Introduction
Cervical cancer is a common malignant tumor in women
(1). According to a statistical
report released in 2019, the number of estimated new cases was
13,170, and the estimated cervical cancer-associated deaths will
increase to 4,250 in the USA in 2019 (2). Meanwhile, in China, a total of 98,900
women were predicted to be diagnosed with cervical cancer, and
~30,500 cervical cancer-associated deaths were predicted in 2015
(3). Although the prognosis of
cervical cancer is not dismal, the 5-year survival rate for
patients with metastasisis 17%, and it is ~50% for those with
regional cervical cancer (2);
however, only 15% of patients are diagnosed with metastasis
(2).
Currently, the primary therapy for regional cervical
cancer is surgical resection followed by radiotherapy or
chemotherapy, while palliative therapy for recurrent or metastatic
cancer does not improve clinical survival time (4). The benefit from all these therapies is
usually short lived and only effective in some patients (4). Recently, target therapy and
immunotherapy have been introduced as new therapies for patients
with cancer, particular for patients with late-stage cancer. For
example, the chimeric antigen receptor T-cell (CAR-T) therapy drug
KYMRIAH® improved the prognosis in patients with acute
lymphoblastic leukemia (5). However,
the major issue regarding this novel therapy is that a critical
protein has to be expressed in or on the surface of tumor cells.
The single chain antibody fragment targeting CD19 molecule was
essential for the clinical outcome as the CAR-T therapy drug
recognized and bound to CD19 (5).
The antibody drug KEYTRUDA® is another successful
example for patients with cancer (6). PD-1 serves an inhibitory role in
activation of T cells, and KEYTRUDA® blocks the
inhibitory signaling via targeting PD-1, allowing T-cell
activation. Therefore, elucidation of the molecular mechanism and
identifying key molecules, such as PD-1,involved in tumor formation
and progression is critical and necessary for new therapies.
Genetic mutations are important factors contributing
to tumorigenesis (7). ALOX12B
encodes a lipoxygenase, and is responsible for the conversion of
arachidonic acid to 12R-hydroxyeicosatetraenoic acid 8). The
ALOX12B gene is located at chromosome 17p13.1, and contains
15 exons (8). Lipoxygenases are
reported to be associated with inflammation, skin disorders and
tumorigenesis (9). Mutations in the
ALOX12B gene mainly result in non-bullous congenital
ichthyosiform erythroderma (10,11).
ALOX12B has also been reported to inhibit immune cytolytic activity
in breast and renal cell tumors (12), and single nucleotide polymorphisms
(SNPs) in ALOX12B are associated with increased risk of lung
and breast cancer (13,14). Agarwal et al (15) reported that inhibition of ALOX12B
directly reduced the proliferation of the vulvar epidermoid
carcinoma A431 cell line. Hence, ALOX12B serves important roles in
the carcinogenesis of tumors, but, to the best of our knowledge,
there are no studies investigating the role of ALOX12B in cervical
cancer.
The aim of the present study was to investigate the
role of the ALOX12B gene in cervical cancer by knocking down
this gene in cervical cancer cells and systemically studying its
role both in vitro and in vivo. Additionally, the
underlying molecular mechanism of ALOX12B in cervical cancer was
explored using western blot analysis.
Materials and methods
Cell culture
Human Ca-Ski and C33A cells were purchased from the
American Type Culture Collection and cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere
with 5% CO2 at 37°C. C33A cells are derived from
HPV-negative cervical cancer, while Ca-Ski cells are derived from
HPV 16-positive cervical cancer (16).
Reverse transcription-quantitative
(RT-q) PCR assay
RNA was extracted from tumor cells, including C33A
and Ca-Ski cells, using an RNAeasy™ kit (Beyotime Institute of
Biotechnology) according to the manufacturer's instructions. RNA
was reverse transcribed into complementary DNA (cDNA) using 1 µg
RNA and PrimeScript™ 1st strand cDNA Synthesis kit (Takara Bio,
Inc.). qPCR was carried out using SYBR Green mix (Shanghai Yeasen
Biotechnology Co., Ltd.) using an ABI 7000 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and PrimeScript™ RT
Master mix (Takara Bio, Inc.). The thermocycling protocol was as
follows: 95°C for 2 min, followed by 40 cycles at 95°C for 15 sec
and 60°C for 15 sec. GAPDH was used as the internal control. The
relative quantity of the target gene was calculated using the
2−∆∆Cq method (17). The
primer sequences for each gene are shown in Table I.
 | Table I.Sequence of the primers used for
quantitative PCR. |
Table I.
Sequence of the primers used for
quantitative PCR.
| Gene name | Sequence,
5′-3′ | Fragment size,
bp |
|---|
| ALOX12B |
| 196 |
|
Forward |
GCCTGTTGGACTGCAAACATT |
|
|
Reverse |
GTGACGGGGAACTTGTCTGG |
|
| GAPDH |
| 217 |
|
Forward |
TCATCATCTCTGCCCCCTCT |
|
|
Reverse |
GGGCCATCCACAGTCTTCTG |
|
Western blotting
Total protein was extracted using a protein
extraction kit (Shanghai Yeasen Biotechnology Co., Ltd.) according
to the manufacturer's protocol, and the protein concentration was
determined using an ultraviolet spectrophotometer (Onedrop1000;
Shanghai Genechem Co., Ltd.). A total of 10 µg protein/lane was
loaded on a 12% polyacrylamide gel, resolved using SDS-PAGE and
transferred to polyvinylidene fluoride (PVDF) membranes (Sangon
Biotech Co., Ltd.). The PVDF membrane was blocked with 5% non-fat
milk for 2 h at room temperature. Subsequently, the PVDF membrane
was incubated with the corresponding primary antibody against the
target protein overnight at 4°C followed by incubation with the
secondary antibody at room temperature for 1 h. The protein band
was detected using a Beyo ECL Star kit (Beyotime Institute of
Biotechnology). The antibodies used were as follows: Anti-PI3K
(ab70912; 1:100), anti-MEK1 (ab32091; 1:1,500), anti-ERK1 (ab32537;
1:600), anti-cdc25 (ab111830; 1:2,000) (all Abcam), anti-C-fos
(554C1a; 1:500; Santa Cruz Biotechnology, Inc.), anti-ALOX12B
(PA5-23608; 1:800; Invitrogen; Thermo Fisher Scientific, Inc.),
anti-GAPDH (ab181602; 1:10,000; Abcam), goat anti-mouse IgG
antibody (ab97035; 1:2,000) and goat anti-rabbit IgG antibody
(ab7090; 1:5,000) (both Abcam).
