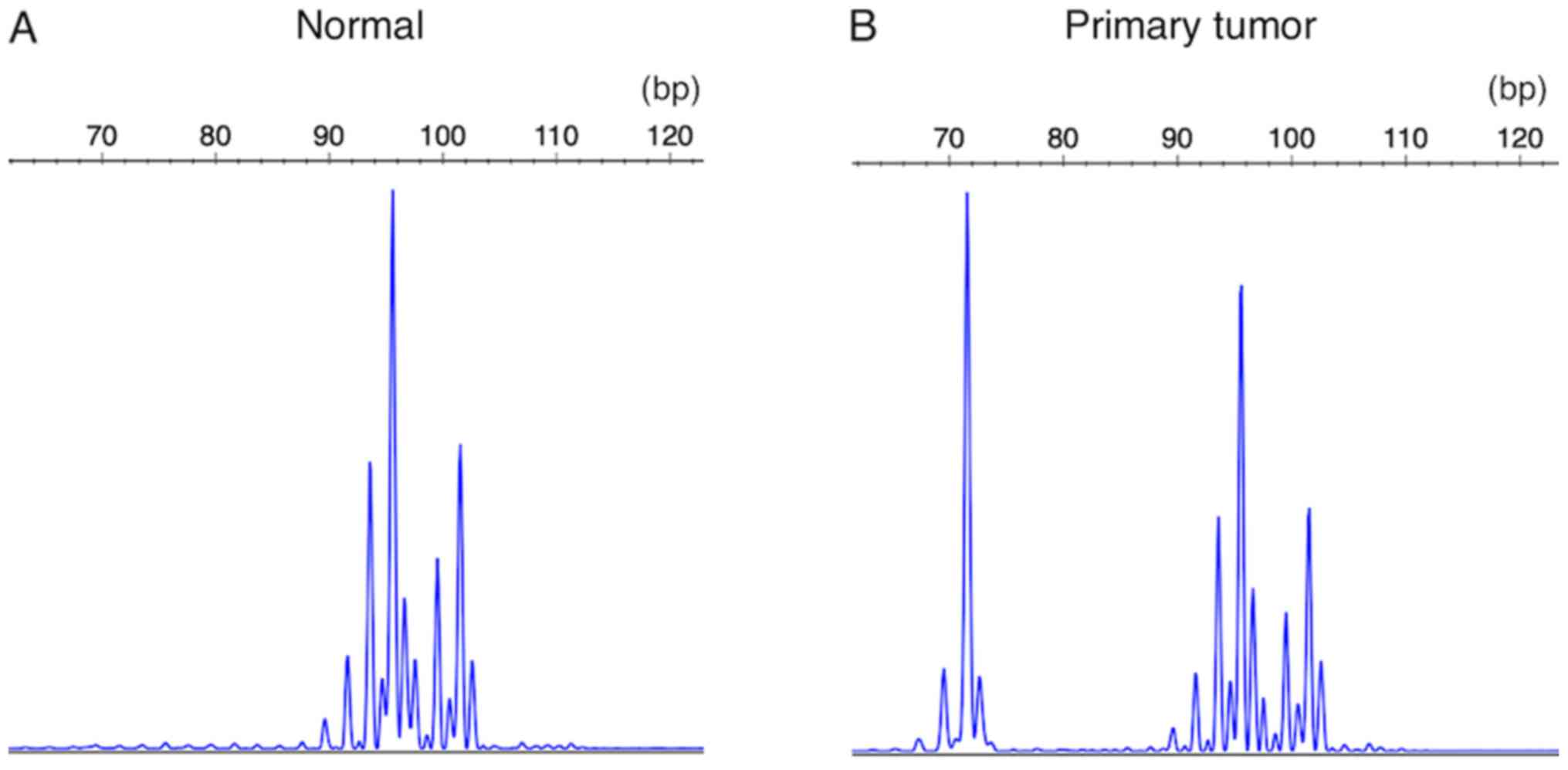
Introduction
Metastasis is an important event that defines
patients' prognosis during cancer treatment. Cancer cells acquire a
variety of phenotypes that allow them to adapt to the distinct
tissue microenvironment during the metastatic process. Mutational
and epigenetic changes in cancer cells strongly characterize these
events. In colorectal cancer (CRC), KRAS, NRAS, BRAF, and
PIK3CA mutations occur as oncogenic drivers, which are also
expected to play an essential role in these metastatic events.
The liver is the most frequently invaded metastatic
organ in CRC, and several studies have demonstrated mutational
signatures associated with CRC liver metastasis (1). CRC liver metastasis, even in multiple
metastatic cases, can be potentially cured by hepatic resection.
Lung metastasis occasionally occurs without liver metastasis;
however, mutational signatures associated with single lung
metastasis are not well known. Single lung metastasis is also
curable by partial resection of the lung. A better understanding of
the metastatic potential of primary CRCs would be significantly
beneficial in the treatment of CRC patients.
The mutational signature differences between primary
and metastatic lesions, especially those associated with single
lung metastasis or multiple metastases, has remained unclear. A
comparison of the clinicopathological and mutational profiles
associated with multiple/single lung metastases in CRC could
unravel the fundamental mechanisms underlying tumor metastasis and
help to identify early detection biomarkers.
The current study aimed to investigate the
clinicopathological and molecular features of tumor heterogeneity
by comparing the mutation status between the primary tumor and
corresponding metastatic lesions in order to detect factors
associated with multiple tumor metastases (which are usually
associated with worse prognosis).
