Introduction
Liver cancer is the fifth and ninth most commonly
diagnosed cancer globally in men and women, respectively (1). It was the fourth leading cause of
cancer-associated mortality worldwide and was estimated to cause
~800,000 global deaths in 2015 (2).
Liver cancer is frequently diagnosed at an advanced stage and is
often characterized by poor prognosis (3). Consequently, only a small proportion of
patients are eligible for potentially curative therapies (4). Various therapeutic strategies are
currently used in the management of liver cancer, including
surgery, liver transplantation, chemotherapy, radiotherapy and
immunotherapy (5–12). However, liver cancer is frequently
unresponsive to chemotherapy and radiotherapy, making its clinical
outcomes poor.
The existence of chemically modified RNA species has
been documented during the past decades (13). One of the most common chemical
modifications on RNA molecules is the N6-methyladenosine (m6A)
modification on mRNAs and long non-coding RNAs, which serve a
crucial role in gene expression (14). This reversible RNA modification is
catalyzed by the adenosine methyltransferases family of enzymes
(15) and is reversed by
demethylases (16). Members of the
m6A methyltransferase family include methyltransferase like
(METTL)3, METTL14, WT1 associated protein (WTAP), RNA binding motif
protein 15 and vir like m6A methyltransferase associated (15). The m6A demethylase group of enzymes
includes the fat mass and obesity-associated protein (FTO) and AlkB
homolog 5 (16). Additionally, it
has been demonstrated that m6A can be selectively recognized by
proteins, including heterogeneous nuclear ribonucleoprotein C,
heterogeneous nuclear ribonucleoprotein A2/B1, YTH
N6-methyladenosine RNA binding protein (YTHDF)2, YTHDF1 and
eukaryotic translation initiation factor 3 subunit A (14). FTO has been demonstrated to modulate
multiple RNA modifications, including m6A and
N6,2-O-dimethyladenosine (17), and
it has been recently reported that the activity of this enzyme is
oncogenic in certain types of cancer (18,19).
Furthermore, FTO has been implicated in the induction of resistance
to chemo-radiotherapy in cervical cancer by influencing the
function of β-catenin via mRNA demethylation (20). Additionally, FTO upregulation has
been associated with breast cancer progression via the PI3K/AKT
signaling pathway (21). Overall,
the aforementioned studies suggest that FTO may have various
oncogenic roles in numerous types of cancer by modulating different
cell signaling pathways.
The role of m6A methylation in the development of
liver cancer has been explored in a few studies. For example, it
has been reported that through a m6A-YTHDF2-dependent mechanism,
METTL3 promotes tumorigenicity and metastasis of liver cancer in
vitro and in vivo (22).
Additionally, it has been proposed that WTAP serves an important
role in the progression of liver cancer via m6A-HuR-dependent
epigenetic silencing of the ETS proto-oncogene 1 (23). However, there is a lack of similar
studies on the role of FTO in liver cancer. The aim of the present
study was to explore the biological functions of FTO and its
clinical relevance in liver cancer.
Materials and methods
Human samples, tissue microarray and
cell lines
A total of 330 liver cancer tissues and 187 adjacent
non-cancerous tissues (≥5 cm from the edge of the tumor tissue),
were obtained from 330 patients at Zhejiang Provincial People's
Hospital (Hangzhou, China). Written informed consent was obtained
from all participants. The collected tissues were analyzed by FTO
immunohistochemistry (IHC) and microarray analysis. The liver
cancer tissue microarray was purchased from Shanghai BioChip Co.,
Ltd, and was performed according to the manufacturer's protocol.
Ethical approval for the present study was obtained from the Ethics
Committee of Zhejiang Provincial People's Hospital. The enrolled
patients consisted of 268 males and 62 females, with a median age
of 56 years (range, 25–91 years) at the time of surgery. Patient
follow-up was performed for ≥5 years and the survival time was
calculated from the date of surgical intervention to death. The
human liver cancer HepG2 cell line was purchased from the American
Type Culture Collection. HepG2 cells were cultured in DMEM
supplemented with 10% FBS (both Gibco; Thermo Fisher Scientific,
Inc.), 100 µg/ml penicillin and 0.1 mg/ml streptomycin at 37°C with
5% CO2 in a humidified incubator.