Package of lentiviral vector carrying
small hairpin RNA(shRNA) targeting ALOX12B and transduction
Three shRNA fragments targeting the ALOX12B
gene were designed, synthesized and transferred into the expression
vector pPLK-GFP (pPLK-shALOX12B-GFP). The sequences of each shRNA
fragment are shown in Table II. In
parallel, a non-targeting sequence was used as the negative control
(NC). Subsequently, a lentiviral vector carrying shALOX12B
(LvshALOX12B) was prepared as follows: 10 µg pspax2 plasmid
(Invitrogen; Thermo Fischer Scientific, Inc.), 5 µg pMD2.G plasmid
(Invitrogen; Thermo Fisher Scientific, Inc.) and 15 µg
pPLK-shALOX12B-GFP were transferred into 293T cells (RanYanBio Co.,
Ltd.) and cultured in DMEM containing 10% FBS (both Gibco; Thermo
Fisher Scientific, Inc.) at 37°C with 5% CO2 for 72 h.
Subsequently, the supernatant was collected, and the lentiviral
particles were concentrated from the supernatant by centrifugation
at 14,000 × g at 4°C for 40 min.
 | Table II.shRNA target sequences for
ALOX12B. |
Table II.
shRNA target sequences for
ALOX12B.
| Name | Target sequence,
5′-3′ |
|---|
| shALOX12B-1 |
GATACACCGTCCAGATCAAT |
| shALOX12B-2 |
GCACGCGGATCCCAGACAA |
| shALOX12B-3 |
CCGATATGTCACTATAGTCAT |
| NC |
GTTCTCCGAACGTGTCACGTT |
Cervical cancer cells were treated with LvshALOX12B
or NC (scrambled) for 6 h and then cultured for 72 h. The
transduction efficiency was detected under a fluorescence
microscope (magnification, ×100; Nikon Corporation). The knockdown
efficiency of ALOX12B was determined by qPCR assay as
aforementioned.
Construction of plasmid expressing
ALOX12B and transfection
The coding sequence for ALOX12B (NM_001139.3) was
synthesized and cloned into the pcDNA3.1 vector (Addgene, Inc.).
The recombinant plasmid pcDNA3.1-ALOX12B (to overexpress ALOX12B)
was confirmed by DNA sequencing. Subsequently, thepcDNA3.1-ALOX12B
plasmid was transfected into tumor cells using Lipofectamine
reagent (40802ES02; Shanghai Yeasen Biotechnology Co., Ltd.). The
transfection efficiency was detected using western blotting for
detection of ALOX12B expression levels, as aforementioned. The time
interval between transfection and subsequent experimentation was 48
h.
Cell proliferation assay using Cell
Counting Kit-8 (CCK-8)
A total of 6,000 lentiviral-infected C33A or Ca-Ski
cells/well were seeded into a 96-well plate. After 24, 48, 72 or 96
h of culture, 10 µl CCK-8 (Beyotime Institute of Biotechnology) was
added into each well and cells were cultured at 37°C with 5%
CO2 for another 4 h according to the manufacturer's
protocol. Subsequently, the absorbance at 450 nm was determined
using a microplate reader. Each assay was repeated independently at
least three times.
Colony forming assay
A total of 100 C33A or Ca-Ski cells/well were seeded
into a 24-well plate. After 15 days, the cells on the plate were
fixed with 4% paraformaldehyde at room temperature for 30 min and
stained with 0.1% crystal violet at room temperature for 15 min.
The colony number was counted manually under a light microscope
(magnification, ×40). Each assay was repeated independently at
least three times.
Wound healing assay
A total of 2×104 C33A or Ca-Ski
cells/well were seeded into a 24-well plate and cultured to 90%
confluence. Next, a 10-µl sterile tip was used to make a scratch in
the middle of each well. The debris was washed with PBS, and fresh
DMEM without FBS was added. After 24 h, the plate was observed, and
images were captured under a light microscope (magnification,
×100), and the width of each scratch was recorded. Each assay was
repeated independently at least three times.
Cel cycle analysis
A total of 1×105 C33A or Ca-Ski cells
treated with lentiviral vector for 72 h were harvested and washed
with cold PBS followed by fixing in 70% ethyl alcohol. Next, 0.5 ml
1X staining buffer (Beyotime Institute of Biotechnology) with 10 µl
propidium iodide (PI) reagent (Beyotime Institute of Biotechnology)
was used to resuspend the tumor cells, which were then cultured for
30 min at 37°C. The cell cycle distribution was analyzed using
fluorescence-activated cell sorting (Beckman Coulter, Inc.).
In vivo tumor growth assay
Nanchang Royo Biotech Co., Ltd. performed the
following experiments: 20 female BALB/c-nu mice (6 weeks old; 20–30
g) were obtained from Shanghai SLAC Laboratory Animal Co., Ltd. and
divided equally into NC and shALOX12B groups. Mice were kept in
groups of 4 or 5 in individual cages and provided with sterilized
food and water ad libitum. Mice were maintained in
conditions with a 12-h light-dark cycle at 22°C and 55% humidity.
After in vitro culture, ~2×107 C33A cancer cells
were inoculated subcutaneously into the right flank. Tumor growth
was monitored consecutively for 35 days. Subcutaneous tumor volume
(V) was measured twice a week and calculated as follows:
V=(LxW2)/2, where L is the length and W is the width of
the tumor. At the 35th day, mice were anesthetized according to the
guidelines involving the use of diethyl ether approved by the
Laboratory Animal Ethics Committee of Nanchang Royo Biotech Co.,
Ltd. (approval no. RYE2019011104); a sterile gauze soaked in 99.5%
diethyl ether was placed in a 500 ml beaker and mice were placed
into the beaker for 5 min. Subsequently, the mice were sacrificed
by cervical dislocation. If the tumor volume was >1,500
mm3 or if ulcers occurred, the study was terminated
prematurely.
Statistical analysis
All data are displayed as the mean ± standard
deviation, and were analyzed using SPSS version 16.0 (SPSS Inc.).