Materials and methods
Case selection and histological
evaluation
A total of 2,912 cases of CRC were surgically
resected at the Juntendo University Hospital (Tokyo, Japan) between
2003 and 2017. We collected data and tissues from 31 CRC cases with
lung metastasis (20 with multiple metastases and 11 with single
metastasis) from the pathological record. The following
clinicopathological factors were evaluated: Gender, age, tumor
location, tumor size, histological type, lymphovascular invasion,
tumor budding, poor differentiated cluster, perineural invasion,
cancer stroma, depth of invasion, lymph node metastasis, distant
metastasis, and tumor-node-metastasis (TNM) stage. TNM staging was
determined using the 8th UICC TNM staging system of tumors of the
colon and rectum (2). The presence
of tumor budding (TB) and poorly differentiated cluster (PDC) were
evaluated at the invasion front as previously described. TB was
counted in the area with the highest density and classified as
follows: BD1: 0–4; BD2: 5–9; and BD3: ≥10 (×200 magnification).
Furthermore, PDC was classified into three groups: G1, G2, and G3,
when they have a maximum number of <5, 5–9, ≥10 PDC,
respectively; the counting was done in the highest density area at
×200 magnification (3). All patients
were followed-up every three months after surgery. The survival
periods were determined as survival times after diagnosis. The mean
follow-up time was 69.7 months (the range was 18–178 months).
Next-generation sequencing (NGS)
A CRC sample with heterochronous multiple
metastases, including brain metastasis, was subjected to NGS using
the Ion Ampliseq Cancer Hotspot Panel v2 (Thermo Fisher Scientific,
Inc.). The details of this NGS analysis are described in our
previously published article (4).
The patient with the aforementioned sample had experienced liver
metastasis on three occasions (in total, six lesions: Three lesions
at the first metastasis, two lesions at the second, and one lesion
at the third metastasis) followed by lung and brain metastases
during the 101 months after initial surgery. Due to poor sample
quality, only four recent samples (liver, lung, and two samples
from the brain metastasis) from this patient were examined by NGS.
Both tumoral and the corresponding non-tumoral DNA were extracted
using the QIAamp FFPE tissue kit (Qiagen).
Sanger sequences
Genomic DNA was extracted as previously described
(4). Sanger sequencing for
APC (Exon 16), KRAS (Exon 2),
NRAS (Exons 2 and 3), GNAS (Exon 8),
BRAF (Exon 15), CRAF (Exons 3, 11, and
14), CTNNB1 (Exon 3), PIK3CA (Exons 9
and 20), and TP53 (Exons 2,4,5,6,7, and 8) was
performed for all the cases. Furthermore, telomerase reverse
transcriptase (TERT) promoter mutations were also
identified. Primer sequences are described in Table SI. Polymerase chain reaction (PCR)
products were cut from the gel and purified by using DNA, RNA, and
protein purification kits (MACHEREY-NAGEL). Purified PCR products
were sequenced with dideoxynucleotides (BigDye Terminator v3.1;
Applied Biosystems; Thermo Fisher Scientific, Inc.), and specific
primers were purified using a BigDye X Terminator Purification kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) and analyzed
with a capillary sequencing machine (3730×l Genetic Analyzer;
Applied Biosystems; Thermo Fisher Scientific, Inc.). The sequences
were then examined by using Sequencing Analysis software version
3.5.1 software (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Mutations were evaluated by two of the authors (Y.Y. and
T.S.) and registered if the mutation peak height reached 20% of the
normal peak height. All mutations were verified by sequencing the
sense and antisense strands.
Immunohistochemistry
Tissue sections (4 µm) prepared from formalin-fixed
and paraffin-embedded tissues were subjected to
immunohistochemistry (IHC). Monoclonal antibodies against
β-catenin (clone 14, 1:200 dilution; BD Biosciences) and p53
(clone 1801, 1:1 dilution; BioGenex) were used. Antigen retrieval
was performed by heating in an autoclave in Tris-EDTA buffer (pH
6.0) for β-catenin and in Tris-EDTA buffer (pH 9.0) for p53.
The sections were incubated at 4°C overnight to react with primary
antibodies. Immunohistochemical staining was performed using an
Envision kit (Dako) with a substrate-chromogen solution.
β-catenin nuclear staining (labeling index) was evaluated in
three randomly selected areas by counting the number of positive
cells among at least 500 tumor cells at ×400 magnification. We
judged the tissue as positive for the overexpression of p53 when
the number of positive cells >10% of the total cell count
(Fig. S1) (4). Slides were evaluated by two independent
investigators (Y.Y and T.S.) without prior knowledge of the
clinicopathological data. Discrepancies were resolved by
reevaluation to reach a consensus.
Microsatellite instability
Microsatellite instability (MSI) analysis was
performed using five markers (Bethesda panel: BAT25, BAT26, D5S346,
D2S123, and D17S250) (5). Samples
with two or more altered markers were classified as MSI-high
(MSI-H), samples with one altered marker were classified as MSI-low
(MSI-L), and samples without altered markers were classified as
microsatellite stable (MSS). We used the primer sets for highly
fragmented DNA extracted from the FFPE tissue (6).
Survival analysis and statistical
analysis
Correlations between clinicopathological factors and
genetic alterations were analyzed by the Fisher's exact test,
chi-squared test, and Student's t-test. To elucidate the prognostic
impact of each factor, we performed Kaplan-Meier survival analysis
and log-rank tests. P<0.05 was considered to indicate a
statistically significant difference. These statistical analyses
were performed with JMP® ver.12 software (SAS Institute
Inc.).
