Introduction
Glioblastoma (GBM) is the most incurable primary
brain tumor, with the median survival of patients with GBM being
12–14 months (1). The 5-year
survival rate was 28% in patients diagnosed between 1985 and 2005,
and it is estimated that 138,054 patients had a GBM diagnosis in
the United States in 2010 (2,3).
Therapies targeting glioblastoma are limited, resulting in poor
patient prognosis (3). The standard
therapy is the combination of radiation and chemotherapy using
temozolomide following surgical resection of the tumor. However,
despite treatment, patients often experience tumor recurrence,
leading to poor survival rates (4).
Cancer stem cells (CSCs) are considered as a major cause of tumor
relapse and malignancy (5). CSC
characteristics include their self-renewal capacity, persistent
proliferation and tumorigenicity when orthotopically injected into
immunodeficient mice (6,7). GBM stem cells (GSCs) with a phenotype
of resistance to chemotherapy are present in low number in tumor
bulks and are considered as a major cause of recurrence following
standard therapy (8). GSC-targeted
therapy has decreased the recurrence rate and improved tumor
clearance (9–11). Recently, based on transcriptomic
profiling, GBM has been classified into the proneural (PN),
mesenchymal (MES) and classical subtypes (12). The outcome of therapeutic approaches
and prognosis of patients is associated with the GBM subtype, and
patients with MES GBM have a poorer prognosis than those with
non-MES GBM due to the tumor immune microenvironment (12). Therefore, defining the subtype of GBM
and targeting GSCs are crucial for successful therapy.
The dihydropyrimidinase-related protein (DRP)
family, also known as collapsin-response mediator proteins, was
first identified as a cytosolic protein family that mediates
neurite outgrowth, growth cone collapse and axon guidance via
semaphorin 3A signaling during neuronal development (13–15). The
DRP family is composed of five members (DRP1-5). DRP1
overexpression decreases invasion in lung cancer (16). Furthermore, inhibition of DRP2
phosphorylation reportedly enhances paclitaxel on-target activity
in ovarian cancer (17). In
addition, inhibition of DRP3 promotes lung cancer metastasis. Also,
DRP4 is induced by TP53 and regulates energy metabolism in
adipocytes and non-small cell lung cancer (18,19).
DRP5 is a recently classified member of the DRP protein family that
is expressed in neuroendocrine lung cancer and glioblastoma
(16). DRP5 can stabilize Notch
receptors and promote Notch signaling by preventing ubiquitination
and lysosomal degradation of Notch receptors, contributing
therefore to cell proliferation in GBM (20). Furthermore, DRP5 expression is
associated with cancer cell growth rate and tumorigenicity in
osteosarcoma, and with neurological autoimmune disease (21,22).
Overall, defining the GBM subtype and targeting GSCs
is crucial for a successful therapy. Therefore, functional markers
for each subtype-specific GSCs should be defined. Public gene
expression databases are useful for finding candidate genes that
may serve as GSC markers. The present study analyzed these
databases to identify specific functional markers of GSCs.
Materials and methods
Cells and cell culture conditions
The human GBM cell lines U87MG (RRID, CVCL_0022,
glioblastoma of unknown origin), LN229 (RRID, CVCL_0393), LN18
(RRID, CVCL_0392), T98G (RRID, CVCL_0556), A172 (RRID, CVCL_0131)
and A1207 (RRID, CVCL_8481) were purchased from the American Type
Culture Collection. Normal human astrocytes (NHAs) were purchased
from ScienCell Research Laboratories, Inc. Human GBM cells and NHAs
were cultured in high-glucose DMEM supplemented with 10% FBS, 1%
penicillin/streptomycin and 2 mM L-glutamine (all from HyClone; GE
Healthcare Life Sciences) and placed at 37°C in a humidified
incubator containing 5% CO2 and 95% humidity. PN-GSCs
[528NS, GSC11 (RRID, CVCL_DR55) and GSC23 (RRID, CVCL_DR59)],
MES-GSCs (GSC20) and non-defined GSCs (GSC28) were kindly provided
by Dr Ichiro Nakano of Ohio State University (528NS) and Dr Erik
Sulman of The University of Texas M.D. Anderson Cancer Center
(GSC11, GSC23, GSC20 and GSC28). GSCs were cultured in DMEM/F12
(HyClone; GE Healthcare Life Sciences) supplemented with 0.04% B27
(Invitrogen; Thermo Fisher Scientific, Inc.), 20 ng/ml epidermal
growth factor (EGF; R&D Systems, Inc.), 20 ng/ml basic
fibroblast growth factor (bFGF; R&D Systems, Inc.), 1%
penicillin/streptomycin (HyClone; GE Healthcare Life Sciences) and
2 mM L-glutamine (HyClone; GE Healthcare Life Sciences) and placed
at 37°C in a humidified incubator containing 5% CO2 and
95% humidity. Mycoplasma testing was performed using reverse
transcription-quantitative PCR (RT-qPCR). All human cell lines were
authenticated using short tandem repeat profiling.
Plasmids, transfection and lentivirus
infection
The DRP5 short hairpin RNA (shRNA) target sequence
5′-CAGGACTCACTGTCCAATCTAC-3′ was selected and screened for
off-target complementarity using NCBI-Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The shRNA
was cloned into the commercial pLKO.1-puro lentiviral vector
(Sigma-Aldrich; Merck KGaA). A total of 2×106 293FT
cells (Invitrogen; Thermo Fisher Scientific, Inc.) were seeded on
100Φ cell culture plates for transfection. After 24 h, 4 µg
pLKO.1-shNT-puro, 4 µg pLKO.1-shDRP5-puro and second-generation
lentiviral packaging vectors (3 µg pCMV-dR8.91 and 1 µg VSV-G) were
transfected into 293FT cells using Lipojet™ in vitro
transfection reagent (SignaGen Laboratories) to produce lentiviral
particles. Lentiviruses were concentrated using the Lenti-X™
Concentrator (Takara Bio, Inc.) and resuspended into 400 µl PBS. A
total of 1.5×106 528NS cells were seeded on 100Φ cell
culture plates for infection. After 24 h, 528NS cells were infected
with 200 µl lentiviral particles. Cells infected with the
lentiviral particles were selected with Puromycin (3 µg/ml) during
a 1-week incubation. Subsequently, pLKO.1-shNT-puro
lentivirus-infected 528NS cells were renamed 528NS-puro and
pLKO.1-shDRP5-puro lentivirus-infected 528NS cells were renamed
528NS-DRP5 knockdown (KD). Knockdown efficiency was confirmed by
western blotting.
