Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide (1–4). In 2019, there were almost 145,600 newly
diagnosed patients, and more than 50,000 patients died from the
disease in the United States (4).
Surgery and adjuvant chemotherapy are the main treatments for CRC.
The 5-year survival rate following surgical resection of colorectal
metastases has increased from 25 to 55%, but most patients relapse
(5,6). Therefore, further investigation on the
underlying mechanisms involved in the development and progression
of CRC may provide novel directions for CRC treatment.
Calcium-activated chloride channel (CLCA) regulators
are proteins with a symmetrical homocysteine motif in the terminal
tail of the amino group (7). The
loci of the human CLCA genes are at chromosome 1p31-1p22 (8). Research has demonstrated that CLCA
protein members influence a wide range of biological processes,
including cell differentiation, adhesion, apoptosis and airway
inflammation (7–9). Recent studies have revealed that the
expression levels of CLCA proteins, such as CLCA1, CLCA2 and CLCA4,
are abnormal in a variety of cancer types (10–16), and
could therefore be potential cancer predictors for patients. CLCA1
inhibits the proliferation of CRC cells and has been associated
with a favorable prognosis of patients with CRC (10). CLCA2 is a p53-inducible inhibitor of
cell proliferation and could be a marker of differentiated
epithelium that is downregulated with tumor progression (11). Low CLCA2 expression promotes cell
proliferation and metastasis by driving the
epithelial-to-mesenchymal transition signaling pathway (12,13).
CLCA4 has a similar structure to CLCA1 and CLCA2 (17), and loss of CLCA4 expression has been
observed in hepatocellular carcinoma (14), breast cancer (15) and bladder cancer (16), facilitating tumor cell growth and
metastasis via epithelial-to-mesenchymal transition. However, to
the best of our knowledge, the role of CLCA4 in the prognosis of
patients with CRC has not been well clarified. The present study
aimed to measure the expression levels of CLCA4 during the
development of CRC and assess its association with the survival of
patients with different types of tumor, including CRC.
Materials and methods
Microarray data information and CLCA4
gene identification
The gene chip detection technique has been employed
for more than 10 years (18). The
National Center for Biotechnology Information (NCBI)-Gene
Expression Omnibus (GEO) is a free database of microarray/gene
profiles (https://www.ncbi.nlm.nih.gov/geo). For the present
study, an original microarray dataset, GSE49355 (19), was downloaded from the NCBI-GEO
database. The microarray data of GSE49355 is based on the GeneChip
Human Genome U133 and includes 21 patients with advanced CRC
(submission date: September 1, 2013).
Oncomine platform analysis
The transcriptional levels of CLCA4 in CRC specimens
and normal controls were analyzed using the Oncomine platform
(https://www.oncomine.org) (20). Additionally, the threshold of the
P-values, fold-change and gene bank in cancerous tissues compared
with non-cancerous type-matched tissues were evaluated.
GEPIA analysis
As an interactive web that includes 9,736 tumors and
8,587 normal samples from TCGA and the GTEx projects, the online
database Gene Expression Profiling Interactive Analysis (GEPIA) was
used to analyze the expression of CLCA4 in both colon and rectal
cancers using Colon Adenocarcinoma and Rectum Adenocarcinoma
datasets of TCGA (21), compared to
Match TCGA normal data.
The Human Protein Atlas (THPA)
database analysis
THPA, as a database containing images from
immunohistochemical (IHC)-based tissue microarrays (TMAs; 46 normal
human tissues and 20 types of human cancer) for 11,250 human
proteins, was accessed to analyze CLCA4 protein expression in CRC
tissues and non-cancerous colorectal tissues, and the images and
expression levels of CLCA4 were downloaded from the website
(22). Furthermore, Kaplan-Meier
survival analysis was performed using log-rank tests via THPA
website to analyze the association between CLCA4 expression and the
overall survival rate of tumor patients originally sourced from The
Cancer Genome Atlas (TCGA) database (cancer.gov/tcga). The auto ‘Best expression cut off’
was set as cut off for high and low expression of CLCA4 (CRC: High
>5.31, low ≤5.31; breast cancer: High >0.05, low ≤0.05; head
and neck cancer: High >0.81, low ≤0.81; stomach cancer: High
>0.02, low ≤0.02).
IHC-based TMA analysis
To assess the protein expression levels of CLCA4 in
CRC samples, a TMA (cat. no. HLinAde075Met01) containing colon
cancer (n=8) and rectal cancer (n=6) tissues, and their paired
non-cancerous colon and rectal tissues was used. To further explore
the role of CLCA4 expression during the development of CRC, a TMA
(cat. no. HColAde080CD01) with colorectal normal (n=4), adenoma
(n=11) and carcinoma (n=11) tissues was used. The tissue specimens
for both TMAs were original collected from Taizhou Hospital of
Zhejiang Province (Taizhou, China) and tissue collection was
approved by its Ethics Committee in accordance with the principles
of the Declaration of Helsinki. Both of the aforementioned TMAs
were provided by Shanghai Outdo Biotech. IHC was performed to
detect CLCA4 protein expression in TMAs using the standard
technique (23). Briefly, the
sections (4 μm) were incubated with a primary antibody against
CLCA4 (dilution, 1:100; cat. no. 35684; Signalway Antibody LLC)
overnight at 4°C after blocking endogenous peroxidase and proteins,
and were subsequently incubated with HRP-labeled anti-rabbit
secondary antibody (ready to use) for 1 h at room temperature using
UltraSensitive™ SP IHC kit (cat. no. KIT-9710; Maxim Biomedical,
Inc.). At the end of the experiment, the slides were scanned using
a NanoZoomer 2.0 HT slide scanner (Hamamatsu Photonics K.K.). The
intensity and percentage of positively stained cells were analyzed
by experienced pathologists who were blinded to the clinical and
pathological data. CLCA4 expression among tissues was analyzed
using IHC scores calculated using the following formula: Final
score=intensity score (0, no staining; 1, weak; 2, moderate; and 3,
strong) × percentage score (1, 1–25% positive; 2, 26–50% positive;
3, 51–75% positive; and 4, 76–100% positive).