Small interfering RNA (siRNA)
transfection
HepG2 cells (1×106 cells/well) were
seeded into 6-well plates and cultured with DMEM supplemented with
10% FBS for 48 h. Subsequently, cells were transfected with a
mixture of siRNA (100 nM) and Lipofectamine® 2000
reagent (Thermo Fisher Scientific, Inc.), according to the
manufacture's protocol. The sequences of siRNAs used were as
follows: siRNA-FTO-1, 5′-GGATGACTCTCATCTCGAA-3′; siRNA-FTO-2,
5′-GCTGAAATATCCTAAACTA-3′; siRNA-FTO-3, 5′-GTCACGAATTGCCCGAACA-3′;
and control siRNA, 5′-UUCUCCGAACGUGUCACGU-3′. Cells were collected
24 or 48 h post-transfection for subsequent experimentation.
Western blot analysis
Western blot analysis was performed as previously
described (24). The primary
antibodies used in the present study were anti-FTO (1,1000;
ab124892; Abcam) and anti-β-actin (1:200; ab115777; Abcam).
Immunoreactive products were visualized using the ChemiDoc™ Touch
Imaging system (Bio-Rad Laboratories, Inc.) and semi-quantified by
densitometry using ImageJ software (version 1.50; National
Institutes of Health).
Cell proliferation assay
The cell proliferation assays were performed using
the Cell Counting Kit-8 (CCK-8; cat. no. CK04; Dojindo Molecular
Technologies, Inc.) according to the manufacturer's protocol
(23). Briefly, 200 µl HepG2 cells
or siRNA-FTO HepG2 cells (2×104) were seeded into
96-well cell culture-treated plates. Cells were then cultured for
48, 72 or 96 h, after which 10 µl CCK-8 was directly added into the
culture medium in each well. Subsequently, cells were incubated at
37°C for 2 h, and the absorbance was read at 450 nm using a
microplate reader. Cell proliferation was measured in five wells
for each experimental group.
Colony formation assay
A total of 1×102 HepG2 or siRNA-FTO HepG2
cells were seeded into each well of a 6-well cell culture plate and
cultured for 7 days. Subsequently, cells were washed with PBS and
fixed with 4% paraformaldehyde, and then stained with 0.5% crystal
violet. Cells were observed under an inverted light microscope
(magnification, ×10).
Apoptosis analysis
HepG2 or siRNA-FTO HepG2 cells (1×105
cells/well) were seeded in 24-well plates and cultured for 48 h.
After being washed with PBS twice and trypsinization, cells were
resuspended in 500 µl binding buffer supplemented with 5 µl Annexin
V-FITC (Nanjing KeyGen Biotech Co., Ltd.) and 5 µl propidium iodide
according to the manufacturer's protocol. Finally, the fluorescence
intensity of the samples was determined by flow cytometry (EPICS
XL-MCL; Beckman Coulter, Inc.). The number of apoptotic cells in
each sample was analyzed using FCS Express version 3.0 (De Novo
Software).
Transwell assay and wound healing
assay
Matrigel (BD Biosciences) was thawed at 4°C
overnight and diluted with DMEM. A total of 60 µl diluted Matrigel
was added in the upper chambers of a 24-well Transwell insert and
incubated at 37°C for 30 min. A total of 200 µl HepG2 cells or
siRNA-FTO HepG2 cells (2×104 cells/well) in serum-free
DMEM were seeded into the upper chamber. DMEM supplemented with 10%
FBS (600 µl) was added into the lower chamber and incubated for 24
h at 37°C. Non-migrating cells on the top of the Transwell insert
were removed using a cotton swab. The cell migration rate was
analyzed using methanol and 0.3% crystal violet staining. Wound
healing experiments were performed to investigate the effect of FTO
on the migration ability of HepG2 cells. The initial HepG2 or
siRNA-FTO HepG2 cell seeding density was 2×105
cells/cm2. A scratch wound was made using a 10-µl
pipette tip. The cells were washed twice with PBS and then
incubated with serum-free DMEM for 24 or 48 h at 37°C. The cells
were visualized under an inverted light microscope. The amount of
wound healing was quantified using ImageJ software (version 1.50;
National Institutes of Health).