Western blotting
Cells from the aforementioned cell lines were lysed
using RIPA lysis buffer (150 mM sodium chloride, 1% NP-40, 0.1% SDS
and 50 mM Tris pH 7.4) containing 1 mM β-glycerophosphate, 2.5 mM
sodium pyrophosphate, 1 mM sodium fluoride, 1 mM sodium
orthovanadate and protease inhibitor (Roche Diagnostics). Cell
lysis was performed through two-time sonication (one cycle
sonication condition, 20 kHz; amplitude 20%; 3 sec on, 2 sec off,
total 15 sec; 4°C; total energy input, 8 J) and the lysed cells
were incubated at 4°C for 3 h and centrifuged at 21,000 × g at 4°C
for 20 min to obtain the supernatant. Total protein concentration
was quantified using Bradford assay reagent (Bio-Rad Laboratories,
Inc.) according to the manufacturer's protocol. A total of 10 µg
protein/lane was separated by 10% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes (Pall Life Sciences). Membranes
were blocked with 5% non-fat milk for 1 h at 25°C and incubated for
12 h at 4°C with either rabbit anti-DRP5 (1:500; cat. no.
HPA072387; Atlas Antibodies) or mouse anti-β-actin (1:10,000; cat.
no. A5316; Sigma-Aldrich; Merck KGaA). Membranes were subsequently
incubated for 2 h at 25°C with horseradish peroxidase-conjugated
goat anti-rabbit (cat. no. 31460) or anti-mouse (cat. no. 31430)
IgG secondary antibodies (both 1:5,000; Pierce; Thermo Fisher
Scientific, Inc.), and protein bands were visualized using the
SuperSignal West Pico Chemiluminescent Substrate (Pierce; Thermo
Fisher Scientific, Inc.).
In vitro limiting dilution assay
(LDA)
528NS cells infected with pLKO.1-puro (control) or
pLKO.1-shDRP5-873 lentivirus were plated in 96-well plates with a
decreasing number (20, 10, 5 and 1) cells/well, with 24 wells used
for each cell number. The cells were cultured in DMEM/F12
supplemented with 0.2% B27, 20 ng/ml bFGF and 20 ng/ml EGF. The
medium was replaced every 3 days with fresh bFGF and EGF.
Neurospheres were counted after 13 days using a light microscope
(CKX53; Olympus Corporation). The experiment was performed in
duplicate. Extreme limiting dilution analysis was performed using
the ELDA software (http://bioinf.wehi.edu.au/software/elda/).
Orthotopic glioma cell
implantation
Ten female BALB/c nude mice (4–5 weeks old; average
weight, 15 g), were purchased through Orient Bio, Inc. Mice were
maintained in a 12-h light/12-h dark cycle at 23±2°C and 55±5%
humidity, and they had constant access to food and water. For
orthotopic implantation, cells were resuspended in PBS, and 3 µl of
5×104 528NS-puro and 528NS-DRP5 KD cells were
stereotactically injected into the brains of 5 BALB/c nude mice
each (coordinates, 2 mm right and 1 mm rostral from bregma, and 3
mm depth from the surface of the skull) as previously described
(23). Anesthesia was performed
using Zoletil® (30 mg/kg) and Rompun® (10
mg/kg).
Mice generally exhibited neurological problems after
35–40 days. Therefore, after 40 days mice were anesthetized by
intraperitoneal injection of Avertin (250 mg/kg; Sigma-Aldrich;
Merck KGaA). Once mice lost consciousness, their body temperature
was determined using an infrared thermometer. When body temperature
decreased by >2°C compared, mice were perfused with PBS and 4%
paraformaldehyde (PFA; Sigma-Aldrich; Merck KGaA) for tissue
fixation (48 h at 4°C). Anesthetized mice were subsequently
decapitated prior to brain collection. Brain was cut into two
pieces and stored in 4% PFA until paraffin blocks were made. Animal
experiments were approved by the Korea University Institutional
Animal Care & Use Committee and performed according to the
governmental and institutional guidelines and Korean regulations
(approval no. KUIACUC-2018-0017).
Hematoxylin and eosin (H&E)
staining
Fixed brain tissues were embedded in paraffin,
sectioned into 4-µm thick slides and mounted on glass slides. For
deparaffinization, slides were incubated at 65°C in a dry oven for
20 min. For hydration, the slides were serially dipped into xylene,
absolute ethanol, 95, 80 and 70% ethanol and washed with distilled
water. After deparaffinization and hydration, slides were stained
with hematoxylin (Merck KGaA) for 5 min and rinsed with tap water.
Subsequently, tissue slides were dipped 10–15 times in acidic
alcohol and rinsed with tap water. All slides were stained with an
eosin solution (Merck KGaA) for 30 sec, followed by washing with
distilled water. Finally, the stained slides were serially
dehydrated with 95% ethanol, absolute ethanol and xylene, and
mounted with mounting solution (SP15-100; Fisher Chemical; Thermo
Fisher Scientific, Inc.). Tumor area was captured and quantified by
measuring the pixel intensity of the stained tumor area using a
dissecting light microscope (magnification, ×10) and ImageJ
software (v1.8.0_112; National Institutes of Health). Only four
slides per group were analyzed due to the loss of one sample. After
measurement, the mean of DRP5-KD pixels was normalized to the mean
of control pixels.
Immunofluorescence
A total of 5×104 cells/well (528NS,
GSC11, A172 and NHA cells) were seeded on cover slips in a 4-well
plate. After 24 h, cells were washed with ice-cold PBS and fixed
with 4% PFA for 20 min at 25°C, followed by permeabilization with
0.3% Triton X-100 in PBS and blocking with 3% bovine serum albumin
for 1 h at 25°C. The cells were stained with DRP5 antibody (1:100;
cat. no. HPA072387; Atlas Antibodies) for 12 h at 4°C. Cells were
washed with PBS three times for 5 min each and incubated with Alexa
594-conjugated secondary antibody (1:500; cat. no. A37240;
Invitrogen; Thermo Fisher Scientific, Inc.) at 25°C for 2 h. After
DAPI staining (Sigma-Aldrich; Merck KGaA) for 10 min at 25°C,
slides were washed three times with PBS, mounted in ProLong™ Gold
Antifade Mountant (P36930; Invitrogen; Thermo Fisher Scientific,
Inc.), and stored at 4°C in the dark. Fluorescence was detected
using a confocal microscope (magnification, ×40).
