
pathways, which should remain largely unaffected during treatment
of BCa. Analysis of complex signaling networks and genes, which are
differentially expressed after treatment, can provide valuable
input before progressing to further preclinical as well as clinical
trials.
We have screened a library of small molecules
(analogs of Withaferin A) and observed significant growth
inhibition and induction of apoptosis in MIBC cells with ASR488
treatment. To further explore the mechanism of action of ASR488 in
regulating the growth of BCa, we analyzed the gene expression
profiling using RNA-seq.
We observed that ASR488 treatment significantly
affected the expression of key regulatory genes, such as CPEB and
IL11. Depletion of CPEB1 expression levels has been explicitly
linked with increased metastatic potential in different cancer
types (21,22). CPEB1/2 downregulate TWIST1
expression, which is considered one of the main inducers of EMT
(23). It is also been shown that
skin and lung cells were able to circumvent the M1 crisis stage of
senescence in CPEB knockdown cells by undergoing telomere erosion,
and its reintroduction restored the senescence-like phenotype. The
knockdown was also followed with recommencement of cellular growth
and fewer mitochondria. These cells had reduced respiration and
reactive oxygen species (ROS) and resembled transformed cells by
having normal ATP levels, and enhanced rates of glycolysis
(24). We also observed that
mitochondrial translational elongation and telomere C strand
synthesis was significantly affected in GO and reactome enrichment
analysis, respectively. Additionally, CPEB knockdown cells have p53
mRNA with an unusually short poly(A) tail, which ultimately
resulted in a significant decrease (greater than 50%) in p53
protein levels (24). Our reactome
analysis indicates that ASR488 treatment significantly affected p53
stabilization and the p53 dependent DNA damage checkpoint. Overall,
these findings indicate that regulation of mitochondrial processes
and p53 stabilization might be possible mechanisms for cell growth
arrest in ASR488-treated BCa cells.
Another significantly upregulated gene in our study,
IL11, has been shown to be dysregulated in human gastric (25), colon (26), breast (27), and bladder cancers (28). Unlike IL6, the role of IL-11 in
various inflammation-associated cancers is not well studied.
Interestingly, IL-11 has generally been considered as an
anti-inflammatory cytokine, which is in contrast with the
well-studied pro-inflammatory function of IL-6. Although
aggressiveness of several cancer types has been attributed to
increased IL11 levels, a decrease in IL11 has been specifically
recognized as a factor contributing to carcinogenesis of the
bladder. Wu et al (28) have
shown that the expression of IL-11 was downregulated in human BCa
cell lines and transitional cell carcinoma (TCC) when it was
compared with primary human bladder cell culture. The same study
also demonstrated that the BCa patients samples had reduced urinary
levels of IL-11 in comparison to healthy subjects (28). In our study, another important
signaling immune pathway (the TGFβ pathway) was significantly
downregulated in KEGG analysis. It has been demonstrated that
levels of EMT markers, such as vimentin, slug, and twist, are
downregulated in TGFβ knockout mice, and abrogation of TGFβ pathway
depletes tumorigenic and invasive potential in an induced mouse BCa
model (1). As discussed in an
earlier section, there is also a proven direct link between CPEB
expression and downregulation of twist1, CPEB overexpression
combined with downregulation of TGFβ signaling during ASR488
treatment could reduce the metastatic potential of BCa cells.
Another interesting observation from the GO
enrichment analysis was the significant downregulation of ATPase
activity in ASR488-treated BCa cells. ATPase is considered as an
important ion transporter that is involved in signal transduction.