Results
Clinicopathological differences
between CRC with multiple metastases and that with single lung
metastasis
Clinicopathological differences between CRC with
multiple metastases and that with single lung metastasis are
summarized in Table I. In this
study, the incidence of the single lung metastasis was 0.38% and
the cases in which the first metastatic focus was observed in the
lung was 0.52% among the multiple metastases group (multiple
metastases: 4, single lung metastasis: 11). All the cases with
multiple metastases were tubular adenocarcinoma, whereas 3 of 11
cases with single metastasis were mucinous adenocarcinoma. CRCs,
which eventually caused multiple metastases, were located evenly
from the right of the rectal origin of primary tumors, whereas none
of the CRCs with single lung metastasis arose from the right-sided
colon, although this difference was not statistically significant
(P=0.06). CRC with multiple metastases more frequently showed
vascular invasion, but not lymphatic invasion, than those with
single lung metastasis. Furthermore, CRC with multiple metastases
was associated with strong tumor budding (P=0.04). The presence of
PDC did not affect single or multiple metastatic states. CRC
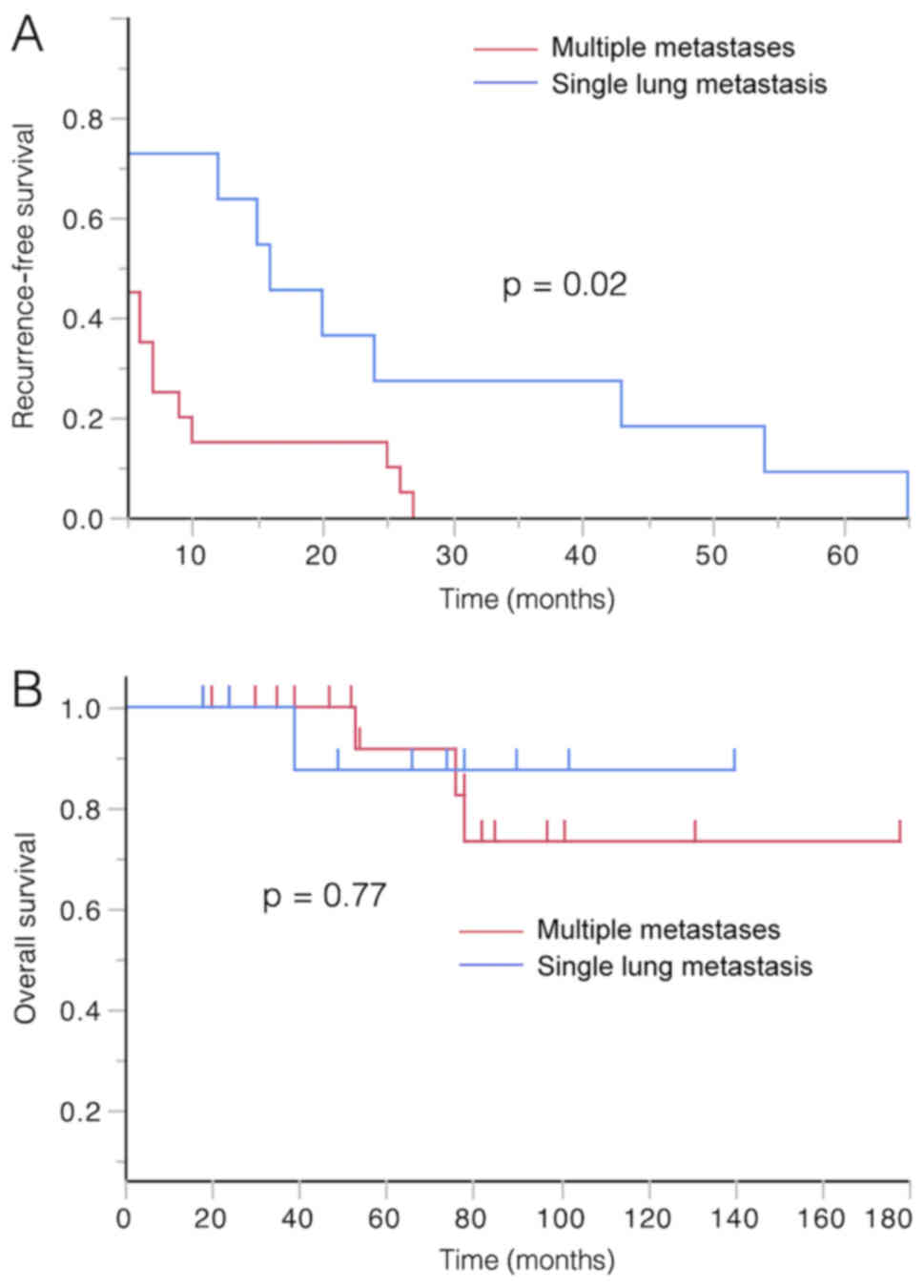
patients with multiple metastases presented shorter recurrence-free
survival rates compared with those with single lung metastasis with
statistical significance (P=0.02). However, there was no
significant difference between these two groups regarding overall
survival rates. The impact of KRAS, TP53, and APC
mutation signatures and IHC of p53 overexpression status on
clinicopathological factors were also assessed. TP53 and
APC mutations seemed to be associated with the presence of
lymphovascular invasion and strong tumor budding, respectively,
although these differences were not statistically significant
(Tables II and III). The overexpression of p53 tended to
be associated with vascular invasion, but this association was not
statistically significant (Table
IV). No significant association was found with KRAS
mutation (Table SII).
 | Table I.Clinicopathological differences
between colorectal cancer with multiple metastases and single
metastasis. |
Table I.
Clinicopathological differences
between colorectal cancer with multiple metastases and single
metastasis.