RT-qPCR
RT-qPCR was performed to quantify mRNA levels. Total
RNA was isolated from cells (NHA, 528NS, GSC11, GSC23, GSC20,
GSC28, U87MG, T98G, LN18, LN229, A1207, A172, 528NS-puro and
528NS-shDRP5 KD) using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. A total of 1 µg RNA pre-treated with RNase-free DNase was
used as a template to synthesize cDNA using the RevertAid First
Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RT-qPCR analysis was
performed using SYBR Premix Ex Taq (Takara Bio, Inc.) and CFX096
(Bio-Rad Laboratories, Inc.) using the following thermocycling
conditions: Initial denaturation at 95°C for 30 sec, followed by 50
cycles at 95°C for 5 sec and 60°C for 30 sec for annealing and
elongation. Target gene expression levels were normalized to 18S
ribosomal (r)RNA and quantified using the 2−ΔΔCq method
(24). The sequences of the primers
used were as follows: DRP1 forward, 5′-CTACCACGCCCGACTACTTG-3′ and
reverse 5′-TCCTCTATCCCGTTGACACC-3′; DRP2 forward,
5′-CACAGCGAGGGAGACTTAGG-3′ and reverse 5′-CAGGGACCTCTTCGTCCTCT-3′;
DRP3 forward, 5′-CAAGACGCTGGATTTCGATGC-3′ and reverse
5′-ACGGTCACTCTTGTCCTTGGG-3′; DRP4 forward,
5′-ATCAGTCGGGGTTCAGCCTAT-3′ and reverse
5′-GGAGAGAGAGGTGATGTTGGA-3′; DRP5 forward,
5′-TAAGGAGGCACTGGATTTGG-3′ and reverse 5′-GCCGAGATACTGGACACGTT-3′;
18S rRNA forward, 5′-CAGCCACCCGAGATTGAGCA-3 and reverse
5′-TAGTAGCGACGGGCGGTGTG-3′; Nestin forward,
5′-AACAGCGACGGAGGTCTCTA-3′ and reverse 5′-TTCTCTTGTCCCGCAGACTT-3′;
and SOX2 forward, 5′-CAAGATGCACAACTCGGAGA-3′ and reverse,
5′-CGGGGCCCGTATTTATAATC-3′.
Patient dataset analysis and gene set
enrichment analysis (GSEA)
Microarray datasets from The Cancer Genome
Atlas-GBM/low grade glioma (TCGA-GBM/LGG) database of the National
Cancer Institute (https://cancergenome.nih.gov) and Gene Expression
Omnibus (https://www.ncbi.nlm.nih.gov/geo/) were collected and
classified into the ‘DRP5-high’ or ‘DRP5-low’ groups according to
the higher or lower than average expression values. GSE4536 is a
gene expression dataset comprising serum-free and serum-cultured
GBM cells, whereas GSE67089 is a PN- and MES-subtype GSC gene
expression dataset (25,26). A total of five stemness-associated
gene sets for GSEA were downloaded from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp):
‘Kyoto Encyclopedia of Genes and Genomes_Notch signaling pathway’,
‘Hallmark_Hedgehog signaling pathway’, ‘Hallmark_Wnt/β-catenin
signaling’, ‘Hallmark_IL6_JAK_STAT3 signaling’ and
‘Hallmark_TNFA_VIA_NFKB signaling’. GSEA was performed using the
GenePattern public server (https://cloud.genepattern.org) and the aforementioned
datasets. GSEA was run using default options except for the
collapse dataset set as ‘False’.
Statistical analysis
Single sample GSEA (ssGSEA; ssGSEAProjection
v10.0.3; http://www.genepattern.org/) was
performed to obtain enrichment scores for PN, MES and GSC-high gene
sets in each patient from the TCGA-GBM database. Kaplan-Meier
survival analyses were performed using GraphPad Prism 6 (GraphPad
Software, Inc.). Subsequently, Pearson's correlation coefficient (r
value) was determined by comparing each enrichment score with the
level of DRP5 expression. For statistical significance, all
analyses were performed using a two-tailed unpaired Student's
t-test, except for comparison of DRP5 expression with tumor grade
and histology, for which a one-way ANOVA followed by the
Tukey-Kramer test was used for multiple comparisons. Student's
t-test was performed using Microsoft Office Excel (Microsoft
Corporation) and ANOVA was performed using GraphPad Prism 6. Data
were presented as the mean ± standard error of the mean. P<0.05
was considered to indicate a statistically significant
difference.
Results
DRP5 identification and specific
expression in GSCs
The GSE4536 and GSE67089 datasets (GBM
patient-derived cancer stem cell microarray dataset) were analyzed
to identify a gene list unique to GSCs. The data were compared with
gene expression levels of non-GSCs and NHAs, and the gene list was
classified into ‘GSC-high’ and ‘GSC-low’ groups (P<0.05;
Fig. 1A). Genes in the GSC-high
group were analyzed to identify GSC-specific genes based on the
prognosis of patients and mRNA expression levels from TCGA-GBM/LGG
microarray database. DRP5 was chosen as a predicted GSC-specific
gene due to its high mRNA expression in GSCs and in high grade and
aggressive brain tumors (Fig. 1B).
Furthermore, the prognosis of patients with GBM with high DRP5
expression was lower than that in patients with low DRP5 expression
(Fig. 1C).