Statistical analysis
All statistical tests were conducted using SPSS
version 19.0 (IBM Corp.). For the data from GSE49355 dataset and
IHC score analysis, 2 related samples comparison were performed
with Wilcoxon signed-rank test, and k independent samples
comparisons were analyzed with Kruskal-Wallis test followed by
Mann-Whitney U tests and Bonferroni's was used to correct multiple
comparisons. P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of lower CLCA4 mRNA
expression in CRC and metastatic tissues
The NCBI-GEO is a free database of microarray/gene
profiles and next-generation sequencing, from which differentially
expressed genes between CRC and normal tissues were identified in
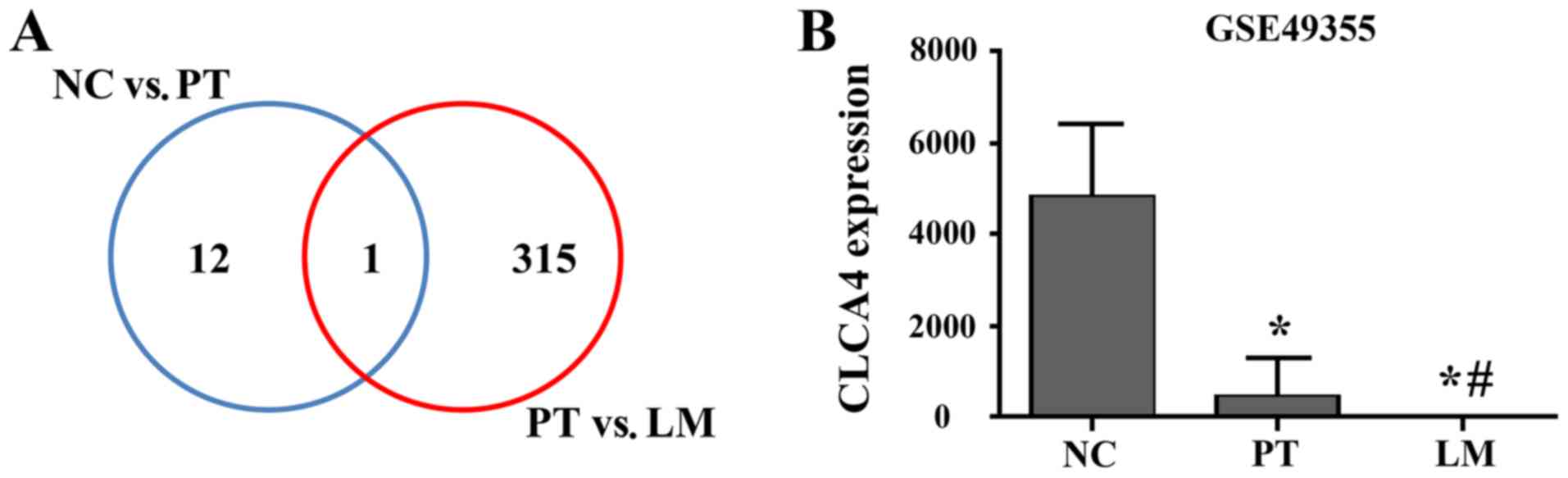
the GSE49355 dataset. CLCA4 was identified as the only
differentially expressed gene between normal colon tissues and
primary tumor tissues, and between primary tumors and liver
metastases (Fig. 1A). As presented
in Fig. 1B, CLCA4 was highly
expressed in normal colon tissues, while its expression was
significantly downregulated in primary tumor tissues (P<0.05
versus non-cancerous colorectal tissues). Furthermore, CLCA4 mRNA
expression was further downregulated in liver metastatic tissues
(P<0.05 versus non-cancerous colorectal tissues and versus
primary CRC tissues).
Lower CLCA4 mRNA expression in CRC
tissues
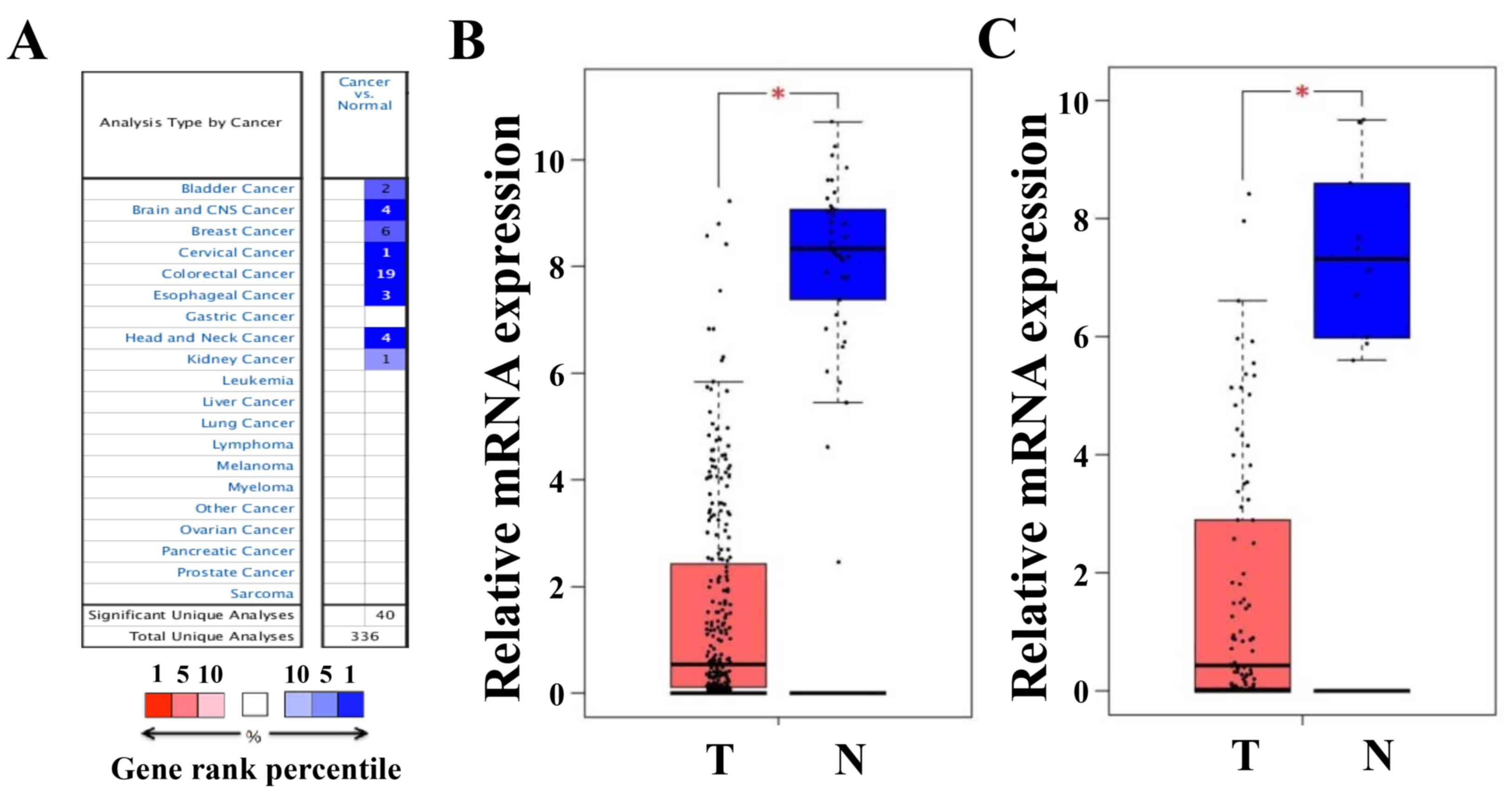
The Oncomine platform was used to analyze the
transcriptional levels of CLCA4 in CRC and other cancer specimens,
and in normal controls. As presented in Fig. 2A, CLCA4 mRNA expression was markedly