| Variable | Multiple
(n=20) | Single (n=11) | P-value |
|---|
| Sex, male/female
(n) | 13/7 | 8/3 |
>0.99a |
| Age, years (mean ±
SD) | 58.6±11.6 | 60.2±7.4 | 0.69b |
| Tumor site,
right/left/rectum (n) | 5/7/8 | 0/2/9 | 0.06a |
| Histological type,
tublar/mucinous (n) | 20/0 | 8/3 | 0.04a |
| Ly, 0/1a/1b/1c
(n) | 9/4/7/0 | 5/4/2/0 | 0.50a |
| V, 0/1a/1b/1c
(n) | 3/8/9/0 | 7/1/3/0 | 0.03a |
| Tumor budding,
G1/2/3 (n) | 14/0/6 | 10/1/0 | 0.04a |
| Poorly
differentiated cluster, G1/2/3 (n) | 4/12/4 | 1/9/1 | 0.62a |
| PN, no/yes (n) | 15/5 | 11/0 | 0.13a |
| Cancer stroma,
sci/int/med (n) | 2/12/6 | 1/8/2 | 0.85a |
| Depth of invasion,
T1/T2/T3/T4a (n) | 0/3/14/3 | 0/2/8/1 |
>0.99a |
| Lymph node
metastasis, 0/1/2/3 (n) | 6/11/2/1 | 4/6/1/0 |
>0.99a |
| TNM
I/II/IIIA/IIIB/IV (n) | 2/1/6/2/9 | 1/1/5/1/3 | 0.86a |
| Size, mm (mean ±
SD) | 44.4±18.8 | 37.7±13.3 | 0.26b |
| Survival period,
months (mean ± SD) | 72.8±46.8 | 64.0±11.4 | 0.77c |
| Recurrence-free
period, months (mean ± SD) | 6.7±9.0 | 22.6±22.3 | 0.02c |
| Mutation rate at
the primary site (n) |
|
|
|
|
KRAS mutated/wild | 12/8 | 7/4 |
>0.99a |
|
TP53 mutated/wild | 9/11 | 3/8 | 0.45a |
|
APC mutated/wild | 8/12 | 3/8 | 0.70a |
|
BRAF mutated/wild | 0/20 | 0/11 |
>0.99a |
|
CRAF mutated/wild | 0/20 | 0/11 |
>0.99a |
|
NRAS mutated/wild | 0/20 | 0/11 |
>0.99a |
|
GNAS mutated/wild | 0/20 | 0/11 |
>0.99a |
|
PIK3CA
mutated/wild | 0/20 | 0/11 |
>0.99a |
|
TERT promoter
mutated/wild | 0/20 | 0/11 |
>0.99a |
|
Immunohistochemistry (n) |
|
|
|
| p53
overexpression positive/negative | 11/9 | 4/7 | 0.46a |
 | Table II.Association between TP53
mutation status and clinicopathological factors. |
Table II.
Association between TP53
mutation status and clinicopathological factors.
|
| TP53, n | P-value |
|---|
|
|
|
|
|---|
| Variable | Wild | Mutated | χ2 | Fisher |
|---|
| Poorly
differentiated clusters |
|
|
|
|
|
G1/2/3 | 3/13/3 | 3/7/2 | 0.80 | >0.99 |
| Budding grade |
|
|
|
|
|
G1/2/3 | 15/1/3 | 11/1/0 | 0.34 | 0.36 |
| PN |
|
|
|
|
|
No/yes | 16/3 | 10/2 |
| >0.99 |
| Ly |
|
|
|
|
|
No/yes | 11/8 | 4/8 |
| 0.17 |
| V |
|
|
|
|
|
No/yes | 8/11 | 2/10 |
| 0.14 |
| Pathologic
type |
|
|
|
|
|
tub/muc | 18/1 | 10/2 |
| 0.95 |
| Location |
|
|
|
|
|
Right/left/rectum | 3/5/11 | 2/4/6 | 0.90 |
|
| pStage |
|
|
|
|
|
I/II/IIIA/IIIB/IV | 2/1/7/2/6 | 1/0/4/1/6 | 0.73 | 0.93 |
 | Table III.Correlation between APC
mutaion status and clinicopathological factors. |
Table III.
Correlation between APC
mutaion status and clinicopathological factors.
|
| APC, n | P-value |
|---|
|
|
|
|
|---|
| Variable | Wild | Mutated | χ2 | Fisher |
|---|
| Poorly
differentiated clusters |
|
|
|
|
|
G1/2/3 | 17/1/2 | 9/1/1 | 0.91 | >0.99 |
| Budding grade |
|
|
|
|
|
G1/2/3 | 18/0/2 | 6/1/4 | 0.06 | 0.06 |
| PN |
|
|
|
|
|
No/yes | 17/3 | 9/2 |
| >0.99 |
| Ly |
|
|
|
|
|
No/yes | 11/9 | 4/7 |
| 0.27 |
| V |
|
|
|
|
|
No/yes | 7/13 | 3/8 |
| 0.49 |
| Pathologic
type |
|
|
|
|
|
tub/muc | 17/3 | 11/0 |
| 0.25 |
| Location |
|
|
|
|
|
Right/left/rectum | 4/4/12 | 1/5/5 | 0.30 | 0.35 |
| pStage |
|
|
|
|
|
I/II/IIIA/IIIB/IV | 2/1/9/2/6 | 1/1/2/1/6 | 0.60 | 0.51 |
 | Table IV.Association between p53
immunohistochemistry and clinicopathological factors. |
Table IV.
Association between p53
immunohistochemistry and clinicopathological factors.
|
| p53 overexpression,
n | P-value |
|---|
|
|
|
|
|---|
| Variable | Positive | Negative | χ2 | Fisher |
|---|
| Poorly
differentiated clusters |
|
|
|
|
|
G1/2/3 | 4/8/3 | 2/12/2 | 0.44 | 0.51 |
| Budding grade |
|
|
|
|
|
G1/2/3 | 12/0/3 | 12/1/3 | 0.51 | >0.99 |
| PN |
|
|
|
|
|
No/yes | 12/3 | 14/2 |
| 0.65 |
| Ly |
|
|
|
|
|
No/yes | 7/8 | 8/8 |
| >0.99 |
| V |
|
|
|
|
|
No/yes | 3/12 | 7/9 |
| 0.25 |
| Pathologic
type |
|
|
|
|
|
Tub/muc | 13/2 | 15/1 |
| 0.60 |
| Location |
|
|
|
|
|
Right/left/rectum | 3/5/7 | 2/4/10 | 0.67 | 0.69 |
| pStage |
|
|
|
|
|
I/II/IIIA/IIIB/IV | 1/1/6/1/6 | 2/1/5/2/6 | 0.51 | >0.99 |
KRAS mutation signatures according to
metastatic events (Fig. 1)
KRAS mutations were found in 12 out of 20
cases with multiple metastases at the primary sites, and in 7 out
of 11 cases with single metastasis at the primary sites.