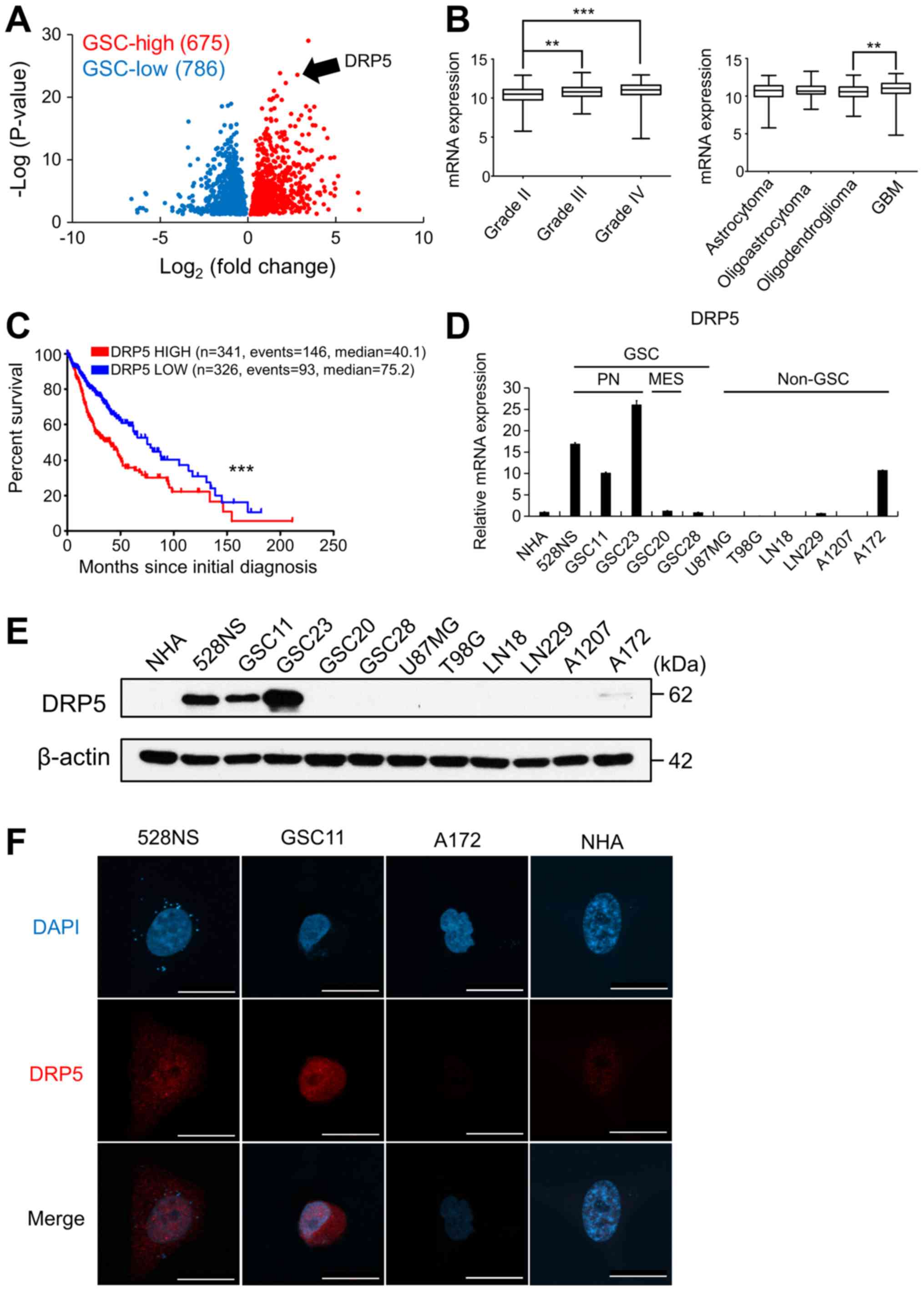 | Figure 1.DRP5 expression in GSCs and its
clinical significance. (A) Identification of DRP5 as a GSC-specific
upregulated gene using GSE4536 and GSE67089 datasets. GSC-high and
GSC-low cut-off values are ±2-fold change. (B) DRP5 expression in
patients in TCGA-GBM/LGG according to tumor grade and histology.
(C) Kaplan-Meier survival analysis of patients in TCGA-GBM/LGG
according to the expression levels of DRP5. (D) Reverse
transcription-quantitative PCR analysis analyzing relative DRP5
mRNA expression in PN- and MES-subtype GSCs and non-GSC glioma
cells compared with NHAs. Data were analyzed using the
2−ΔΔCq method, and human 18S ribosomal RNA was used as
the internal control. (E) Western blot analysis of DRP5 protein in
GSCs and non-GSC glioma cells compared with NHAs. Human β-actin was
used as the loading control. (F) Immunofluorescence images
displaying DRP5 (red) in NHAs, two GSCs (528N and GSC11), and the
non-GSC A172 glioma cells. Scale bar, 20 µm. **P<0.01;
***P<0.001. DRP5, dihydropyrimidinase-related protein 5; GBM,
glioblastoma; GSC, GBM stem cell; TCGA, The Cancer Genome Atlas;
PN, proneural; MES, mesenchymal; NHA, normal human astrocytes;
DAPI, 4′,6-diamidino-2-phenylindole. |
To further analyze DRP5 expression in GSCs, RT-qPCR
and western blotting were performed in GSCs and non-GSCs compared
with NHAs (Fig. 1D and E,
respectively). The results demonstrated that mRNA and protein
expression of DRP5 in GSCs was elevated compared with non-GSCs and
NHAs. Among GSCs, the PN-subtype GSCs (528NS, GSC11 and GSC23)
exhibited higher DRP5 expression compared with the MES-subtype GSCs
(GSC20) and the non-defined GSCs (GSC28). In addition, A172 non-GSC
GBM cells exhibited similar expression levels of DRP5 compared with
GSC11, but protein expression of DRP5 in A172 cells was markedly
lower than those in the PN-subtype GSCs (Fig. 1D and E).
DRP5 was mainly localized in the nucleus of GSCs