lower in multiple types of cancer tissues, including CRC, than in
normal type-matched tissues. The decreases in CLCA4 expression in
CRC tissues from 19 Oncomine datasets were presented in Table I (24–28).
Furthermore, analyses of CLCA4 expression from the Colon
Adenocarcinoma and Rectum Adenocarcinoma datasets of TCGA through
the GEPIA website further confirmed the decrease in CLCA4
expression in both colon and rectal cancer tissues (both P<0.05
versus non-cancerous colorectal tissues; Fig. 2B).
 | Table I.Oncomine analysis of chloride channel
accessory 4 expression in colorectal cancer. |
Table I.
Oncomine analysis of chloride channel
accessory 4 expression in colorectal cancer.
| Cohort | Data type | Sample (n) | Fold-change | P-value |
|---|
| Hong Colorectal
(24) | mRNA | Colorectal Carcinoma
(70) vs. Normal (12) | −594.53 |
3.06×10−37 |
| Kaiser Colon
(25) | mRNA | Rectosigmoid
Adenocarcinoma (10) vs. Normal (5) | −89.63 |
1.88×10−10 |
|
| mRNA | Rectal Adenocarcinoma
(8) vs. Normal (5) | −107.16 |
6.54×10−9 |
|
| mRNA | Colon Mucinous
Adenocarcinoma (13) vs. Normal (5) | −94.18 |
3.12×10−11 |
|
| mRNA | Cecum
Adenocarcinoma (17) vs. Normal (5) | −86.41 |
1.38×10−12 |
|
| mRNA | Colon
Adenocarcinoma (41) vs. Normal (5) | −54.90 |
5.91×10−14 |
| Skrzypczak
Colorectal (26) | mRNA | Colon
Adenocarcinoma (45) vs. Normal (24) | −57.90 |
3.59×10−14 |
|
| mRNA | Colorectal
Carcinoma (36) vs. Normal (24) | −29.41 |
8.46×10−13 |
| Skrzypczak 2
Colorectal (26) | mRNA | Colorectal
Carcinoma (5) vs. Normal (10) | −214.32 |
4.80×10−9 |
|
| mRNA | Colon Adenoma (5)
vs. Normal (10) | −281.61 |
2.00×10−7 |