KRAS mutation signatures were maintained throughout the
metastatic events in most cases. In two cases (Case #M19 and #S2),
clones different from the primary sites were detected at the
metastatic sites, and in one case (Case #M20), clones with
KRAS mutation were first detected at the metastatic site,
whereas no KRAS mutation was detected in the primary tumor.
Both metastatic brain lesions in Case #M5 harbored KRAS
mutations similar to that of the primary tumor, and only two out of
the other seven metastatic lesions contained KRAS
mutations.
TP53 mutation signatures and p53
immunohistochemistry, and the relationship with metastatic events
(Fig 1)
TP53 mutations were found at the primary
sites in 9 of 20 cases with multiple metastases, and in 3 of 11
cases with single metastasis at the primary sites. In one patient
(Case #M14), aTP53 mutation was detected in the latest
metastatic lesion, despite the absence of a TP53 mutation at
the primary site. In another patient (Case #S7) with single
metastasis, a TP53 mutation was detected only in the primary
site. In most cases, TP53 mutation signatures were preserved
throughout the multiple metastatic events. Regarding a patient with
metastatic brain lesions (Case #M5), five out of seven metastatic
lesions contained TP53 mutations, in addition to the brain
metastatic tumors. The overexpression of p53 was observed in 11 of
20 multiple metastatic cases at the primary site (55.0%), 9 of
which had a TP53 mutation. Eight of 9 cases with a mutation
in the primary site showed overexpression of p53 in all the
metastatic sites; however, TP53 mutations were not preserved
in all of the metastatic lesions (Cases #M1, #M5, and #M7). In Case
#M5, the overexpression of p53 was not observed in two metastatic
lesions of the liver, one of which contained a TP53
mutation. In Case #M14, p53 overexpression was detected only in
metastatic lesions. A TP53 mutation was absent in the
primary site and detected only in the second metastatic site in the
lung. In patients with single metastasis, the overexpression of p53
was observed in 4 of 11 cases (36.3%), 3 of which had a TP53
mutation in the primary tumors. In one of the three cases with
single lung metastasis, the overexpression of p53 was also observed
at the metastatic site without a mutation (Case #S7). We also
observed the overexpression of p53 both at the primary and
metastatic sites in three cases without a TP53 mutation
(Case #M6, #M11, and #S3). There were no statistically significant
differences between multiple metastases and single lung metastasis
in terms of mutation ratio and overexpression of p53 (P=0.45 and
P=0.46, respectively).
APC mutation signatures according to
the metastatic events (Fig. 1)
APC mutations were detected in 8 out of 20
cases with multiple metastases at the primary sites, and in 3 out
of 11 cases with single metastasis at the primary sites. All
mutations were considered frameshift or nonsense, and the mutation
signatures were preserved in most cases. In one case (Case #M11),
frameshift and nonsense mutations were preserved from the primary
tumor to the three metastatic lesions. In another case (Case #M13),
APC mutation was detected only in one of two metastatic
sites without mutation at the primary site. Regarding the case with
metastatic brain lesions (Case #M5), six out of seven metastatic
lesions contained APC mutations in addition to the brain
metastatic tumors.
Clinicopathological and molecular
differences between CRC with multiple lung metastases and that with
single lung metastasis
There were two cases of multiple lung metastases
without metastasis to other organs (Case #M10 and #M16). The
primary tumors were located in the rectum in both cases, and both
tumors harbored a KRAS mutation. Furthermore, these two
cases tended to show histologically higher PDC grade, BD grade, and
vascular invasion compared with those with single lung metastasis.
It seemed that the primary tumors that can eventually cause
multiple lung metastases have higher metastatic potential compared
with single lung metastatic cases. However, there were no
significant differences between any clinicopathological and
molecular characteristics of multiple lung metastases and those of
single lung metastasis, although the study was limited by the small
sample size (Table SIII).
Sanger sequencing for other genes
Sanger sequencing was performed for TERT, CTNNB1,
BRAF, CRAF, NRAS, PIK3CA, and GNAS. No hot spot
mutations were detected in these genes.
Wnt signal activation in metastatic
CRC
Due to the frequency of APC (40.0% in
multiple metastatic cases, 27.3% in single lung metastatic cases)
and CTNNB1 (0%) mutations, the mutations in these series of
cases seemed to be relatively rare compared with reported values
(7−9). The status of Wnt signal activation was assessed by
β-catenin nuclear staining. Four out of 20 cases of multiple
metastases did not show β-catenin nuclear staining, whereas one of
them harbored an APC mutation. The average β-catenin nuclear
labeling index was 34.3% in multiple metastatic cases and 40.7% in
single lung metastatic cases (Table
SIV).
Status of microsatellite instability
in metastatic CRC
The status of MSI was also assessed for the primary
tumors. MSI-L was found in only two of the multiple metastatic
cases (Case #M7 and #M16) and the remaining tumors were classified
as MSS (Fig. 2).
Prognostic impacts of
clinicopathological factors and metastatic state
TNM stage at the time of the primary surgery
significantly affected the patients' overall survival rate (OSR)
and time to the first recurrence (TFR). TFR was significantly
shorter for the patients who eventually experienced multiple
metastases than for patients with single metastasis (Fig. 3A; P=0.02). However, although OSR was
slightly worse for patients who experienced eventual multiple
metastases, this finding was not statistically significant
(Fig. 3B; P=0.77).
Discussion
It has been reported that lung metastasis occurs in
0.67–15.8% of CRC (10,11). In this study, the incidence of single
lung metastasis was 0.38%, and the cases in which the first
metastatic focus was observed in the lung was 0.52% of the multiple
metastases group. The incidence rate in our study seems to be lower
than previously reported values. However, the previous studies
probably included ‘suspected cases’ examined only by computed
tomography (CT) or plain radiography, and the present study only
analyzed cases that were surgically resected and histologically
proven as metastasis. These differences might have influenced the
difference in the incidence. However, our findings suggest that the
first metastatic event can be observed in the lung in approximately
0.9% of CRC cases.