(Fig. 1F). A previous study reported
that a carboxyl-terminal truncated isoform of DRP5 is localized to
the nucleus compared with a full-length DRP5 that is localized in
the cytoplasm and enhances cancer cell proliferation (27). It was therefore hypothesized that
DRP5 in GSCs may be a carboxyl-terminal truncated isoform with
biological functions that differ from the ones of full-length
DRP5.
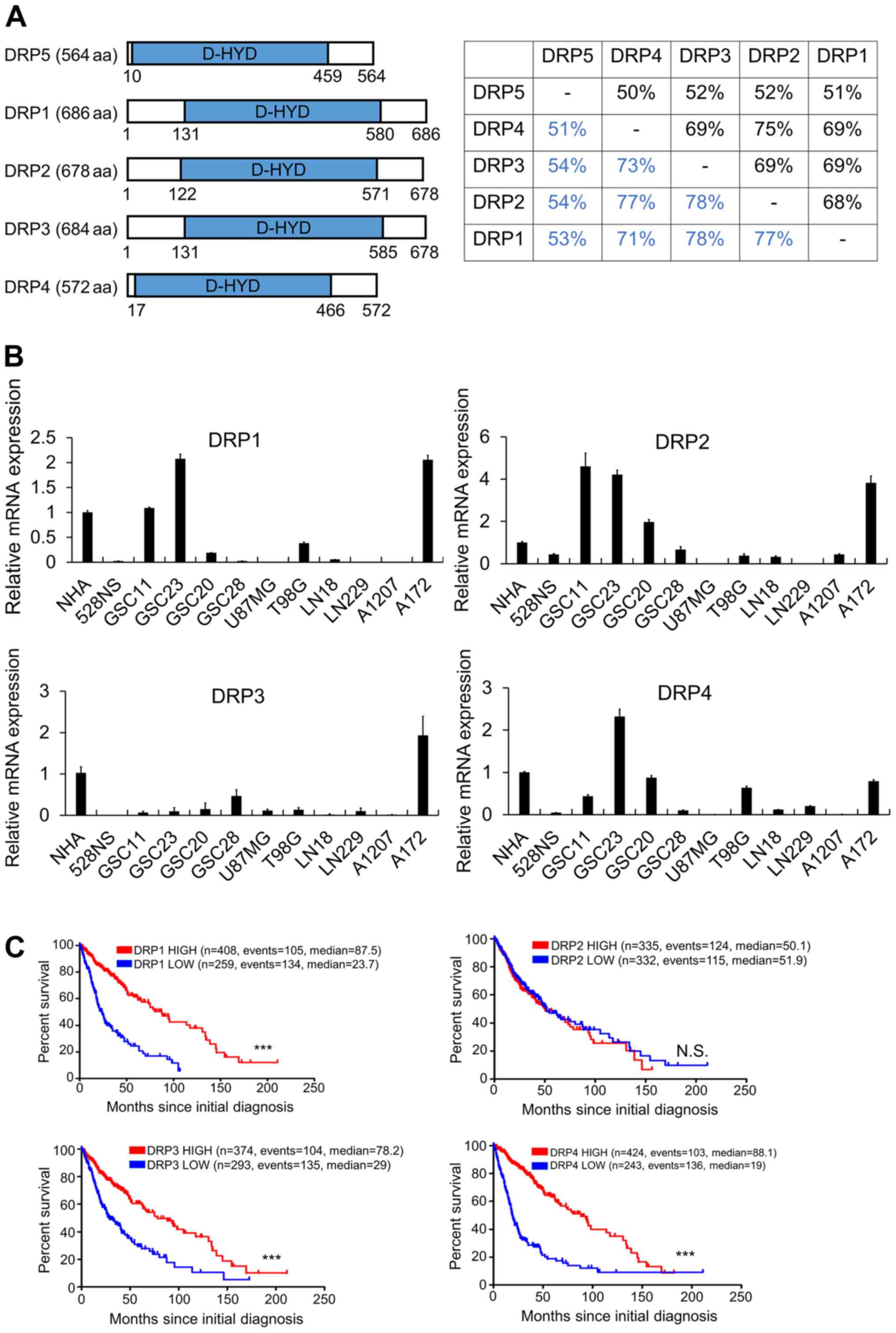
DRP5 is structurally and functionally
distinct from other DRPs
It was hypothesized that DRP5 functions by
interacting with other DRPs in GSCs, suggesting that DRPs might
function as a complex (28). DRPs
have a conserved D-hydantionase dihydropyrimidinase (D-HYD) domain;
however, the amino acid sequence of DRP5 is different to that of
the other DRPs (Fig. 2A). Although
expression levels of DRP1-4 were higher in some GSCs and in A172
cells than those in NHA cells, high expression levels of DRP1-4 in
patients with GBM were not associated with poor prognosis compared
with low expression levels (Fig. 2B and
C). A previous study reported that DRP5 has several amino acid
substitutions at the core catalytic D-HYD domain and does not
possess dihydropyrimidinase enzymatic activity (29). It was therefore hypothesized that
DRP5 may serve a specific role in GSCs.
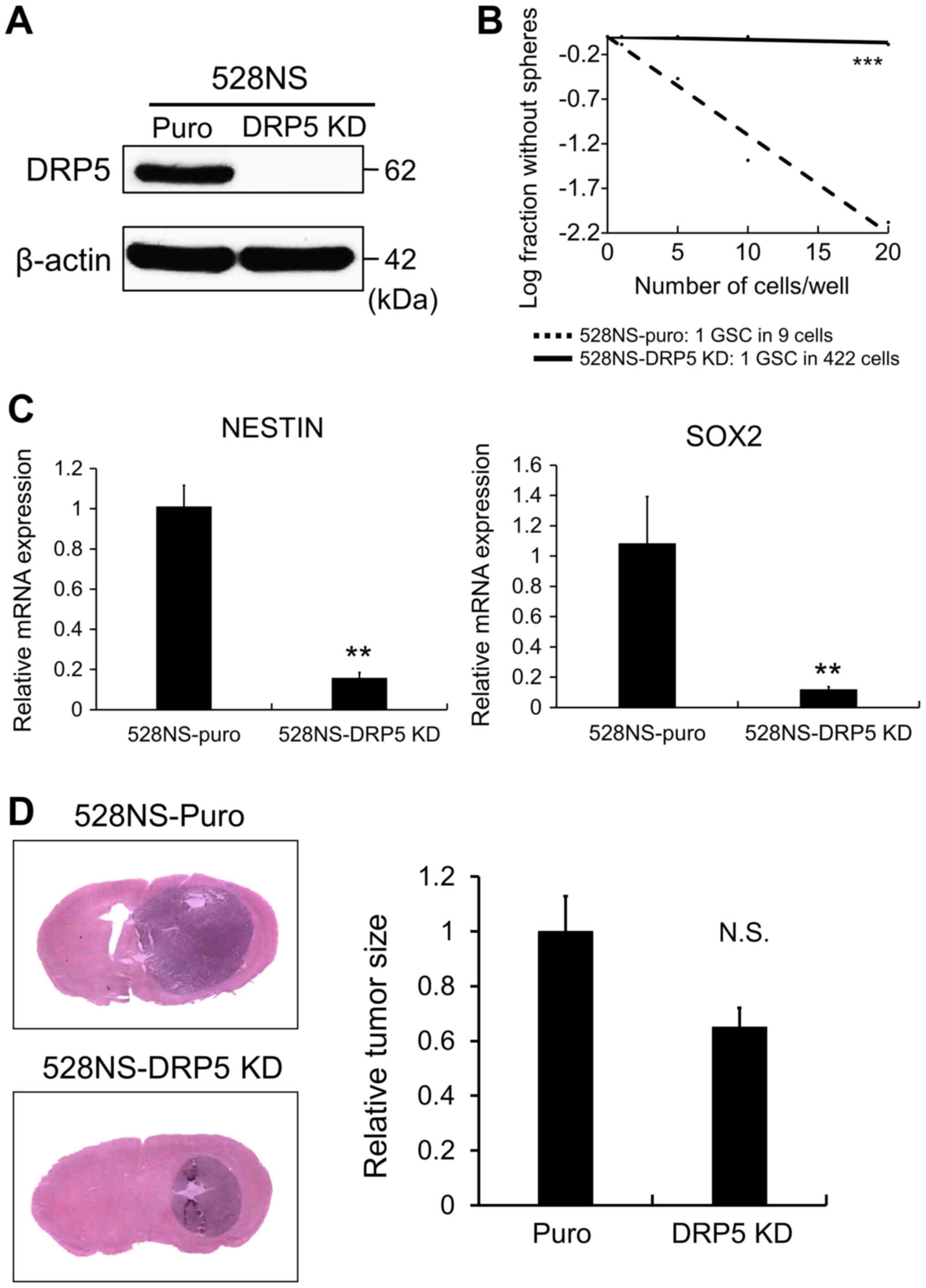
DRP5 maintains GSC properties
To investigate the role of DRP5 in GSCs,
shRNA-mediated KD of DRP5 was performed in 528NS GSCs, since DRP5
expression was higher in PN-GSCs than in other GSCs. Following
knockdown, the effect of DRP5 loss was evaluated on cancer stemness
and tumorigenesis. DRP5 KD was confirmed in 528NS GSCs by western
blotting (Fig. 3A). Subsequently, an
LDA was performed to compare the tumor sphere-forming ability
between GSCs lacking DRP5 and control cells. The results
demonstrated that DRP5 KD significantly decreased the
sphere-forming ability (Fig. 3B).