The difference between two groups was compared using an unpaired
Student's t-test. The differences among multiple groups were
compared using a one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Effective ALOX12B-knockdown in
cervical cancer C33A cells
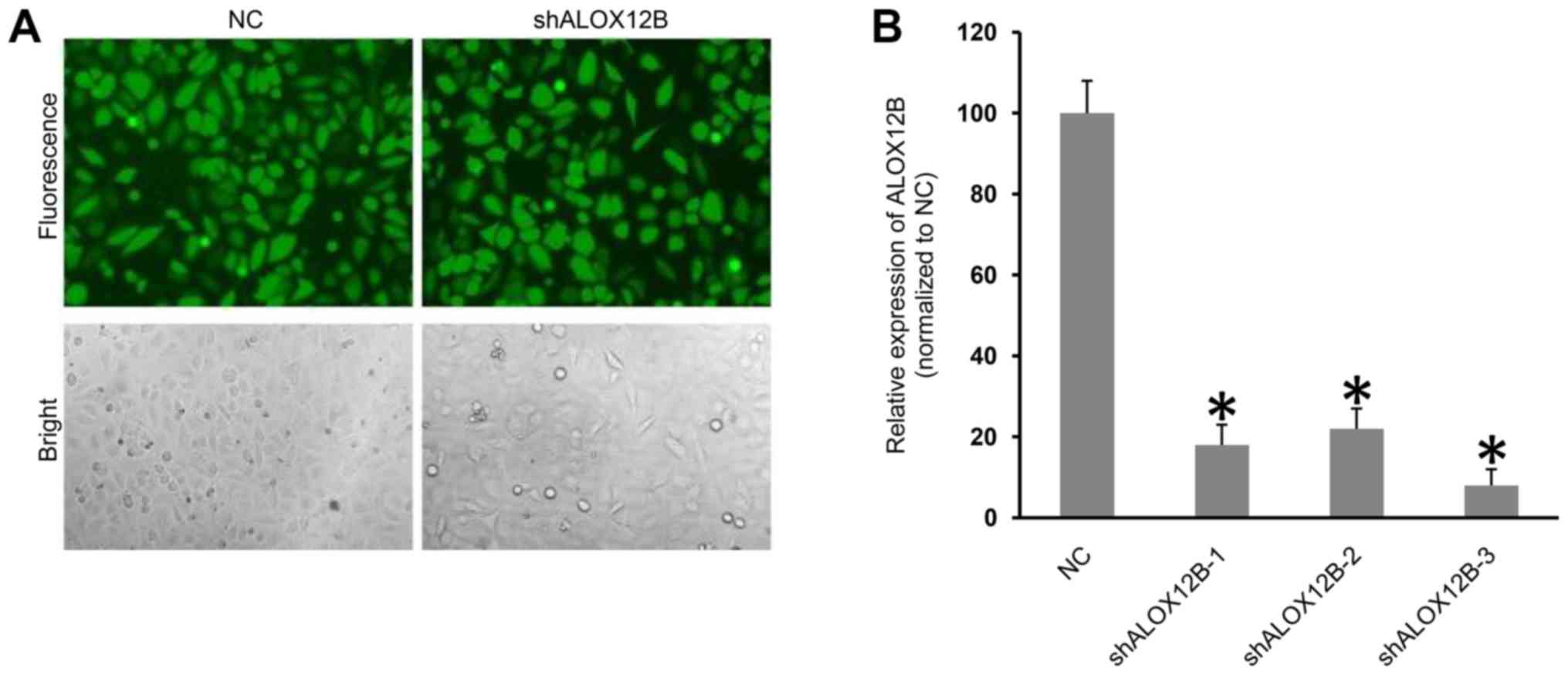
LvshALOX12B was used to transfect C33A cells, and
the transfection efficiency was monitored by GFP expression. As
shown in Fig. 1A, the transfection
efficiency was high after transduction for 72 h, according to GFP
expression. Subsequently, qPCR technology was used to determine the
expression levels of the ALOX12B gene in C33A cells. qPCR
analysis demonstrated that the knockdown efficiency of each shRNA
was >75% compared with that of the NC (Fig. 1B). The knockdown efficiency of
shALOX12B-3 was ~90%; therefore, shALOX12B-3 was chosen for
functional analysis.
Knockdown of ALOX12B inhibits the
proliferation and colony formation of C33A cells
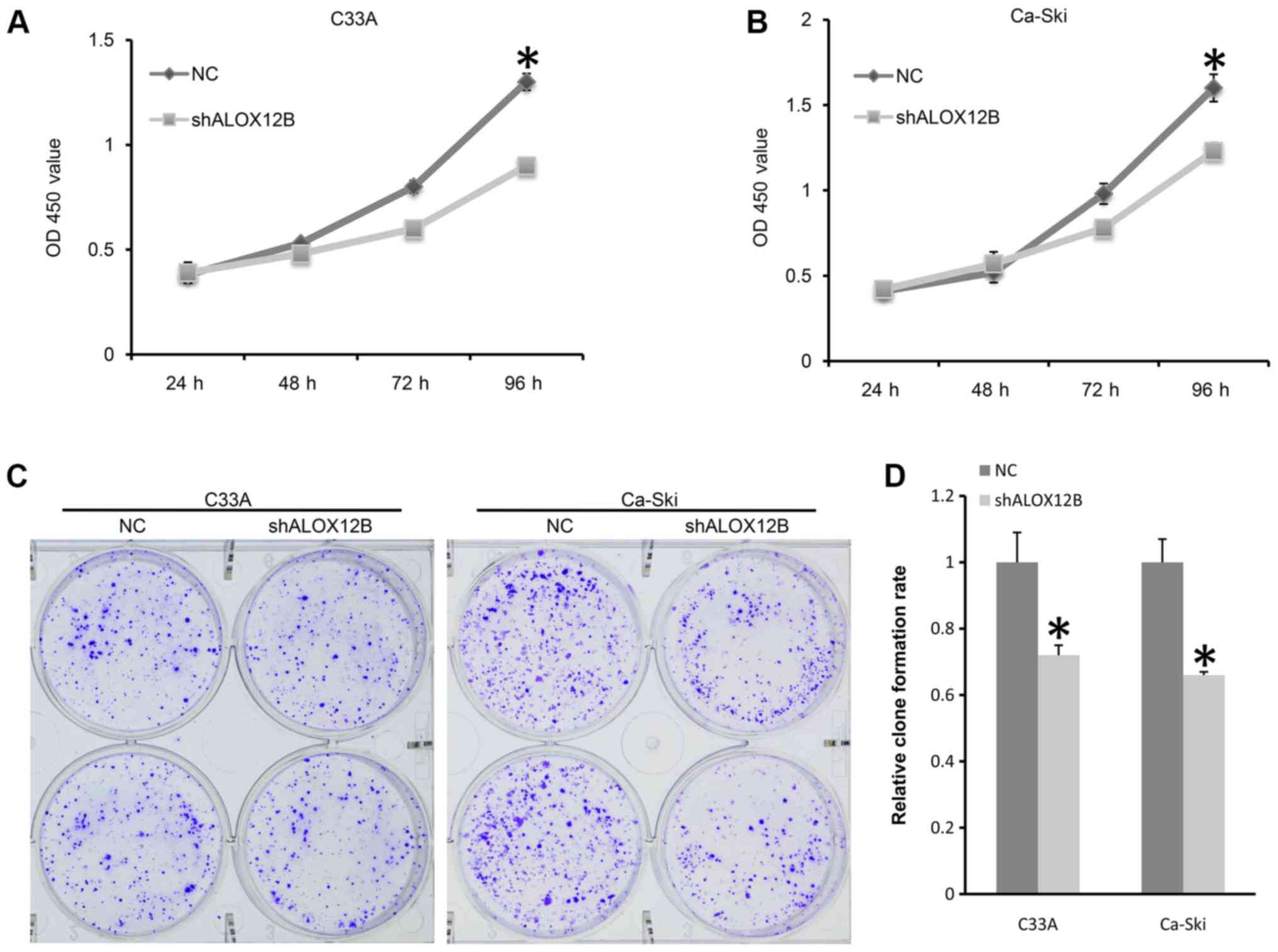
A CCK-8 assay was designed to detect the effect of
shALOX12B-3 on the proliferation of C33A and Ca-Ski cells. As shown
in Fig. 2A and B, the proliferation
rate of C33A treated with shALOX12B-3 was 69% compared with that of
the NC group, while the proliferation rate was 76% in Ca-Ski cells
after transfection for 96 h. Colony forming ability also reflects
the proliferative potential. The relative colony formation rate in
the NC group was significantly higher compared with that of the
shALOX12B-3 group in C33A cells (P<0.05; Fig. 2C and D). For Ca-Ski cells, the
relative colony formation rate in the NC group was also
significantly higher compared with that of the shALOX12B-3 group
(P<0.05; Fig. 2C and D). These
results suggested that ALOX12B participates in the proliferation
and growth of cervical cancer cells.