In this study, all CRC cases of right-sided origin
belonged to the multiple metastatic group, which is consistent with
the finding that right-sided primary CRCs show worse prognosis than
left-sided primary tumors (7,8,12). In contrast, it was unexpected that
the single lung metastatic group contained three cases of mucinous
carcinoma, and none were found in the multiple metastatic group.
Mucinous carcinoma, except for those with MSI-H, tends to show
worse prognosis than conventional adenocarcinoma (13–16). We
examined MSI status in the cases, but MSI-H was not found in any of
the cases. Therefore, the reason for the single metastasis of the
aggressive mucinous carcinoma to the lung, but not to other organs,
is unclear.
We included PDC and TB as pathological factors and
evaluated if these factors are associated with metastatic status,
as a recent study demonstrated that a grading system using PDC and
TB in neoplastic cells is a strong predictor of nodal metastases
and adverse outcome in colon cancer (17). Strong TB was frequently observed in
the group with multiple metastases and this finding was
statistically significant; however, PDC did not affect the
metastatic state. This finding provides us with a possible
therapeutic strategy for multiple metastases in CRC patients with
strong TB. Furthermore, the impact of KRAS, TP53, and
APC mutation signatures on clinicopathological factors was
also assessed. To date, the impact of the mutation signatures on
PDC and TB has not been reported. This study revealed that
TP53 and APC mutations were able to indicate the
presence of lymphovascular invasion; however, none of the genetic
alterations was related to PDC and TB.
A recent study demonstrated that right-sided primary
microsatellite stable CRC is associated with shorter survival rate
and increased mutation ratio (18).
In this study, CRCs with multiple metastases were almost evenly
distributed in the right/left/rectal regions, whereas none of the
CRCs with single lung metastasis arose from the right-sided colon.
The occurrence of single lung metastasis was associated with the
location of the primary tumors. However, the location of the
primary tumor did not affect the eventual multiple metastases in
this series of CRCs, which almost completely comprised MSS with
only two cases of MSI-L.
Clonal evolution plays an essential role in the
metastatic process of CRC. Regarding this point, different clones
of KRAS mutations were identified at the metastatic lung
sites in two cases (Case #M19 and #S2). However, a drastic change
in genetic alterations could not be detected in any of the genes
analyzed in this study. This is consistent with previous findings
indicating that the rate of epigenetic change has been estimated to
be orders of magnitude higher than that of genetic alterations and
is considered the primary determinant of clonal evolution (19–21).
Mutated clones were observed in the metastatic lesion, one case
each from multiple metastatic and single lung metastatic tumors,
despite the presence of the wild-type alleles in the primary tumor.
However, KRAS, TP53, and APC mutation signatures seemed to
be preserved throughout the metastatic events in cases with
detected mutations. Therefore, the analysis of cell-free DNA or DNA
derived from circulating tumor cells to detect the mutations in the
primary tumor may be helpful for the early detection of CRC
metastasis.
In this series, APC mutations were detected
in 11 cases (8 in the primary tumors from multiple metastatic cases
and 3 in the primary tumor of the single metastatic case), and
CTNNB1 mutations including in-frame deletions were not found
in any of the cases. The frequency of APC mutations has been
shown in up to 90% of colorectal adenocarcinomas, and the frequency
of CTNNB1 mutations was reported to be less than 10% in
colorectal adenocarcinoma (7,18,22).
However, the prognostic value has not been clearly shown. In
contrast, it has been shown that CRC with wild type APC has
a worse prognosis than those with APC mutations (23). Although the frequency of APC
mutations seemed lower than the reported value, but this is
probably due to the fact that this study involved only metastatic
tumors.
In addition, we came across a case (Case #M5) with
brain metastasis in CRC. Brain metastasis in patients with CRC is
reported to be rare, and little is known regarding the mutations
involved in this process. A previous study analyzed molecular
profiling in 30 cases of metastatic brain samples out of 2010
samples with metastatic CRCs and demonstrated that brain metastasis
showed the highest KRAS mutation rate (24). In this study, both lesions of brain
metastasis contained KRAS mutations similar to that detected
in the primary tumor, whereas only two out of seven metastatic
tumors from the same patient contained this type of mutation. Our
findings are consistent with previous findings and also provide
evidence that anti-epidermal growth factor receptor therapy is not
effective for the treatment of metastatic brain tumors in CRC.
Regarding liver metastasis, 12 out of 20 cases in
multiple metastatic tumors harbored KRAS mutations in the
primary tumor, and 8 of these 12 patients developed liver
metastasis. Seven out of 14 metastatic liver samples from the eight
patients did not harbor KRAS mutations. Liver metastasis is
associated with TOPO2A gene amplification but not with a
high frequency of KRAS mutation, although TOPO2A gene
amplification was not examined in our series (24). A previous study reported that tumors
with KRAS mutations were more likely to develop lung
metastasis. However, the overall survival did not differ according
to the KRAS status (25). In
this study, 12 out of 20 (60.0%) cases with multiple metastases and
7 out of 11 (63.6%) cases with single lung metastasis harbored a
KRAS mutation at the primary sites.
Furthermore, there was no significant difference
between the two groups with regards to overall survival. Thus,
KRAS mutation status did not affect single/multiple lung
metastases in this study. Moreover, in this study, BRAF
mutations were not detected in many cases, which is consistent with
previous findings (24).
With respect to treatment, the general condition of
the patients allowed the resection of the metastatic lesions on
several occasions in most of the patients with multiple metastases.