Additionally, RT-qPCR was performed to determine whether DRP5 KD
decreased the expression levels of GSC markers, such as Nestin and
SRY-box transcription factor 2 (SOX2). The results revealed that
the expression levels of Nestin and SOX2 were decreased in
528NS-DRP5 KD cells compared with control cells (Fig. 3C). Subsequently, in vivo
experiments using a xenograft mouse model were performed, and
528NS-puro and 528NS-DRP5 KD cells were orthotopically injected
into the brains of immunodeficient mice to investigate
tumorigenicity. The results demonstrated that 528NS-DRP5 KD cells
displayed decreased tumorigenicity compared with 528NS-puro cells,
according to the tumor size evaluated following H&E staining
(Fig. 3D). Taken together, these
findings suggested the importance of DRP5 in maintaining GSC
stemness and tumorigenicity.
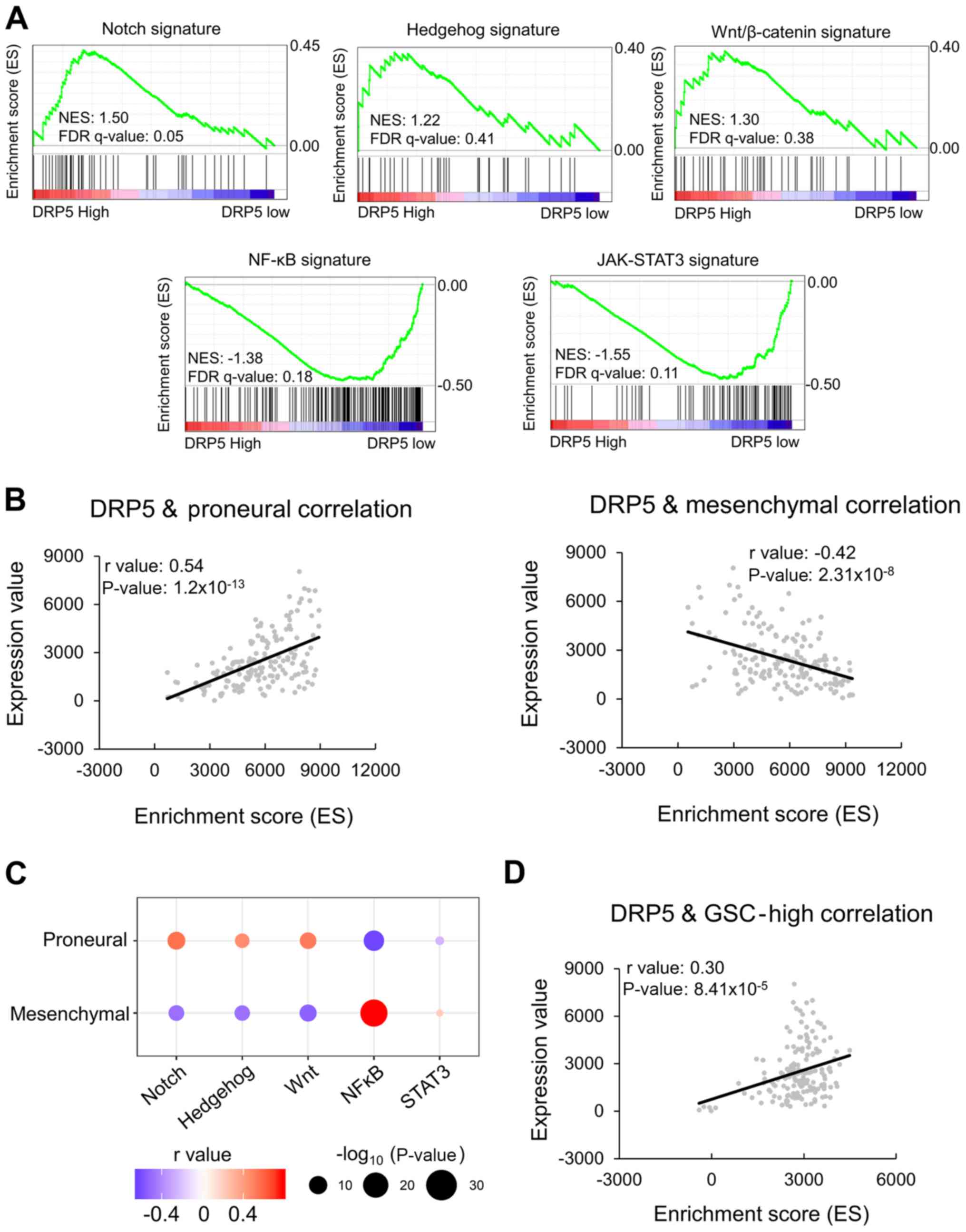
DRP5 expression is positively
correlated with stemness-associated gene signatures
GSEA was performed to confirm the associations
between DRP5 expression patterns in the patient dataset of
TCGA-GBM/LGG and cancer stemness-associated gene signatures. There
are five known stemness-associated gene signatures: Notch,
Hedgehog, Wnt/β-catenin, Janus kinase-signal transducer and
activator of transcription 3 (JAK-STAT3) and NF-κB. An enrichment
plot for each signature was performed and the enriched signature in
the DRP5-high patient group was analyzed. The Notch, Hedgehog and
Wnt/β-catenin signatures were enriched in the DRP5-high group,
whereas the JAK-STAT3 and NF-κB signatures were enriched in the
DRP5-low group (Fig. 4A). Tumor
necrosis factor-α (TNF-α)/NF-κB signaling lead to a PN-MES
transition, resulting in the enrichment of CD44 subpopulations and
radio-resistant phenotypes (30).
Additionally, JAK-STAT3 signaling has previously been associated
with MES differentiation and poor clinical outcomes (31). Therefore, NF-κB and JAK-STAT3
signatures are considered MES-subtype stemness signatures. The
present results revealed that DRP5 expression was positively
correlated with PN-subtype stemness signatures and negatively
correlated with MES-subtype signatures (Fig. 4B). PN-gene signatures were positively
correlated with Notch, Hedgehog and Wnt/β-catenin signatures,
whereas MES-gene signatures were positively correlated with NF-κB
and JAK-STAT3 signatures (Fig. 4C).
Furthermore, DRP5 expression was positively correlated with the
GSC-high gene list in patients with GBM (Fig. 4D). Overall, these results suggested
that DRP5 may be considered as a novel biomarker of PN-subtype
GSCs.