ALOX12B is not essential for cell
migration in cervical cancer
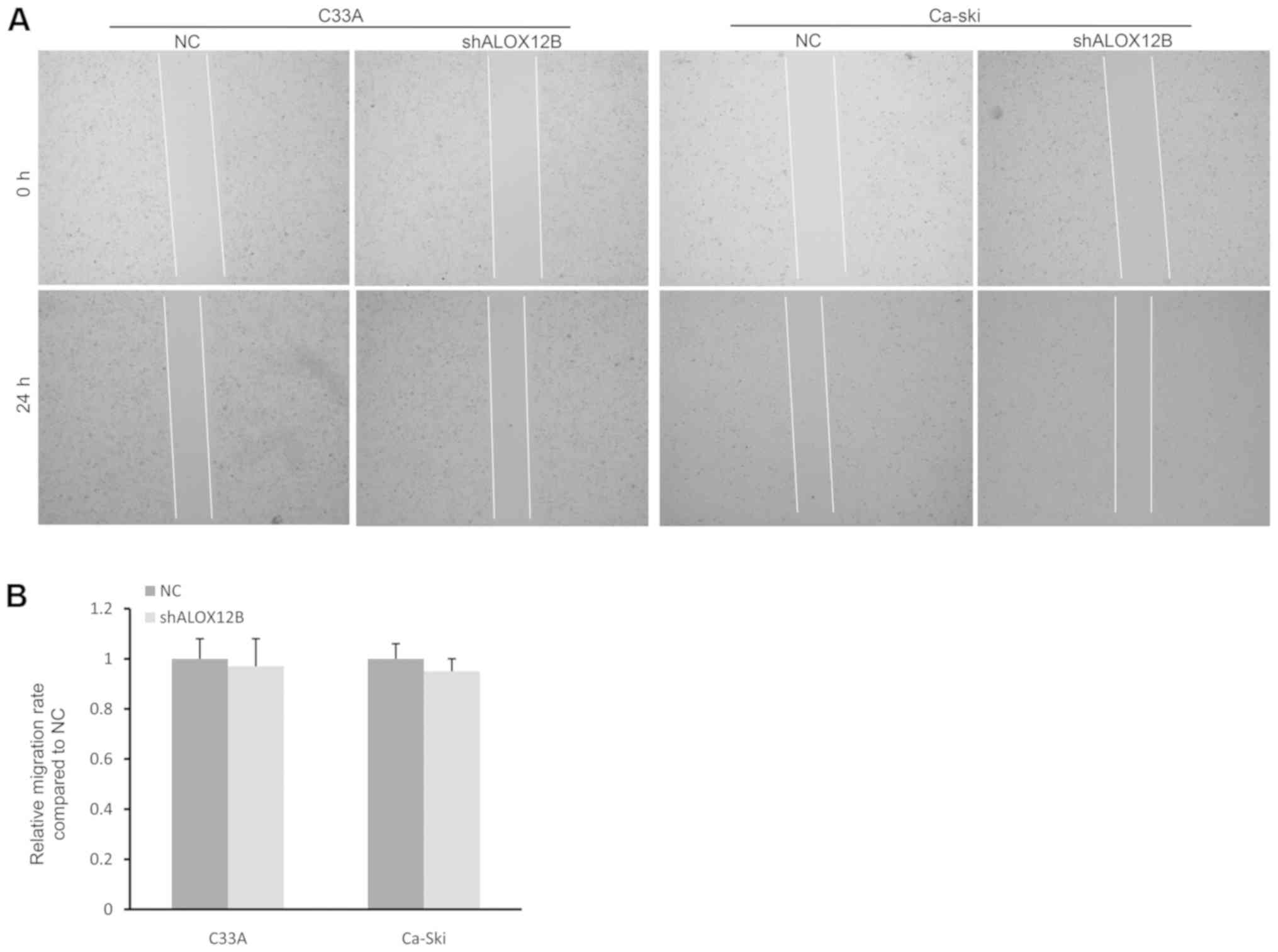
In general, cancer cells have a marked migration
ability in the majority of cancers (18). A wound healing assay was performed to
determine the effect of shALOX12B-3 on the migration of cervical
cancer cells. However, no significant difference was observed at 24
h between the NC group and the shALOX12B-3 group in C33A or Ca-Ski
cells (Fig. 3A and B). The 48-h time
point was also assessed in the wound healing assay, but no
significant differences were observed, as the relative migration
rate in the NC group was equivalent to that in the shALOX12B group
(data not shown). Therefore, it is possible that ALOX12B may not
regulate cell migration in cervical cancer.