It is well established that ATPase expression profile is altered in
various tumors, such as breast cancer (29). Inhibition of ATPase activity
significantly reduced cell proliferation, motility, and invasion in
breast cancer. More recently, downregulation of longevity assurance
homolog 2 of yeast LAG1 (LASS2) has been associated with a poor
prognosis in patients with BCa. LASS2 binds directly to subunit C
of vacuolar H+-ATPase (V-ATPase) and its silencing
resulted in increased ATPase activity, which, in turn activated
secreted matrix metalloproteinase (MMP)-2 and MMP-9, and thus
enhanced cell proliferation, cell survival, and cell invasion in
vitro, as well as increase of BCa growth rate in vivo
(30). This decrease in ATP activity
is important to point out as we have seen that CPEB knockout
results in resumption of cell growth, fewer mitochondria, and
resembled transformed cells by maintaining normal ATP levels by
increasing glycolysis (24). Hence,
with normal ATPase activity or levels, the cells might bypass the
M1 crisis stage of senescence and thus act as transformed cells
with increased proliferative profile. However, an increased CPEB
level and decrease in ATPase activity will be detrimental to the
growth of these cancer cells and, ultimately, lead to
downregulation of metastatic and proliferative capacities.
In summary, using RNAseq data, here we identify
signaling molecules and pathways that are significantly affected
upon ASR488 treatment in MIBC cells. Interestingly, these pathways
are interlinked in a way that reduces the proliferative and
metastatic efficacy of MIBC cells. This study also indicated that
ASR488 might be a potential small molecule for BCa treatment.
However, these results need to be validated in other in
vitro and in vivo BCa models.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CD and MA conceived and supervised the study. AT and
VK performed the experiments, conducted the analysis as well as the
interpretation of the RNASeq data, performed the statistical
analysis and wrote the manuscript. BC and US performed the
statistical analysis and compiled the data into figures and revised
the manuscript. AS conceptualized, designed and performed synthesis
of the compound. AS critically revised the manuscript for its
intellectual content and also gave final approval for the corrected
version of manuscript. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liang Y, Zhu F, Zhang H, Chen D, Zhang X,
Gao Q and Li Y: Conditional ablation of TGF-β signaling inhibits
tumor progression and invasion in an induced mouse bladder cancer
model. Sci Rep. 6:294792016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Erlich A and Zlotta AR: Treatment of
bladder cancer in the elderly. Investig Clin Urol. 57 (Suppl
1):S26–S35. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van den Bosch S and Alfred Witjes J:
Long-term cancer-specific survival in patients with high-risk,
non-muscle-invasive bladder cancer and tumour progression: A
systematic review. Eur Urol. 60:493–500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pectasides D, Pectasides M and Nikolaou M:
Adjuvant and neoadjuvant chemotherapy in muscle invasive bladder
cancer: Literature review. Eur Urol. 48:60–68. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Akand M, Kilic Ö, Harmankaya I, Karabagli
P, Yavas Ç and Ata Ö: Aggressive treatment for urothelial
cancer-complete urinary tract extirpation: Operative feasibility in
two cases. Turk J Urol. 45:393–397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mathes J, Rausch S, Todenhöfer T and
Stenzl A: Trimodal therapy for muscle-invasive bladder cancer.
Expert Rev Anticancer Ther. 18:1219–1229. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roberts JT, von der Maase H, Sengeløv L,
Conte PF, Dogliotti L, Oliver T, Moore MJ, Zimmermann A and Arning
M: Long-term survival results of a randomized trial comparing
gemcitabine/cisplatin and
methotrexate/vinblastine/doxorubicin/cisplatin in patients with
locally advanced and metastatic bladder cancer. Ann Oncol. 17
(Suppl 5):v118–v122. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Padma VV: An overview of targeted cancer
therapy. Biomedicine (Taipei). 5:192015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carter PJ: Potent antibody therapeutics by
design. Nat Rev Immunol. 6:343–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hoelder S, Clarke PA and Workman P:
Discovery of small molecule cancer drugs: Successes, challenges and
opportunities. Mol Oncol. 6:155–176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Imai K and Takaoka A: Comparing antibody
and small-molecule therapies for cancer. Nat Rev Cancer. 6:714–727.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mu Y and Sun D: Lapatinib, a dual
inhibitor of epidermal growth factor receptor (EGFR) and HER-2,
enhances radiosensitivity in mouse bladder tumor Line-2 (MBT-2)
cells in vitro and in vivo. Med Sci Monit. 24:5811–5819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hänze J, Kessel F, Di Fazio P, Hofmann R
and Hegele A: Effects of multi and selective targeted tyrosine
kinase inhibitors on function and signaling of different bladder
cancer cells. Biomed Pharmacother. 106:316–325. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan J, Risacher SL, Shen L and Saykin AJ:
Network approaches to systems biology analysis of complex disease:
Integrative methods for multi-omics data. Brief Bioinform.