Discussion
GSCs are resistant to chemotherapy and radiotherapy
and promote tumor growth following surgical resection of the tumor
(6–9). GSC subtype has been associated with
treatment efficacy and prognosis of patients (13). Numerous strategies have been
developed to eradicate GSCs; however, they have proven ineffective
(32). The failure of these
therapies can partly be explained by the different drug responses
depending on the GSC subtype (33).
In order to development successful treatment options, it is
therefore essential to properly define the GSC subtype of the
cancer.
The present study identified DRP5 using gene
expression database analysis from GBM patient-derived
differentiated and stem-like cancer cells. Notably, DRP5 displayed
PN-GSC-specific expression, and the prognosis of patients from
TCGA-GBM/LGG dataset with high DRP5 expression was lower compared
with patients with low DRP5 expression. However, in TCGA-GBM
dataset, DRP5 expression was not significant in determining patient
prognosis (data not shown). DRP5 may therefore be only one of the
factors that maintain stemness signature, making it hard to explain
how DRP5 expression could determine the aggressiveness of GBM
(34,35). However, several studies reported that
a single factor can affect the progression of low-grade glioma
(36–38). Therefore, high DPR5 expression in
PN-GSCs may promote the malignancy of brain tumors. Furthermore,
DRP5 KD decreased tumorigenicity and numerous stemness-associated
molecular signaling pathways of GSCs, although the tumor size in
mice with DRP5 KD was not statistically different from that in
control mice. In a subcutaneous mouse cancer model, tumor size is
crucial for determining tumorigenicity; however, in an orthotopic
brain cancer model, tumor size is not the only determinant of
tumorigenicity. The major cause of death in brain tumor mouse
models is the development of neurodegenerative disorders due to
increased brain pressure and tumor infiltration in neighboring
nervous tissues (39). Overall, DRP5
seems to maintain GSC characteristics and may therefore be
considered as a functional biomarker of PN-GSCs.
DRP5 is a cytosolic protein involved in normal
neuronal development, including neurite outgrowth and axon
guidance. Dihydropyrimidinase (DHPase) is an enzyme that degrades
pyrimidines, such as uracil and thymine (40). The D-HYD domain is therefore required
for successful enzymatic activity. In the DRP family, except
DHPase, the enzymatic catalytic core amino acids of the D-HYD
domain are substituted, abrogating its enzymatic activity (29). The DRP family is therefore
functionally different from DHPase. DRP5 interacts with other DRP
family genes, including DRP2, DRP3 and DRP4 (41). However, in the GSC lines used in the
present study, especially 528NS, the expression levels of other DRP
family genes were lower than that of DRP5. Therefore, DRP5 may
function alone to regulate the characteristics of GSCs. Although
the truncated form of DRP5 is reported to be translocated to the
nucleus in GBM cells and to promote cell proliferation, the
underlying mechanism of this phenomenon remains unclear (26). The present results demonstrated that
DRP5 was specifically expressed in PN-GSCs and was localized to the
nucleus. Therefore, nuclear DRP5 may promote cell proliferation and
maintain PN-GSC characteristics. The mechanism of DRP5
translocation to the nucleus and the role of nuclear DRP5 require
further investigation. A previous study has demonstrated that DRP5
regulates stemness-associated molecular signaling pathways, such as
the Notch signaling pathway (20).
In addition, the current study revealed that DRP5 expression was
positively associated with Notch, Hedgehog and Wnt/β-catenin
signaling pathways. DRP5 in the nucleus may therefore influence
gene expression by acting as a transcriptional regulator, thereby
regulating GSC stemness and tumorigenicity.
In conclusion, the results from the present study
demonstrated that DRP5 was specifically expressed in the PN-subtype
of GSCs, suggesting that it may be used as a functional biomarker
of GBM derived from PN-GSCs.
Acknowledgements
The authors would like to thank Dr. Man Bock Gu, the
director of BK21 PLUS for Biotechnology in the Department of
Biotechnology at Korea University, for the support of publication
cost.
Funding
The present study was supported by the National
Research Foundation (grant no. 2017R1E1A1A01074205).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on request.
GSE4536 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4536)
and GSE67089 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67089)
are available in the GEO database.
Authors' contributions
MGP, SS and HK conceived the study and the
experiments. MGP and SWH performed the experiments. MGP and SHC
captured and analyzed the staining images. MGP performed the
statistical analysis. MGP and HK wrote and corrected the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Animal experiments were performed in the
specific-pathogen-free facility with the approval of the Korea
University Institutional Animal Care & Use Committee (approval
no. KUIACUC-2018-0017) and according to the governmental and
institutional guidelines and Korean regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mazaris P, Hong X, Altshuler D, Schultz L,
Poisson LM, Jain R, Mikkelsen T, Rosenblum M and Kalkanis S: Key
determinants of short-term and long-term glioblastoma survival: A
14-year retrospective study of patients from the Hermelin Brain
Tumor Center at Henry Ford Hospital. Clin Neurol Neurosurg.
120:103–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Porter KR, McCarthy BJ, Freels S, Kim Y
and Davis FG: Prevalence estimates for primary brain tumors in the
United States by age, gender, behavior and histology. Neuro Oncol.
12:520–527. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dobelbower MC, Burnett Iii OL, Nordal RA,
Nabors LB, Hyatt MD and Fiveash JB: Patterns of failure for
glioblastoma multiforme following concurrent radiation and
temozolomide. J Med Imaging Radiat Oncol. 55:77–81. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Neman J and Jandial R: Decreasing glioma
recurrence through adjuvant cancer stem cell inhibition. Biologics.
4:157–162. 2010.PubMed/NCBI
|
|
6
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lathia JD, Mack SC, Mulkearns-Hubert EE,
Valentim CL and Rich JN: Cancer stem cells in glioblastoma. Genes
Dev. 29:1203–1217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen J, Li Y, Yu TS, McKay RM, Burns DK,
Kernie SG and Parada LF: A restricted cell population propagates
glioblastoma growth after chemotherapy. Nature. 488:522–526. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie Q, Mittal S and Berens ME: Targeting
adaptive glioblastoma: An overview of proliferation an invasion.
Neuro Oncol. 16:1575–1584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kanabur P, Guo S, Simonds GR, Kelly DF,
Gourdie RG, Verbridge SS and Sheng Z: Patient-derived glioblastoma
stem cells respond differentially to targeted therapies.
Oncotarget. 7:86406–86419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jackson M, Hassiotou F and Nowak A:
Glioblastoma stem-like cells: At the root of tumor recurrence and a
therapeutic target. Carcinogenesis. 36:177–185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Q, Hu B, Hu X, Kim H, Squatrito M,
Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al: Tumor
evolution of glioma-intrinsic gene expression subtypes associates
with immunological changes in the microenvironment. Cancer Cell.