It is reported that the resection of liver and lung metastases
provides good long-term survival (26). All patients who succumbed from the
disease (multiple metastasis: 3, single lung metastasis: 1) could
not continue with the postoperative adjuvant chemotherapy or
undergo subsequent surgery due to poor performance status and the
side effects of the drugs.
Finally, CRC is known, in general, to initially
spread to the liver, and then to the lung and the brain. It was
reported that synchronous liver and pulmonary metastases occur in
45 to 70% of patients with CRC (9).
Besides, lung metastasectomy in patients with previously resected
liver metastases showed a significantly better five-year survival
(27). Closer observation of the
liver and lung metastases is needed to improve the prognosis of
patients. The rationales for comparing single lung metastasis and
multiple lung metastases in CRC in the current study are as
follows: (1) clinicians need to
carefully follow-up the patients who experienced early relapse, as
they have a higher risk of multiple metastasis in the near future.
(2) lung metastasis from CRC is
usually encountered after liver metastasis. In the case of single
lung metastasis after CRC, the possibility of primary pulmonary
adenocarcinoma with enteric differentiation needs to be ruled out
(28,29). Frequently conserved mutations in
TP53, APC, and KRAS, together with p53 IHC findings,
could help to distinguish metastasis from primary pulmonary
adenocarcinoma with enteric differentiation.
In conclusion, early relapses in CRC patients could
be a sign of eventual multiple metastases, although this may not
affect the overall survival of CRC patients. Drastic mutational
changes seem rare during metastatic events in CRC patients.
A few limitations can be considered in this study.
First, the numbers of the cases were too small to draw definitive
conclusions. The sample number should be the same in each group.
However, based on available pathological records, we found only 31
CRC cases with lung metastasis (20 with multiple metastases and 11
with single metastasis) from amongst 2,912 cases of CRC. Therefore,
it is not possible to increase the number of cases. More sample
accumulation is necessary to find more persuasive correlations or
differences. Second, we verified mutation findings only in
TP53 but not in APC and KRAS. We employed p53
IHC to verify the mutation findings, because it is well known that
p53 IHC antibody is able to detect mutated p53 as overexpression.
However, there are no commercially available IHC antibodies that
can efficiently detect mutated APC and KRAS, making it difficult to
confirm the mutation findings.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by a
Grant-in-Aid for General Scientific Research from the Ministry of
Education, Science, Sports and Culture (grant no. 17K08704 to TY;
Tokyo, Japan).
Availability of data and materials
All datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TS, TH and TY planned this study and diagnosed the
surgical specimens. YY, YA, NY and ST performed the experiments and
analyzed the data. YY and TS wrote the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Juntendo University, Tokyo, Japan (approval no.
17-214), and all patients provided written informed consent prior
to enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
APC
|
adenomatous polyposis coli
|
|
BRAF
|
B-Raf proto-oncogene, serine/threonine
kinase
|
|
CRAF
|
Raf-1 proto-Oncogene, serine/threonine
kinase
|
|
CTNNB1
|
catenin β-1
|
|
GNAS
|
guanine nucleotide binding protein, α,
stimulating complex locus
|
|
KRAS
|
KRAS proto-oncogene, GTPase
|
|
NGS
|
next-generation sequencing
|
|
NRAS
|
NRAS proto-oncogene, GTPase
|
|
PDC
|
poorly differentiated cluster
|
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate
3-kinase catalytic subunit α
|
|
TB
|
tumor budding
|
|
TOPO2A
|
DNA topoisomerase II α
|
|
TP53
|
tumor protein p53
|
|
TERT
|
telomere reverse transcriptase
|
References
|
1
|
Taki K, Ohmuraya M, Tanji E, Komatsu H,
Hashimoto D, Semba K, Araki K, Kawaguchi Y, Baba H and Furukawa T:
GNAS(R201H) and Kras(G12D) cooperate to promote murine pancreatic
tumorigenesis recapitulating human intraductal papillary mucinous
neoplasm. Oncogene. 35:2407–2412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brierley JD, Gospodarowicz MK and
Wittekind C: TNM classification of malignant tumors, 8th
edition. Wiley-Blackwell. 2017.
|
|
3
|
Lee VWK and Chan KF: Tumor budding and
poorly-differentiated cluster in prognostication in stage II colon
cancer. Pathol Res Pract. 214:402–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Akazawa Y, Saito T, Hayashi T, Yanai Y,
Tsuyama S, Akaike K, Suehara Y, Takahashi F, Takamochi K, Ueyama H,
et al: Next-generation sequencing analysis for gastric
adenocarcinoma with enteroblastic differentiation: Emphasis on the
relationship with hepatoid adenocarcinoma. Hum Pathol. 78:79–88.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boland CR, Thibodeau SN, Hamilton SR,
Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA,
Fodde R, Ranzani GN and Srivastava S: A national cancer institute
workshop on microsatellite instability for cancer detection and
familial predisposition: Development of international criteria for
the determination of microsatellite instability in colorectal
cancer. Cancer Res. 58:5248–5257. 1998.PubMed/NCBI
|
|
6
|
Umetani N, Sasaki S, Watanabe T, Ishigami
H, Ueda E and Nagawa H: Diagnostic primer sets for microsatellite
instability optimized for a minimal amount of damaged DNA from
colorectal tissue samples. Ann Surg Oncol. 7:276–280. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jass JR, Young J and Leggett BA: Evolution
of colorectal cancer: Change of pace and change of direction. J
Gastroenterol Hepatol. 17:17–26. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miyaki M, Iijima T, Kimura J, Yasuno M,
Mori T, Hayashi Y, Koike M, Shitara N, Iwama T and Kuroki T:
Frequent mutation of beta-catenin and APC genes in primary
colorectal tumors from patients with hereditary nonpolyposis
colorectal cancer. Cancer Res. 59:4506–4509. 1999.PubMed/NCBI
|
|
10
|
Parnaby CN, Bailey W, Balasingam A,
Beckert L, Eglinton T, Fife J, Frizelle FA, Jeffery M and Watson
AJ: Pulmonary staging in colorectal cancer: A review. Colorectal
Dis. 14:660–670. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mitry E, Guiu B, Cosconea S, Jooste V,
Faivre J and Bouvier AM: Epidemiology, management and prognosis of
colorectal cancer with lung metastases: A 30-year population-based
study. Gut. 59:1383–1388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boeckx N, Koukakis R, Op de Beeck K, Rolfo
C, Van Camp G, Siena S, Tabernero J, Douillard JY, André T and
Peeters M: Primary tumor sidedness has an impact on prognosis and
treatment outcome in metastatic colorectal cancer: Results from two
randomized first-line panitumumab studies. Ann Oncol. 28:1862–1868.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanemitsu Y, Kato T, Hirai T, Yasui K,
Morimoto T, Shimizu Y, Kodera Y and Yamamura Y: Survival after
curative resection for mucinous adenocarcinoma of the colorectum.