Cell cycle transition is suppressed by
ALOX12B-knockdown
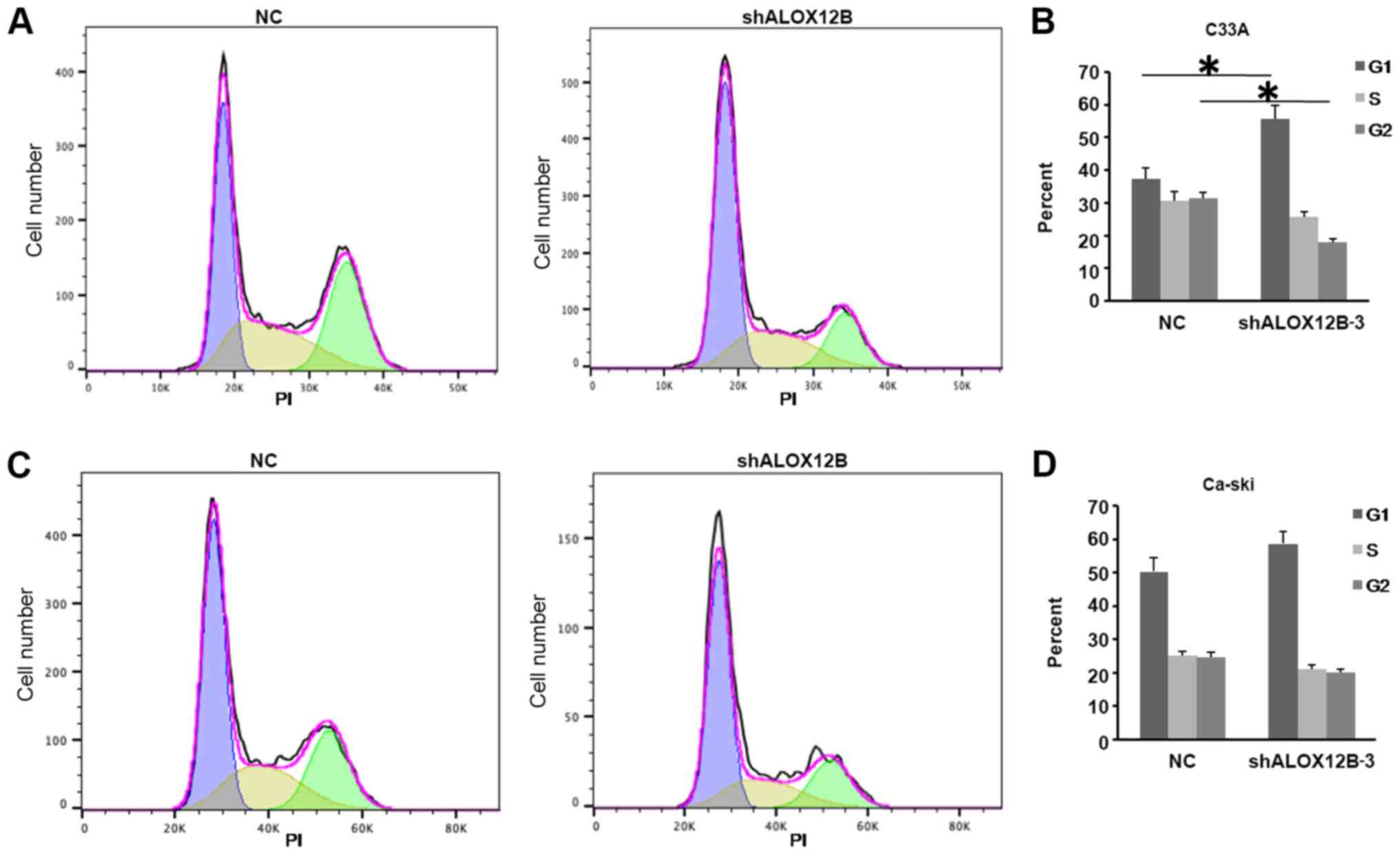
To detect the effect of shALOX12B-3 on cell cycle in
cervical cancer, C33A and Ca-Ski cells were stained with PI dye
after treatment with shALOX12B-3 and NC. As shown in Fig. 4A and B, the cell cycle was arrested
at the G1 phase after shALOX12B was knocked down. The
ratio of C33A cells in the G1 phase increased from 37.4
to 55.7% (NC vs. shALOX12B). By contrast, no significant difference
was observed in Ca-Ski cells. The phase distribution in Ca-Ski
cells showed no significant difference between the shALOX12B-3 and
NC groups (Fig. 4C and D). This
might be attributed to biological variation between different
cells.
Knockdown of ALOX12B delays tumor
growth in a xenograft mouse model
Since ALOX12B contributed to the cell proliferation
and growth of C33A cells, and regulated the cell cycle
distribution, it was hypothesized that ALOX12B could regulate tumor
growth in vivo. C33A cells transfected with shALOX12B-3
showed a significant slower growth in a mouse xenograft model
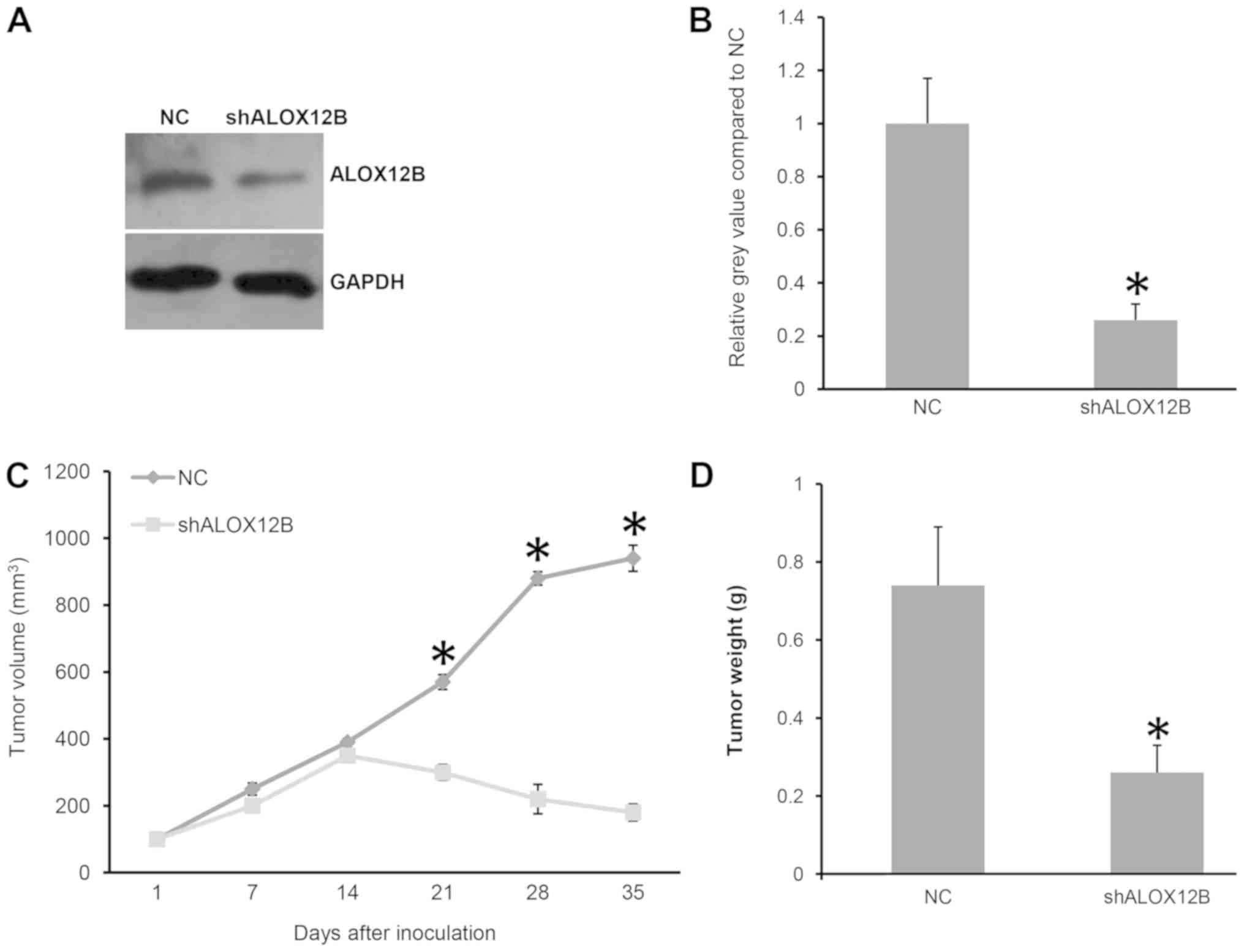
(P<0.05; data not shown). As shown in Fig. 5A and B, the knockdown of ALOX12B was
confirmed by western blotting. It was demonstrated that the tumor
volume in the shALOX12B-3 group was significantly smaller compared
with that of the NC group (180 vs. 940 mm3; P<0.05;
Fig. 5C). The tumor weight in the
shALOX12B-3 group was also significantly lighter compared with that
of the NC group (0.26 vs. 0.74g; P<0.05; Fig. 5D). As expected, ALOX12B was essential
for in vivo tumor growth of cervical cancer.