|
| mRNA | Colon Adenoma
Epithelia (5) vs. Normal (10) | −35.33 |
1.41×10−6 |
|
| mRNA | Colon Carcinoma
Epithelia (5) vs. Normal (10) | −36.68 |
1.25×10−6 |
| Sabates-Bellver
Colon (27) | mRNA | Rectal Adenoma (7)
vs. Normal (32) | −51.08 |
6.65×10−5 |
|
| mRNA | Colon Adenoma (25)
vs. Normal (32) | −28.15 |
3.99×10−9 |
| TCGA
Colorectal | mRNA | Cecum
Adenocarcinoma (22) vs. Normal (22) | −31.51 |
1.08×10−12 |
|
| mRNA | Rectal
Adenocarcinoma (60) vs. Normal (22) | −20.29 |
9.89×10−18 |
|
| mRNA | Colon
Adenocarcinoma (101) vs. Normal (22) | −10.47 |
4.55×10−14 |
|
| mRNA | Colon Mucinous
Adenocarcinoma (22) vs. Normal (22) | −14.69 |
1.93×10−8 |
| Gaedcke Colorectal
(28) | mRNA | Rectal
Adenocarcinoma (65) vs. Normal (65) | −13.91 |
1.03×10−20 |
Lower CLCA4 protein expression in CRC
tissues
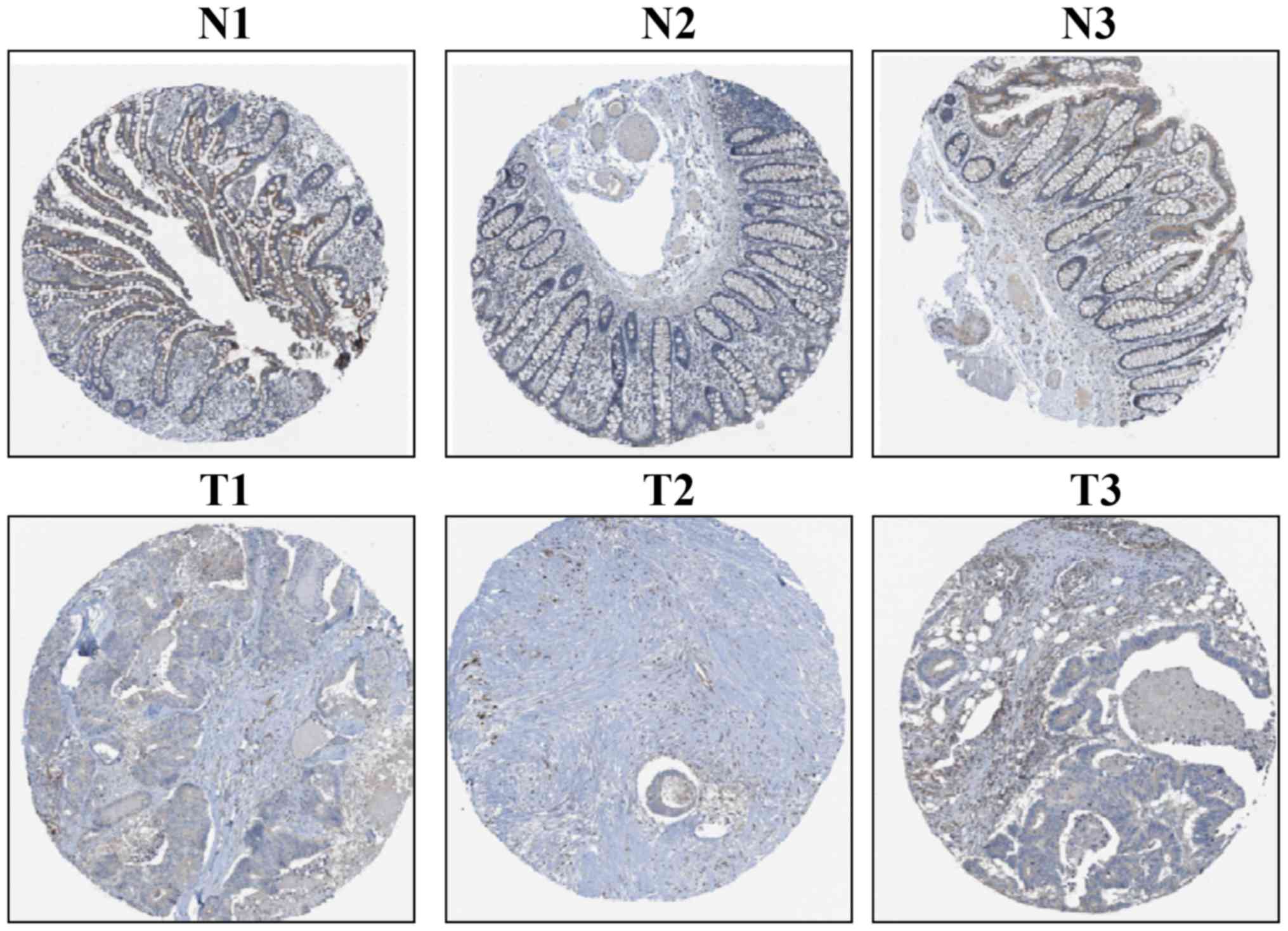
The IHC-based THPA database was used to assess CLCA4
protein expression in CRC tissues. CLCA4 protein expression was
moderate in three non-cancerous colorectal tissues and was weak
(n=4) or not detectable (n=8) in 12 CRC tissues; representative
images are presented in Fig. 3. The
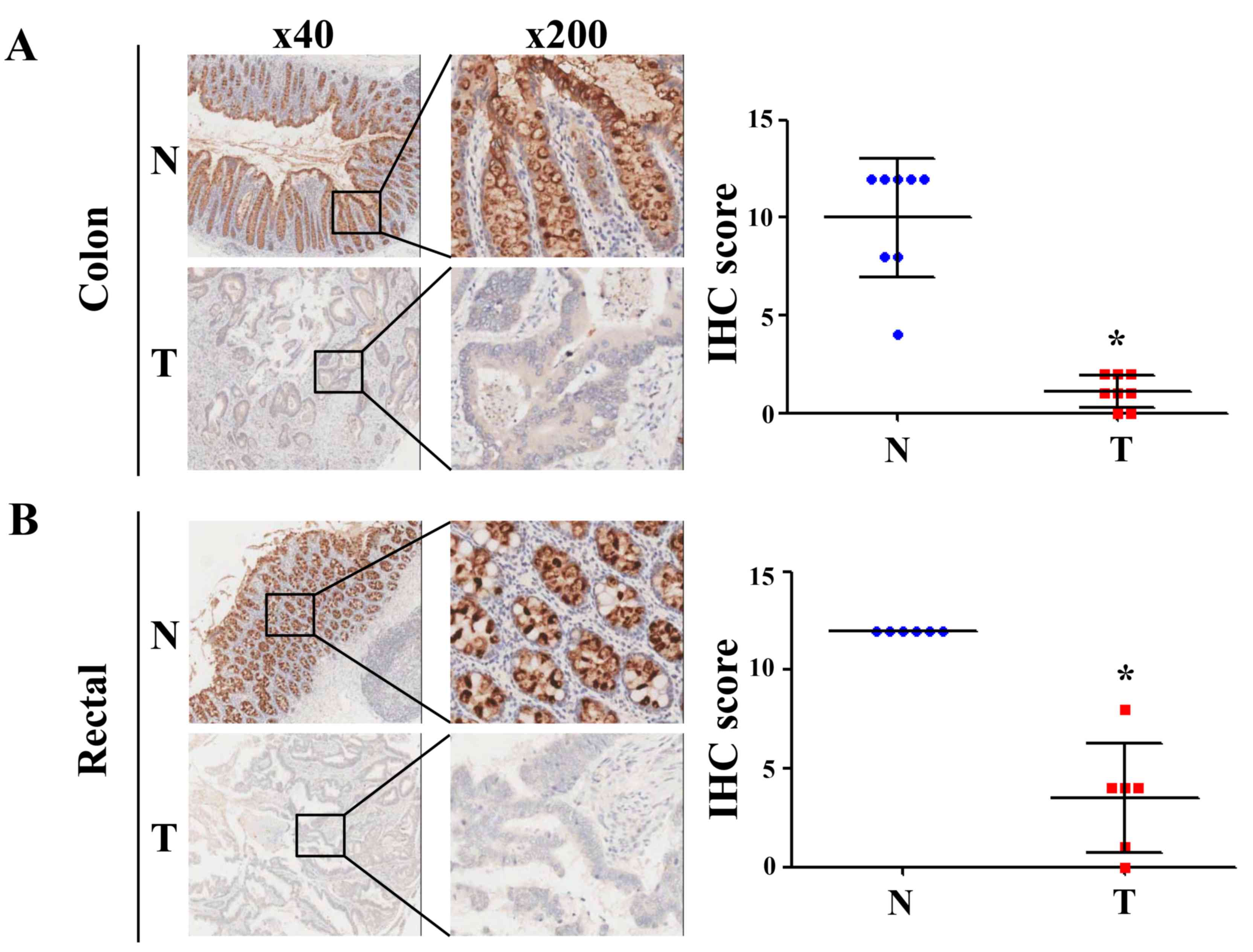
determination of CLCA4 protein expression from the IHC-based TMAs
revealed that CLCA4 protein expression was downregulated in both
colon and rectal cancer tissues (P<0.05 versus paired
non-cancerous colon or rectal tissues; Fig. 4). The clinical characteristics of the
patients with colon and rectal cancer are presented in Table II. The aforementioned experiments
revealed that CLCA4 protein expression was downregulated in CRC
tissues.