Dis Colon Rectum. 46:160–167. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jimi S, Hotokezaka M, Ikeda T, Uchiyama S,
Hidaka H, Maehara N, Ishizaki H and Chijiiwa K: Clinicopathological
features, postoperative survival and prognostic variables for
cancer-related survival in patients with mucinous colorectal
carcinoma. Surg Today. 45:329–334. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamaguchi T, Taniguchi H, Fujita S, Sekine
S, Yamamoto S, Akasu T, Kushima R, Tani T, Moriya Y and Shimoda T:
Clinicopathological characteristics and prognostic factors of
advanced colorectal mucinous adenocarcinoma. Histopathology.
61:162–169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ott C, Gerken M, Hirsch D, Fest P,
Fichtner-Feigl S, Munker S, Schnoy E, Stroszczynski C, Vogelhuber
M, Herr W, et al: Advanced mucinous colorectal cancer:
Epidemiology, prognosis and efficacy of chemotherapeutic treatment.
Digestion. 98:143–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reggiani Bonetti L, Barresi V, Bettelli S,
Caprera C, Manfredini S and Maiorana A: Analysis of KRAS, NRAS,
PIK3CA, and BRAF mutational profile in poorly differentiated
clusters of KRAS-mutated colon cancer. Human Pathol. 62:91–98.
2017. View Article : Google Scholar
|
|
18
|
Yaeger R, Chatila WK, Lipsyc MD, Hechtman
JF, Cercek A, Sanchez-Vega F, Jayakumaran G, Middha S, Zehir A,
Donoghue MTA, et al: Clinical sequencing defines the genomic
landscape of metastatic colorectal cancer. Cancer Cell.
33:125–136.e123. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Greaves M and Maley CC: Clonal evolution
in cancer. Nature. 481:306–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maley CC, Aktipis A, Graham TA, Sottoriva
A, Boddy AM, Janiszewska M, Silva AS, Gerlinger M, Yuan Y, Pienta
KJ, et al: Classifying the evolutionary and ecological features of
neoplasms. Nat Rev Cancer. 17:605–619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Siegmund KD, Marjoram P, Woo YJ, Tavaré S
and Shibata D: Inferring clonal expansion and cancer stem cell
dynamics from DNA methylation patterns in colorectal cancers. Proc
Natl Acad Sci USA. 106:4828–4833. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Giannakis M, Mu XJ, Shukla SA, Qian ZR,
Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, et
al: Genomic correlates of immune-cell infiltrates in colorectal
carcinoma. Cell Rep. 17:12062016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schell MJ, Yang M, Teer JK, Lo FY, Madan
A, Coppola D, Monteiro AN, Nebozhyn MV, Yue B, Loboda A, et al: A
multigene mutation classification of 468 colorectal cancers reveals
a prognostic role for APC. Nat Comm. 7:117432016. View Article : Google Scholar
|
|
24
|
El-Deiry WS, Vijayvergia N, Xiu J,
Scicchitano A, Lim B, Yee NS, Harvey HA, Gatalica Z and Reddy S:
Molecular profiling of 6,892 colorectal cancer samples suggests
different possible treatment options specific to metastatic sites.
Cancer Biol Ther. 16:1726–1737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pereira AA, Rego JF, Morris V, Overman MJ,
Eng C, Garrett CR, Boutin AT, Ferrarotto R, Lee M, Jiang ZQ, et al:
Association between KRAS mutation and lung metastasis in advanced
colorectal cancer. Br J Cancer. 112:424–428. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rajakannu M, Magdeleinat P, Vibert E,
Ciacio O, Pittau G, Innominato P, SaCunha A, Cherqui D, Morère JF,
Castaing D and Adam R: Is cure possible after sequential resection
of hepatic and pulmonary metastases from colorectal cancer? Clin
Colorectal Cancer. 17:41–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wiegering A, Riegel J, Wagner J, Kunzmann
V, Baur J, Walles T, Dietz U, Loeb S, Germer CT, Steger U and Klein
I: The impact of pulmonary metastasectomy in patients with
previously resected colorectal cancer liver metastases. PLoS One.
12:e01739332017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mackinnon AC Jr, Luevano A, de Araujo LC,
Rao N, Le M and Suster S: Cribriform adenocarcinoma of the lung:
Clinicopathologic, immunohistochemical, and molecular analysis of
15 cases of a distinctive morphologic subtype of lung
adenocarcinoma. Mod Pathol. 27:1063–1072. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matsushima J, Yazawa T, Suzuki M,
Takahashi Y, Ota S, Nakajima T, Yoshino I, Yokose T, Inoue T,
Kawahara K and Nakatani Y: Clinicopathological,
immunohistochemical, and mutational analyses of pulmonary enteric
adenocarcinoma: Usefulness of SATB2 and β-catenin immunostaining
for differentiation from metastatic colorectal carcinoma. Hum
Pathol. 64:179–185. 2017. View Article : Google Scholar : PubMed/NCBI
|