ALOX12B regulates the PI3K/ERK1
signaling pathway in cervical cancer
As a novel gene, there is little information
regarding the signaling pathway regulated by ALOX12B. In several
types of cancer, important signaling pathways regulating cell
proliferation include PI3K, Wnt, mTOR and MAPK (19–21). The
expression levels of these molecules were analyzed using western
blotting. As shown in Fig. 6A and B,
after knockdown of ALOX12B in C33A cells, the expression levels of
PI3K, MEK1, ERK1, C-fos and cdc25 were significantly reduced
(P<0.05). By contrast, overexpression of ALOX12B in cells
treated with the plasmid pcDNA-ALOX12B significantly increased the
expression levels of PI3K, MEK1, ERK1, C-fos and cdc25 (P<0.05;
Fig. 6A and C). In vivo,
knockdown of ALOX12B also significantly reduced the expression of
PI3K, MEK1, ERK1, C-fos and cdc25 (P<0.05; Fig. 6A and D). PI3K/ERK1signaling serves
important roles in several types of cancer (19); therefore, it is possible that ALOX12B
promotes the proliferation of cervical cancer cells by regulating
the PI3K/ERK1 signaling pathway.
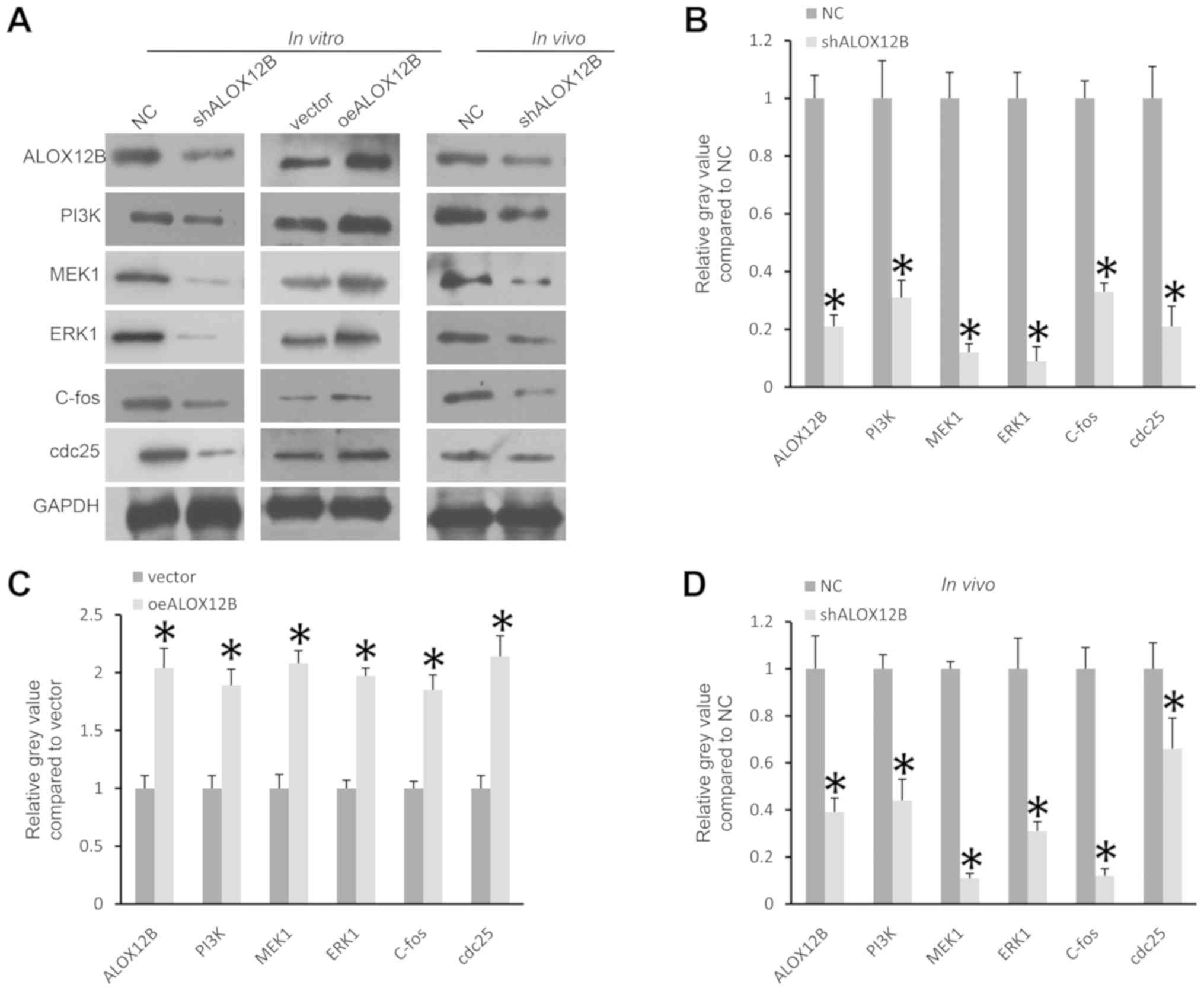 | Figure 6.ALOX12B regulates the expression
levels of PI3K/ERK1 signaling molecules. (A and B) Knockdown of
ALOX12B decreased the expression levels of PI3K, MEK1, ERK1, C-fos
and cdc25 in C33A cells. (A and C) Overexpression of ALOX12B in
C33A cells increased the expression levels of PI3K, MEK1, ERK1,
C-fos and cdc25. (A and D) Knockdown of ALOX12B decreased the
expression levels of PI3K, MEK1, ERK1, C-fos and cdc25 in tumor
tissues in the xenograft tumor model. GAPDH was used as the
internal control. *P<0.05. sh, small hairpin; NC, negative
control; oe, overexpressed. |
Discussion
Cervical cancer is a malignant disease affecting
women worldwide, and poses a great threat to life and life quality
(2,3). According to the most recent report by
the American Association for Cancer Research, both the incidence
and death rate of patients with cervical cancer has remained steady
in recent years (1). This could be
attributed to the failure of traditional therapies such as surgery,
chemotherapy and radiotherapy. This high death rate indicates that
some patients with cervical cancer, particularly those who have
relapsed or have metastasis, do not benefit sufficiently from such
therapies (3). In the past decade, a
series of new biotherapies have been developed, including
immunotherapy and targeted therapy. For example, antibody drugs and
cell immunotherapy have greatly improved the survival of patients
with cancer (4,5). However, to harness the potential
clinical benefit of these therapies, the underlying molecular
mechanisms of a particular cancer type need to be clarified to
prevent treatment failure.