 | Table II.Clinicopathological features of 14
patients with colorectal cancer. |
Table II.
Clinicopathological features of 14
patients with colorectal cancer.
| Characteristic | n (%) |
|---|
| Age, years |
|
|
<65 | 11 (79) |
|
≥65 | 3 (21) |
| Sex |
|
|
Female | 3 (21) |
|
Male | 11 (79) |
| Tumor location |
|
|
Rectum | 6 (43) |
|
Colon | 8 (57) |
| Clinical stage |
|
| I | 0 (0) |
| II | 12 (86) |
|
III | 2 (14) |
| IV | 0 (0) |
| T stage |
|
| T1 | 5 (36) |
| T2 | 0 (0) |
| T3 | 8 (57) |
| T4 | 1 (7) |
| N stage |
|
| N0 | 7 (50) |
| N1 | 5 (36) |
| N2 | 2 (14) |
| M stage |
|
| M0 | 12 (86) |
| M1 | 2 (14) |
| Lymph node
metastasis |
|
|
Yes | 6 (43) |
| No | 8 (57) |
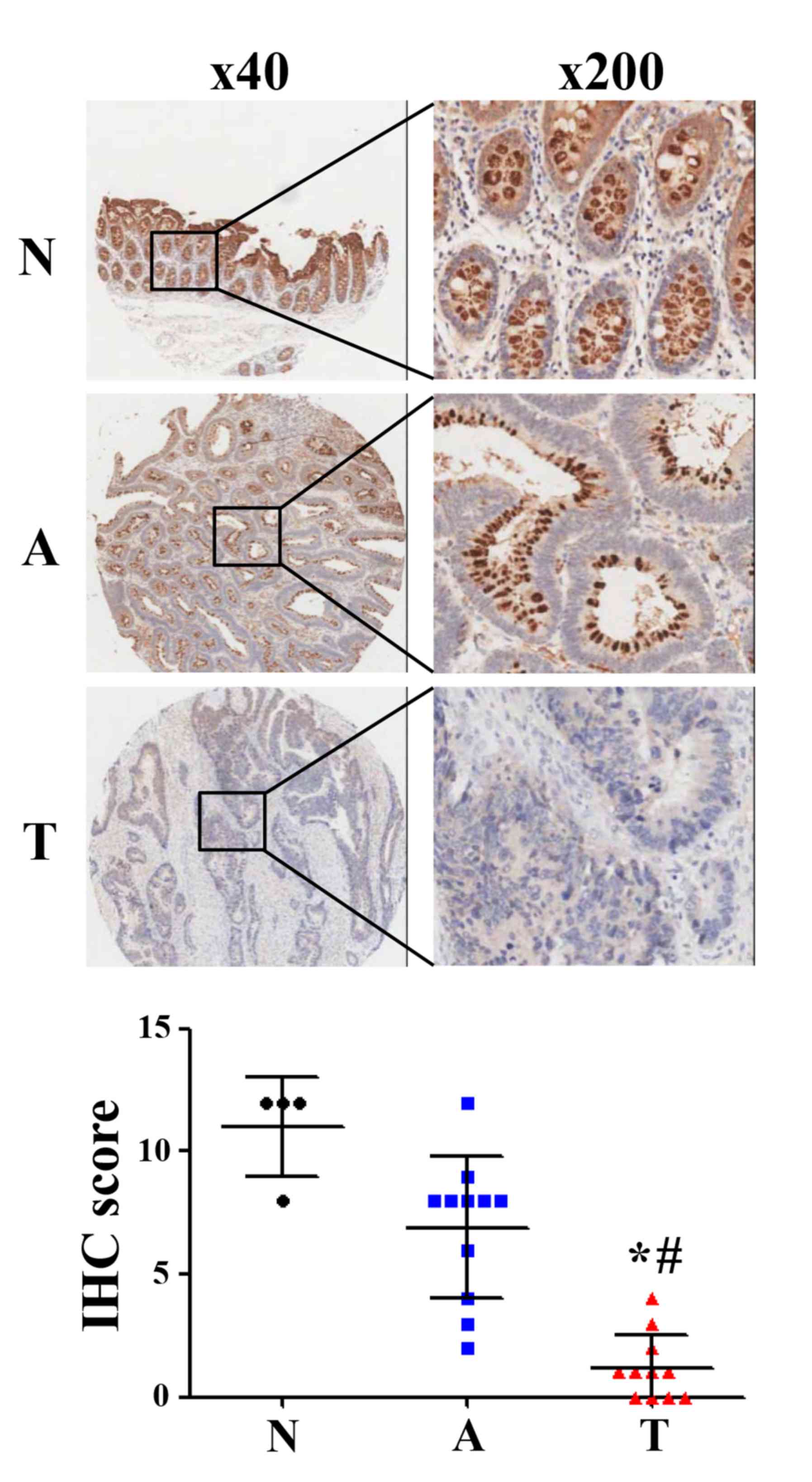
Gradual decrease in CLCA4 expression
during the development of CRC
To further evaluate CLCA4 expression during the
development of CRC, an IHC-based TMA was used to analyze CLCA4
expression in colorectal normal, adenoma and carcinoma tissues. The
results revealed that CLCA4 protein expression was downregulated in
colorectal adenoma tissues (IHC score: 6.91) compared with normal
tissues (IHC score: 11.00), but there was no statistical
significance (P>0.05; Fig. 5).