19:1370–1381. 2018.PubMed/NCBI
|
|
16
|
Tyagi A, Chandrasekaran B, Kolluru V, Rai
S, Jordan AC, Houda A, Messer J, Ankem M, Damodaran C and Haddad A:
Combination of androgen receptor inhibitor and cisplatin, an
effective treatment strategy for urothelial carcinoma of the
bladder. Urol Oncol. 37:492–502. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pal D, Tyagi A, Chandrasekaran B, Alattasi
H, Ankem MK, Sharma AK and Damodaran C: Suppression of Notch1 and
AKT mediated epithelial to mesenchymal transition by Verrucarin J
in metastatic colon cancer. Cell Death Dis. 9:7982018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bellmunt J, Powles T and Vogelzang NJ: A
review on the evolution of PD-1/PD-L1 immunotherapy for bladder
cancer: The future is now. Cancer Treat Rev. 54:58–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sundararajan S and Vogelzang NJ: Anti-PD-1
and PD-L1 therapy for bladder cancer: What is on the horizon?
Future Oncol. 11:2299–2306. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng J, Peng M, Wang Z, Zhou S, Xiao D,
Deng J and Yang X, Peng J and Yang X: Novel application of
metformin combined with targeted drugs on anticancer treatment.
Cancer Sci. 110:23–30. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nagaoka K, Fujii K, Zhang H, Usuda K,
Watanabe G, Ivshina M and Richter JD: CPEB1 mediates
epithelial-to-mesenchyme transition and breast cancer metastasis.
Oncogene. 35:2893–2901. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Grudzien-Nogalska E, Reed BC and Rhoads
RE: CPEB1 promotes differentiation and suppresses EMT in mammary
epithelial cells. J Cell Sci. 127:2326–2338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Y, Tsai YH and Tseng SH: Regulation
of the expression of cytoplasmic polyadenylation element binding
proteins for the treatment of cancer. Anticancer Res. 36:5673–5680.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burns DM and Richter JD: CPEB regulation
of human cellular senescence, energy metabolism, and p53 mRNA
translation. Genes Dev. 22:3449–3460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Putoczki TL, Thiem S, Loving A, Busuttil
RA, Wilson NJ, Ziegler PK, Nguyen PM, Preaudet A, Farid R, Edwards
KM, et al: Interleukin-11 is the dominant IL-6 family cytokine
during gastrointestinal tumorigenesis and can be targeted
therapeutically. Cancer Cell. 24:257–271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ernst M and Putoczki TL: Targeting IL-11
signaling in colon cancer. Oncotarget. 4:1860–1861. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Johnstone CN, Chand A, Putoczki TL and
Ernst M: Emerging roles for IL-11 signaling in cancer development
and progression: Focus on breast cancer. Cytokine Growth Factor
Rev. 26:489–498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu D, Tao J, Ding J, Qu P, Lu Q and Zhang
W: Interleukin-11, an interleukin-6-like cytokine, is a promising
predictor for bladder cancer prognosis. Mol Med Rep. 7:684–688.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khajah MA, Mathew PM and Luqmani YA:
Na+/K+ ATPase activity promotes invasion of
endocrine resistant breast cancer cells. PLoS One. 13:e01937792018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang H, Zuo Y, Ding M, Ke C, Yan R, Zhan
H, Liu J, Wang W, Li N and Wang J: LASS2 inhibits growth and
invasion of bladder cancer by regulating ATPase activity. Oncol
Lett. 13:661–668. 2017. View Article : Google Scholar : PubMed/NCBI
|