The majority of cervical cancer cases are caused by
HPV infection, which is most commonly transmitted through sexual
activity (22). However, genome
sequencing technology has revealed that genetic and epigenetic
factors also serve important roles during the tumorigenesis of
cervical cancer (23). Several genes
have been reported to be associated with cervical cancer. For
example, Chang et al (24)
found that high POUF3 expression accelerated the progression of
cervical cancer. Heterogeneity is common in almost all types of
cancer (25); therefore, it is
difficult to determine the function of a single dominant gene in a
specific type of cancer.
To the best of our knowledge, the present study
demonstrated for the first time that ALOX12B may serve an oncogenic
role in cervical cancer. The in vitro data suggested that
ALOX12B contributed to cell proliferation in cervical cancer.
Normally, cells display an inhibitory effect when they contact each
other; however, tumor cells are known to have no contact-inhibitory
characteristics and can readily proliferate (16). An accelerated cell cycle is often
synchronized with cell proliferation and is common in tumor cells.
As expected, knockdown of ALOX12B resulted in cell cycle arrest in
the G1 phase in C33A cells. This was consistent with the
proliferation-promoting role of ALOX12B in cervical cancer.
However, little difference in cell cycle was observed in Ca-Ski
cells. This may be due to the heterogeneity of tumor cells.
Infection with HPV was a major cause of cervical cancer, and
different strains of HPV caused expression of checkpoint molecules
controlling the cell cycle, such as p53 and p105 (16,26,27).
Metastasis is another common phenomenon affecting
patients with cancer (18).
Generally speaking, wound healing assays are often used to reflect
the aggressive ability of tumor cells in vitro (28); however, ALOX12B-knockdown did not
affect the migration ability of cervical cancer cells in the
present study. It is possible that ALOX12B solely regulates cell
proliferation in cervical cancer. The mouse xenograft model bearing
C33A cells further demonstrated that ALOX12B was essential for
tumor cell growth in vivo. These data suggested that ALOX12B
promoted the progression of cervical cancer. In previous studies,
ALOX12B was shown to be involved in the progression of lung, breast
and epidermoid cancer (13–15); thus, the present study may further
improve our understanding of the role of ALOX12B in cancer.
However, additional experiments are necessary to confirm the role
of ALOX12B in promoting the progression of cervical cancer.
Although ALOX12B displayed little effect on cell migration in
vitro, it may be useful to analyze the effect of ALOX12B on the
metastasis of cervical cancer cells in vivo and to
investigate the expression patterns of ALOX12B in tumor versus
normal tissues.
ALOX12B is a lipoxygenase that is critical for the
synthesis of 12R-hydroxyeicosatetraenoic acid (10). The lipoxygenase family of proteins is
large and comprises dozens of members. Different lipoxygenases were
shown to be associated with inflammation and tumorigenesis via
different signaling pathways (9).
For example, ALOX15 suppresses inflammation by inhibiting the
IL-6/STAT3 signaling pathway in colorectal cancer (29). Additionally, lipoxygenase 5-LOX and
12-LOX have been revealed to be essential for cell proliferation in
pancreatic cancer (30). The most
commonly reported alteration concerning ALOX12B was SNP (8–14). Only
Agarwal et al (15)
demonstrated that ALOX12B modulated the ERK and PI3K-Akt signaling
pathways in A431 cells. As ALOX12B was shown to regulate cell
proliferation in cervical cancer in the present study, the
proliferation-related signaling molecules in C33A cells were
determined. It was found that ALOX12B could regulate the expression
levels of PI3K, MEK1, ERK1, C-fos and cdc25.PI3K is known to be an
important gene in the proliferation of tumor cells, and abnormal
activation of PI3K has been shown in a number of cancer types,
including cervical cancer (31).
MEK1 is a mitogen-activated protein kinase and is often involved in
crosstalk with the PI3K signaling pathway. For example, MEK1
transmits signals from PI3K to ERK1, and activates target genes
such as C-fos and cdc25 (32).
Activation of the PI3K/ERK1 signaling pathway has been observed in
several types of cancer and has been shown to promote tumorigenesis
(33). C-fos and cdc25 are two
common genes that promote tumor progression (27,34). In
addition, ALOX12B was shown to regulate PI3K and ERK signaling
pathways in epidermoid cancer cells (15); therefore, it is possible that ALOX12B
regulates PI3K/ERK1 signaling in cervical cancer.
In summary, the present study demonstrated that
ALOX12B promoted cell proliferation via regulation of the PI3K/ERK1
signaling pathway in cervical cancer. These results may provide the
basis for future targeting of ALOX12B in cervical cancer treatment.