Furthermore, CLCA4 protein expression was significantly
downregulated in colorectal carcinoma tissues (IHC score: 1.18)
compared with colorectal normal tissues and colorecta adenoma
tissues, respectively (P<0.05; Fig.
5). The clinical characteristics of the patients were presented
in Table III. The present results
suggest that a decrease in CLCA4 expression may serve a significant
role in the development of CRC.
 | Table III.Clinicopathological features of 22
patients with colorectal cancer. |
Table III.
Clinicopathological features of 22
patients with colorectal cancer.
| Characteristic | n (%) |
|---|
| Age, years |
|
|
<65 | 19 (86) |
|
≥65 | 3 (14) |
| Sex |
|
|
Female | 9 (41) |
|
Male | 13 (59) |
| Tumor type |
|
|
Colorectal adenocarcinoma | 11 (50) |
|
Colorectal carcinoma | 11 (50) |
| Clinical stage |
|
| I | 5 (22) |
| II | 14 (64) |
|
III | 3 (14) |
| IV | 0 (0) |
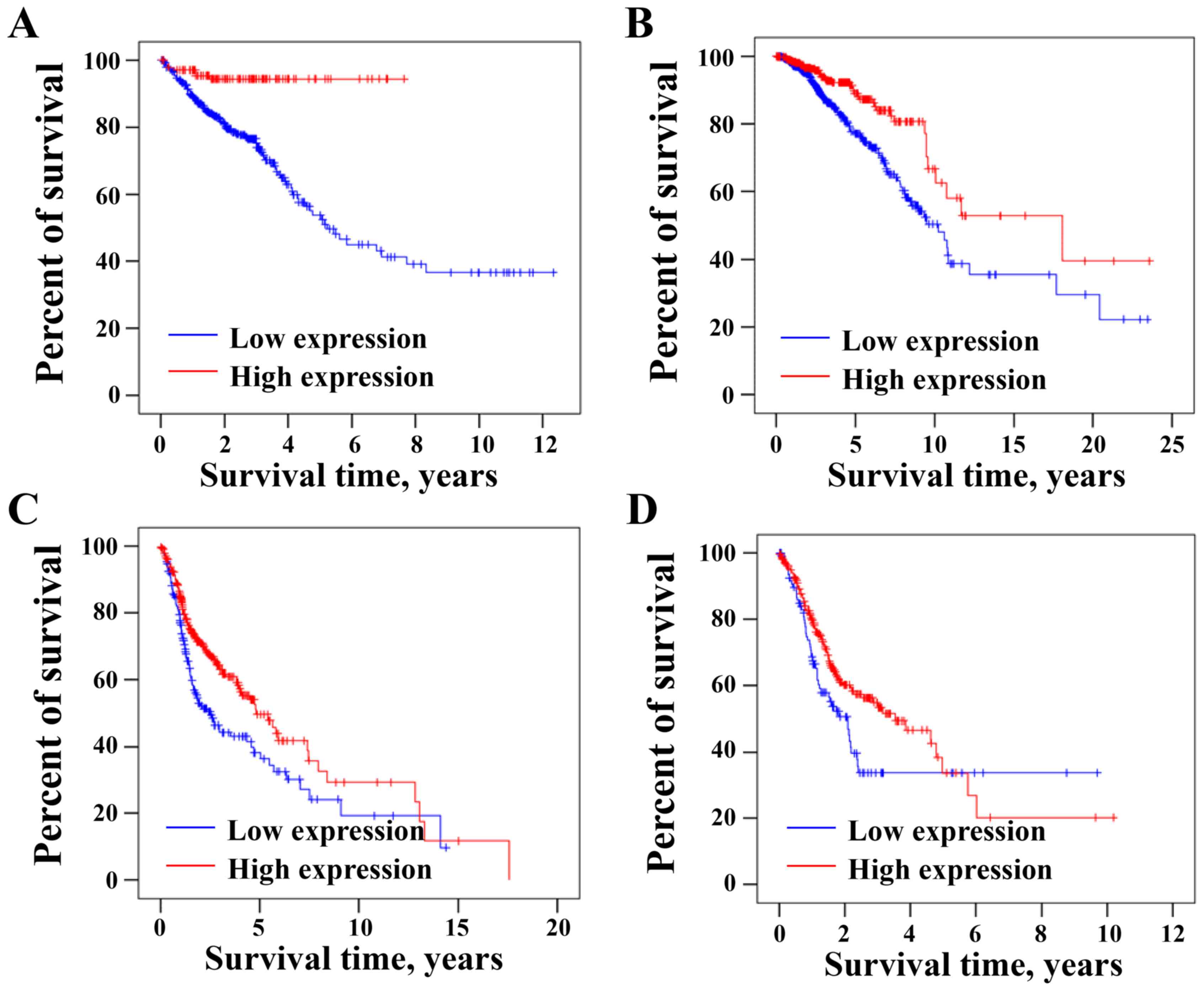
Low CLCA4 mRNA expression is
associated with low overall survival of tumor patients
The association between the expression levels of
CLCA4 and the overall survival of patients with CRC was analyzed
using the online TCGA dataset through THPA database. As presented
in Fig. 6A, overall survival rate of
CRC patients was significantly higher in the high CLCA4 expression
group than that in the low CLCA4 expression group (P<0.05;
cut-off: 5.31). Furthermore, low CLCA4 expression was associated
with the shorter overall survival rate of patients with breast
cancer (P<0.05; cut-off: 0.05; Fig.