32:42–56.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schmidt EF and Strittmatter SM: The CRMP
family of proteins and their role in Sema3A signaling. Adv Exp Med
Biol. 600:1–11. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Quach TT, Honnorat J, Kolattukudy PE,
Khanna R and Duchemin AM: CRMPs: Critical molecules for neurite
morphogenesis and neuropsychiatric diseases. Mol Psychiatry.
20:1037–1045. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Charrier E, Reibel S, Rogemond V, Aguera
M, Thomasset N and Honnorat J: Collapsin response mediator proteins
(CRMPs): Involvement in nervous system development and adult
neurodegenerative disorders. Mol Neurobiol. 28:51–64. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tan F, Thiele CJ and Li Z: Collapsin
response mediator proteins: Potential diagnostic and prognostic
biomarkers in cancers (Review). Oncol Lett. 7:1333–1340. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng Y, Sethi R, Mangala LS, Taylor C,
Goldsmith J, Wang M, Masuda K, Karaminejadranjbar M, Mannion D,
Miranda F, et al: Tuning microtubule dynamics to enhance cancer
therapy by modulating FER-mediated CRMP2 phosphorylation. Nat
Commun. 9:4762018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Y, Jiang Y, Xie D, Liu M, Song N, Zhu
J, Fan J and Zhu C: Inhibition of cell-adhesion protein DPYSL3
promotes metastasis of lung cancer. Respir Res. 19:412018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nagano H, Hashimoto N, Nakayama A, Suzuki
S, Miyabayashi Y, Yamato A, Higuchi S, Fujimoto M, Sakuma I, Beppu
M, et al: p53-inducible DPYSL4 associates with mitochondrial
supercomplexes and regulates energy metabolism in adipocytes and
cancer cells. Proc Natl Acad Sci USA. 115:8370–8375. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Moutal A, Honnorat J, Massoma P,
Desormeaux P, Bertrand C, Malleval C, Watrin C, Chounlamountri N,
Mayeur ME, Besancon R, et al: CRMP5 controls glioblastoma cell
proliferation and survival through Notch-dependent signaling.
Cancer Res. 75:3519–3528. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang L, Liu W, Tang H, Xie X, Zou C, Wang
Y, Gao Z and Yin J: DRP5 is involved in cancer cell growth and
predicts poor prognosis in human osteosarcoma. Cancer Med.
6:982–993. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dubey D, Lennon VA, Gadoth A, Pittock SJ,
Flanagan EP, Schmeling JE, McKeon A and Klein CJ: Autoimmune CRMP5
neuropathy phenotype and outcome defined from 105 cases. Neurology.
90:e103–e110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Petit V, Massonnet G, Maciorowski Z,
Touhami J, Thuleau A, Nemati F, Laval J, Cahteau-Joubert S, Servely
JL, Vallerand D, et al: Optimization of tumor xenograft
dissociation for the profiling of cell surface markers and nutrient
transporters. Lab Invest. 93:611–621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su
Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mao P, Joshi K, Li J, Kim SH, Li P,
Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, et al:
Mesenchymal glioma stem cells are maintained by activated
glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc
Natl Acad Sci USA. 110:8644–8649. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brot S, Malleval C, Benetollo C, Auger C,
Meyronet D, Rogemond V, Honnorat J and Moradi-Ameli M:
Identification of a new CRMP5 isoform present in the nucleus of
cancer cells and enhancing their proliferation. Exp Cell Res.
319:588–599. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tan M, Cha C, Ye Y, Zhang J, Li S, Wu F,
Gong S and Guo G: CRMP4 and CRMP2 interact to coordinate
cytoskeleton dynamics, regulating growth cone development and axon
elongation. Neural Plast. 2015:9474232015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ponnusamy R and Lohkamp B: Insights into
the oligomerization of CRMPs: Crystal structure of human collapsin
response mediator protein 5. J Neurochem. 125:855–868. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bhat KPL, Balasubramaniyan V, Vaillant B,
Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L,
James JD, Goodman LD, et al: Mesenchymal differentiation mediated
by NF-κB promotes radiation resistance in glioblastoma. Cancer
Cell. 24:331–346. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Carro MS, Lim WK, Alvarez MJ, Bollo RJ,
Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al:
The transcriptional network for mesenchymal transformation of brain
tumours. Nature. 463:318–325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Barbato L, Bocchetti M, Di Biase A and
Regad T: Cancer stem cells and targeting strategies. Cells.
8:E9262019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kwon SM, Kang SH, Park CK, Jung S, Park
ES, Lee JS, Kim SH and Woo HG: Recurrent glioblastomas reveal
molecular subtypes associated with mechanistic implications of
drug-resistance. PLoS One. 10:e01405282015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dagogo-Jack I and Shaw AT: Tumor
heterogeneity and resistance to cancer therapies. Nat Rev Clin
Oncol. 15:81–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Patel AP, Tirosh I, Trombetta JJ, Shalek
AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT,
Martuza RL, et al: Single-cell RNA-seq highlights intratumoral
heterogeneity in primary glioblastoma. Science. 344:1396–1401.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dai X, Liao K, Zhuang Z, Chen B, Zhou Z,
Zhou S, Lin G, Zhang F, Lin Y, Miao Y, et al: AHIF promotes
glioblastoma progression and radioresistance via exosomes. Int J
Oncol. 54:261–270. 2019.PubMed/NCBI
|
|
37
|
Qian Z, Ren L, Wu D, Yang X, Zhou Z, Nie
Q, Jiang G, Xue S, Weng W, Qiu Y and Lin Y: Overexpression of
FoxO3a is associated with glioblastoma progression and predicts
poor patient prognosis. Int J Cancer. 140:2792–2804. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo K and Zhuang K: High expression of
PCBP2 is associated with proression and poor prognosis in patients
with glioblastoma. Biomed Pharmacother. 94:659–665. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Martínez-Murillo R and Martínez A:
Standardization of an orthotopic mouse brain tumor model following
transplantation of CT-2A astrocytoma cells. Histol Histopathol.
22:1309–1326. 2007.PubMed/NCBI
|
|
40
|
Huang YH, Ning ZJ and Huang CY: Crystal
structure of dihydropyrimidinase in complex with anticancer drug
5-fluorouracil. Biochem Biophys Res Commun. 519:160–165. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fukada M, Watakabe I, Yuasa-Kawada J,
Kawachi H, Kuroiwa A, Matsuda A and Noda M: Molecular
characterization of CRMP5, a novel member of the collapsin response
mediator protein family. J Biol Chem. 275:37957–37965. 2000.
View Article : Google Scholar : PubMed/NCBI
|