However, the clinical significance of ALOX12B in cervical cancer
remains unclear. The association of ALOX12B with survival and with
clinicopathological factors in patients was not elucidated in the
present study. Additionally, the way in which ALOX12B affects the
PI3K signaling pathway requires further investigation. Another
limitation is that the present study only used a couple of cell
lines, and therefore the role of the ALOX12B gene should be
investigated in more cell lines, which may further support the
present results. Therefore, more work is required to further
support the role of ALOX12B in cervical cancer and to clarify the
link between ALOX12B and PI3K/ERK1. The expression pattern of
ALOX12B in a large cohort of clinical tumor tissues and its
association with clinicopathological factors should be investigated
to confirm its clinical significance.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TJ and LYL conceived and designed the study. QYY and
YML conducted the literature review and interpreted the data. BZ
and TJ analyzed the data. TJ wrote the manuscript. KJT, TJ, BZ and
YML performed the experiments. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Laboratory
Animal Ethics Committee of Nanchang Royo Biotech Co., Ltd.
(approval no. RYE2019011104).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Johnson CA, James D, Marzan A and Armaos
M: Cervical Cancer: An Overview of Pathophysiology and Management.
Semin Oncol Nurs. 35:166–174. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liontos M, Kyriazoglou A, Dimitriadis I,
Dimopoulos MA and Bamias A: Systemic therapy in cervical cancer: 30
years in review. Crit Rev Oncol Hematol. 137:9–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Leahy AB, Elgarten CW, Grupp SA, Maude SL
and Teachey DT: Tisagenlecleucel for the treatment of B-cell acute
lymphoblastic leukemia. Expert Rev Anticancer Ther. 18:959–971.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kwok G, Yau TC, Chiu JW, Tse E and Kwong
YL: Pembrolizumab (Keytruda). Hum Vaccin Immunother. 12:2777–2789.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fang J, Zhang H and Jin S: Epigenetics and
cervical cancer: From pathogenesis to therapy. Tumour Biol.
35:5083–5093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Krieg P, Marks F and Fürstenberger G: A
gene cluster encoding human epidermis-type lipoxygenases at
chromosome 17p13.1: Cloning, physical mapping, and expression.
Genomics. 73:323–330. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mashima R and Okuyama T: The role of
lipoxygenases in pathophysiology; new insights and future
perspectives. Redox Biol. 6:297–310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kurban M, Shimomura Y, Bahhady R, Ghosn S,
Kibbi AG and Christiano AM: Nonsense mutation in the ALOX12B gene
leads to autosomal recessive congenital ichthyosis in a Lebanese
family. J Eur Acad Dermatol Venereol. 24:232–234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rodríguez-Pazos L, Ginarte M, Vega A and
Toribio J: Autosomal recessive congenital ichthyosis. Actas
Dermosifiliogr. 104:270–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumors associated
with local immune cytolytic activity. Cell. 160:48–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shen M, Vermeulen R, Rajaraman P, Menashe
I, He X, Chapman RS, Yeager M, Thomas G, Burdett L, Hutchinson A,
et al: Polymorphisms in innate immunity genes and lung cancer risk
in Xuanwei, China. Environ Mol Mutagen. 50:285–290. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee JY, Park AK, Lee KM, Park SK, Han S,
Han W, Noh DY, Yoo KY, Kim H, Chanock SJ, et al: Candidate gene
approach evaluates association between innate immunity genes and
breast cancer risk in Korean women. Carcinogenesis. 30:1528–1531.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Agarwal S, Achari C, Praveen D, Roy KR,
Reddy GV and Reddanna P: Inhibition of 12-LOX and COX-2 reduces the
proliferation of human epidermoid carcinoma cells (A431) by
modulating the ERK and PI3K-Akt signalling pathways. Exp Dermatol.
18:939–946. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bernard B, Fest T, Prétet JL and Mougin C:
Staurosporine-induced apoptosis of HPV positive and negative human
cervical cancer cells from different points in the cell cycle. Cell
Death Differ. 8:234–244. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Khajah MA, Mathew PM and Luqmani YA:
Inhibitors of PI3K/ERK1/2/p38 MAPK Show Preferential Activity
Against Endocrine-Resistant Breast Cancer Cells. Oncol Res.
25:1283–1295. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taciak B, Pruszynska I, Kiraga L, Bialasek
M and Krol M: Wnt signaling pathway in development and cancer. J
Physiol Pharmacol. 69:692018.
|
|
21
|
Zeng H: mTOR signaling in immune cells and
its implications for cancer immunotherapy. Cancer Lett.
408:182–189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsikouras P, Zervoudis S, Manav B, Tomara
E, Iatrakis G, Romanidis C, Bothou A and Galazios G: Cervical
cancer: Screening, diagnosis and staging. J BUON. 21:320–325.
2016.PubMed/NCBI
|
|
23
|
Le Gallo M, Lozy F and Bell DW:
Next-Generation Sequencing. Adv Exp Med Biol. 943:119–148. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang S, Sun L and Feng G: SP1-mediated
long noncoding RNA POU3F3 accelerates the cervical cancer through
miR-127-5p/FOXD1. Biomed Pharmacother. 117:1091332019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fasterius E, Uhlén M and Al-Khalili
Szigyarto C: Single-cell RNA-seq variant analysis for exploration
of genetic heterogeneity in cancer. Sci Rep. 9:95242019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Crook T, Wrede D, Tidy JA, Mason WP, Evans
DJ and Vousden KH: Clonal p53 mutation in primary cervical cancer:
Association with human-papillomavirus-negative tumours. Lancet.
339:1070–1073. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thomas M, Pim D and Banks L: The role of
the E6-p53 interaction in the molecular pathogenesis of HPV.
Oncogene. 18:7690–7700. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Justus CR, Leffler N, Ruiz-Echevarria M
and Yang LV: In vitro cell migration and invasion assays. J Vis
Exp. 88:510462014.
|
|
29
|
Tian R, Zuo X, Jaoude J, Mao F, Colby J
and Shureiqi I: ALOX15 as a suppressor of inflammation and cancer:
Lost in the link. Prostaglandins Other Lipid Mediat. 132:77–83.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding XZ, Tong WG and Adrian TE:
Cyclooxygenases and lipoxygenases as potential targets for
treatment of pancreatic cancer. Pancreatology. 1:291–299. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bossler F, Hoppe-Seyler K and Hoppe-Seyler
F: PI3K/AKT/mTOR Signaling Regulates the Virus/Host Cell Crosstalk
in HPV-Positive Cervical Cancer Cells. Int J Mol Sci. 20:202019.
View Article : Google Scholar
|
|
32
|
De Luca A, Maiello MR, D'Alessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: Role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Targets. 16 (Suppl
2):S17–S27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Milde-Langosch K: The Fos family of
transcription factors and their role in tumourigenesis. Eur J
Cancer. 41:2449–2461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brenner AK, Reikvam H, Lavecchia A and
Bruserud Ø: Therapeutic targeting the cell division cycle 25
(CDC25) phosphatases in human acute myeloid leukemia - the
possibility to target several kinases through inhibition of the
various CDC25 isoforms. Molecules. 19:18414–18447. 2014. View Article : Google Scholar : PubMed/NCBI
|