6B), head and neck cancer (P<0.05; cut-off: 0.81; Fig. 6C) or stomach cancer (P<0.05;
cut-off: 0.02; Fig. 6D). The current
results suggested that low CLCA4 expression may be an indicator of
poor prognosis in patients with different types of cancer,
including CRC.
Discussion
CRC is the third most prevalent cancer in humans and
poses a significant public health problem worldwide (1–4). CRC
treatment, despite significant improvements, remains unsatisfactory
and CRC prognosis remains poor. In recent decades, systematic
analyses of genomic, transcriptomic and proteomic datasets have
become powerful tools in the discovery and validation of tumor
markers. Therefore, online databases have been analyzed to identify
novel genes involved in the development and progression of CRC. To
further explore the molecular mechanism involved in the development
of CRC, the present study conducted a database analysis and
revealed gradual decreases in CLCA4 expression from non-cancerous
colorectal tissues to primary and metastatic CRC tissues,
suggesting that CLCA4 may act as a tumor suppressor and serve a
significant role in the development and progression of CRC.
CLCA4 was a tumor suppressor that contributes to the
progression of several types of cancer. In liver cancer, CLCA4
downregulation promoted hepatocellular carcinoma cell
proliferation, migration and invasion (14). CLCA4 was aberrantly expressed in
breast cancer, and its abnormal expression inhibits tumor cell
growth (15). In addition, knockdown
of CLCA4 expression causes downregulation of E-cadherin expression
and upregulation of N-cadherin and vimentin expression in breast
cancer cells (15). CLCA4 has been
identified as a potential therapeutic target for the treatment of
oral cancer using bioinformatics analysis (29). In bladder cancer, low CLCA4
expression has been previously associated with larger tumor size,
advanced tumor stage and poor prognosis, and CLCA4 overexpression
profoundly attenuates the proliferation, growth, migratory and
invasive capabilities of bladder cancer cells (16). The aforementioned studies have
demonstrated the essential role of CLCA4 in various types of
tumor.
In the current study, using the online Oncomine
platform, most of the tumor tissues, including those from CRC and
brain, breast and esophageal cancer, had significantly lower CLCA4
transcriptional levels compared with those in associated normal
tissues. CRC tissues exhibited low CLCA4 expression compared with
non-cancerous colorectal tissues from GEPIA datasets. Furthermore,
TCGA database and TMA analyses revealed that the CLCA4 mRNA and
protein expression was downregulated in both colon and rectal
cancer tissues compared with that in non-cancerous colorectal
tissues. Analysis from the Oncomine database indicated that CLCA4
mRNA expression was decreased in various types of tumor tissues.
The present results revealed that a decrease in CLCA4 expression
may be a common event in cancer development, including CRC.
However, the decrease in CLCA4 expression in other types of tumor
tissues should be further evaluated by examining both mRNA and
protein levels, and the number of CRC samples should be
increased.
To assess CLCA4 expression during the development of
CRC, an IHC-based TMA analysis was conducted in the present study.
The results demonstrated that CLCA4 protein expression was
downregulated in colorectal adenoma tissues compared with normal
tissues, but this was not statistical significant, which may have
been due to the limited number of samples. Notably, CLCA4 protein
expression was significantly downregulated in colorectal carcinoma
tissues compared with colorectal normal tissues and colorectal
adenoma tissues, respectively. In general, CLCA4 protein expression
revealed a gradual decrease in CLCA4 expression among colorectal
normal, adenoma and carcinoma tissues, indicating that a decrease
in CLCA4 expression may serve a significant role in the development
of CRC. Therefore, the biological function of CLCA4 during the
development of CRC should be further investigated using
CLCA4-knockout or -knock-in mice in future studies. In addition,
THPA website TCGA survival analysis indicated that low CLCA4
expression was significantly associated with the overall survival
of patients with different types of tumor, including CRC, breast
cancer, head and neck cancer and stomach cancer, indicating that a
decrease in CLCA4 mRNA expression may serve as a prognostic
indicator in patients with tumors, including CRC. Furthermore, by
conducting an IHC-based TMA analysis, CLCA4 protein expression
between primary and metastatic CRC tissues was assessed, but no
significant difference was observed (data not shown), which may
have been due to the limited number of samples or the difficulty in
distinguishing between primary and metastatic CRC tissues.
Therefore, the association between CLCA4 expression and CRC
metastasis requires a more rational design of high-quality studies
to verify the conclusions from the present study. In addition,
since the main structure of CLCA4 is similar with CLCA1 and CLCA2
(17), CLCA4 may have the same
biological function in regulating cell proliferation and metastasis
in CRC, and may regulate these processes by suppressing the
PI3K/AKT pathway. Therefore, the concrete mechanism is still
remaining to be further elucidated.
To summarize, the present study demonstrated that
CLCA4 expression, both in the mRNA and protein levels, was
significantly downregulated in CRC tissues compared with that in
non-cancerous colorectal tissues. Furthermore, CLCA4 expression was
gradually decreased among colorectal normal, adenoma and carcinoma
tissues. Additionally, a decrease in CLCA4 expression was
significantly associated with the overall survival of patients with
different types of tumor, including CRC. Therefore, CLCA4 may serve
as a prognostic marker and a potential therapeutic target for CRC.
However, CLCA4 expression in different CRC cell lines and its
biological functions in cell proliferation and metastasis should be
further explored.
Acknowledgements
Not applicable.
Funding
The present study was sponsored by the National
Natural Science Foundations of China (grant nos. 81774121 and
81703913) and the Natural Science Foundation of Fujian Province,
China (grant no. 2017J01846).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. Microarray dataset of GSE49355
was downloaded from the NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49355).
The data for difference expression analysis of CLCA4 were obtained
from Colon Adenocarcinoma and Rectum Adenocarcinoma datasets of The
Cancer Genome Atlas (TCGA) through Gene Expression Profiling
Interactive Analysis website (http://gepia.cancer-pku.cn/detail.php?gene=CLCA4).
The data for survival analysis were downloaded from TCGA database
(https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
Authors' contributions
LW, WC and JL conceived and designed the study. LW,
WC, JZ and YF performed the data analysis. LW wrote the paper, and
JL reviewed and edited the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Tissue collection was approved by the Ethics
Committee of Taizhou Hospital of Zhejiang Province (Taizhou, China)
in accordance with the principles of the Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
CLCA4
|
chloride channel accessory 4
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
THPA
|
The Human Protein Atlas
|
|
TMA
|
tissue microarray
|
References
|
1
|
Favoriti P, Carbone G, Greco M, Pirozzi F,
Pirozzi RE and Corcione F: Worldwide burden of colorectal cancer: A
review. Updates Surg. 68:7–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brudvik KW, Kopetz SE, Li L, Conrad C,
Aloia TA and Vauthey JN: Meta-analysis of KRAS mutations and
survival after resection of colorectal liver metastases. Br J Surg.
102:1175–1183. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zampino MG, Magni E, Ravenda PS, Cella CA,
Bonomo G, Della Vigna P, Galdy S, Spada F, Varano GM, Mauri G, et
al: Treatments for colorectal liver metastases: A new focus on a
familiar concept. Crit Rev Oncol Hematol. 108:154–163. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Elble RC and Pauli BU: Tumor suppression
by a proapoptotic calcium-activated chloride channel in
mammaryepithelium. J Biol Chem. 276:40510–40517. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patel AC, Brett TJ and Holtzman MJ: The
role of CLCA proteins in inflammatory airway disease. Annu Rev
Physiol. 71:425–449. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Piirsoo M, Meijer D and Timmusk T:
Expression analysis of the CLCA gene family in mouse and human with
emphasis on the nervous system. BMC Dev Biol. 9:102009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang B, Cao L, Liu B, McCaig CD and Pu J:
The transition from proliferation to differentiation in colorectal
cancer is regulated by the calcium activated chloride channel A1.
PLoS One. 8:e608612013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sasaki Y, Koyama R, Maruyama R, Hirano T,
Tamura M, Sugisaka J, Suzuki H, Idogawa M, Shinomura Y and Tokino
T: CLCA2, a target of the p53 family, negatively regulates cancer
cell migration and invasion. Cancer Biol Ther. 13:1512–1521. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiang YY, Li CZ, Sun R, Zheng LS, Peng LX,
Yang JP, Meng DF, Lang YH, Mei Y, Xie P, et al: Along with its
favorable prognostic role, CLCA2 inhibits growth and metastasis of
nasopharyngeal carcinoma cells via inhibition of FAK/ERK signaling.
J Exp Clin Cancer Res. 37:342018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Walia V, Yu Y, Cao D, Sun M, McLean JR,
Hollier BG, Cheng J, Mani SA, Rao K, Premkumar L and Elble RC: Loss
of breast epithelial marker hCLCA2 promotes
epithelial-to-mesenchymal transition and indicates higher risk of
metastasis. Oncogene. 31:2237–2246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Chen M, Xie LK, Liu T, Zou ZW, Li
Y, Chen P, Peng X, Ma C, Zhang WJ and Li PD: CLCA4 inhibits cell
proliferation and invasion of hepatocellular carcinoma by
suppressing epithelial-mesenchymal transition via PI3K/AKT
signaling. Aging (Albany NY). 10:2570–2584. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu Y, Walia V and Elble RC: Loss of CLCA4
promotes epithelial-to-mesenchymal transition in breast cancer
cells. PLoS One. 8:e839432013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou T, Zhou L, Wang L, Kazobinka G, Zhang
X and Chen Z: CLCA4 inhibits bladder cancer cell proliferation,
migration, and invasion by suppressing the PI3K/AKT pathway.
Oncotarget. 8:93001–93013. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Loewen ME and Forsyth GW: Structure and
function of CLCA proteins. Physiol Rev. 85:1061–1092. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Del Rio M, Molina F, Bascoul-Mollevi C,
Copois V, Bibeau F, Chalbos P, Bareil C, Kramar A, Salvetat N,
Fraslon C, et al: Gene expression signature in advanced colorectal
cancer patients select drugs and response for the use of
leucovorin, fluorouracil, and irinotecan. J Clin Oncol. 25:773–780.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based human protein atlas. Nat
Biotechnol. 28:1248–1250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kato T, Hayama S, Yamabuki T, Ishikawa N,
Miyamoto M, Ito T, Tsuchiya E, Kondo S, Nakamura Y and Daigo Y:
Increased expression of insulin-like growth factor-II messenger RNA
binding protein 1 is associated with tumor progression in patients
with lung cancer. Clin Cancer Res. 13:434–442. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hong Y, Downey T, Eu KW, Koh PK and Cheah
PY: A ‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeutics. Clin Exp Metastasis. 27:83–90. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kaiser S, Park YK, Franklin JL, Halberg
RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et
al: Transcriptional recapitulation and subversion of embryonic
colon development by mouse colon tumor models and human colon
cancer. Genome Biol. 8:R1312007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowsk J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5:e130912010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sabates-Bellver J, Van der Flier LG, de
Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA,
Bujnicki JM, Menigatti M, et al: Transcriptome profile of human
colorectal adenomas. Mol Cancer Res. 5:1263–1275. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gaedcke J, Grade M, Jung K, Camps J, Jo P,
Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al: Mutated KRAS
results in overexpression of DUSP4, a MAP-kinase phosphatase, and
SMYD3, a histone methyltransferase, in rectal carcinomas. Genes
Chromosomes Cancer. 49:1024–1034. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bundela S, Sharma A and Bisen PS:
Potential therapeutic targets for oral cancer: ADM, TP53, EGFR,
LYN, CTLA4, SKIL, CTGF, CD70. PLoS One. 9:e1026102014. View Article : Google Scholar : PubMed/NCBI
|