Measurement of total m6A level
Total RNA from HepG2 cells was extracted and
purified using the RNeasy Mini kit (Qiagen GmbH), and the level of
m6A RNA methylation was assessed using the EpiQuik M6A RNA
Methylation Quantification kit (EpiGentek Group, Inc.) according to
the manufacturer's protocol.
IHC analysis
The tissues were fixed in 10% buffered formalin for
6–12 h at room temperature, embedded in paraffin and cut into
4-µm-thick sections. Sections were deparaffinized in xylene,
rehydrated using a gradient of ethanol concentrations (100, 95, 85
and 75%) and boiled in 1 mM TE buffer using a high-pressure cooker
(≥100°C) for 3 min for antigen retrieval. Subsequently, the
sections were blocked with 3% hydrogen peroxide for 15 min at room
temperature to inhibit endogenous peroxidase activity and incubated
with 10% goat non-immune serum (Invitrogen; Thermo Fisher
Scientific, Inc.) for 20 min at room temperature. The sections were
then incubated with anti-FTO antibody (dilution, 1:100; cat. no.
ab124892; Abcam) overnight at 4°C and with goat anti-rabbit IgG
H&L (HRP) (1:2,000; cat. no. ab205718; Abcam) at room
temperature for 15 min. This was followed by development using a
DAB Substrate kit (Dako; Agilent Technologies, Inc.). IHC staining
of FTO was scored by two independent pathologists using a light
microscope (magnification, ×200), based on the intensity and the
proportion of positively stained cells. Specifically, staining
intensity was evaluated with a grading system: 0, negative; 1,
weak; 2, moderate; and 3, strong. The percentage of positive cells
was scored as follows: 0, no staining; 1, 1–25% cells stained; 2,
26–50% cells stained; 3, 51–75% cells stained; and 4, >75% cells
stained. The final score was obtained by multiplying the scores for
intensity and percentage of positive cells.
FTO mRNA expression analysis
UALCAN (http://ualcan.path.uab.edu) is an online tool that
uses The Cancer Genome Atlas RNA-sequencing and clinical data from
31 types of cancer. Additionally, it analyzes the relative
expression levels of a query gene in various tumor sub-groups based
on individual cancer stage, tumor grade, body weight or other
clinicopathological features (25).
In the present study, UALCAN was used to evaluate the mRNA
expression levels of FTO in cancer and normal liver samples.
Statistical analysis
All statistical analyses were conducted using SPSS
v13.0 (SPSS Inc.) and data are presented as the mean ± SD of at
least three individual experiments. An unpaired t-test was used to
compare the differences between two groups. Comparisons among
multiple groups were analyzed using one-way ANOVA followed by
Tukey's post hoc test (all data met the assumption of homogeneity
of variance). A χ2-test was used to assess the
association between FTO expression and clinicopathological
parameters. A multivariate survival analysis was performed to
identify the factors associated with prognosis according to the Cox
proportional hazards regression model. The association between FTO
expression and overall survival (OS) was analyzed by the
Kaplan-Meier method with a log-rank test. P<0.05 (two-tailed)
was considered to indicate a statistically significant
difference.
Results
FTO expression is increased in liver
cancer tissues
To investigate the role of FTO in liver cancer
oncogenesis, the expression profile of FTO was characterized in
liver cancer tissues and was compared with that in adjacent normal
liver tissues by tissue microarrays. FTO scores were calculated
with scores of 0–5 and 6–12 representing the low and high
expression groups, respectively. The results of the IHC analysis
revealed high FTO expression in 330 liver cancer tissues (126/330;
38.2%), which was significantly higher than high FTO expression in
187 normal liver tissues (46/187; 24.6%; χ2=10.765;
P=0.001; data not shown). These results were in agreement with the
outcome of an analysis using UALCAN that revealed that FTO
expression was higher in liver cancer tissues compared with in
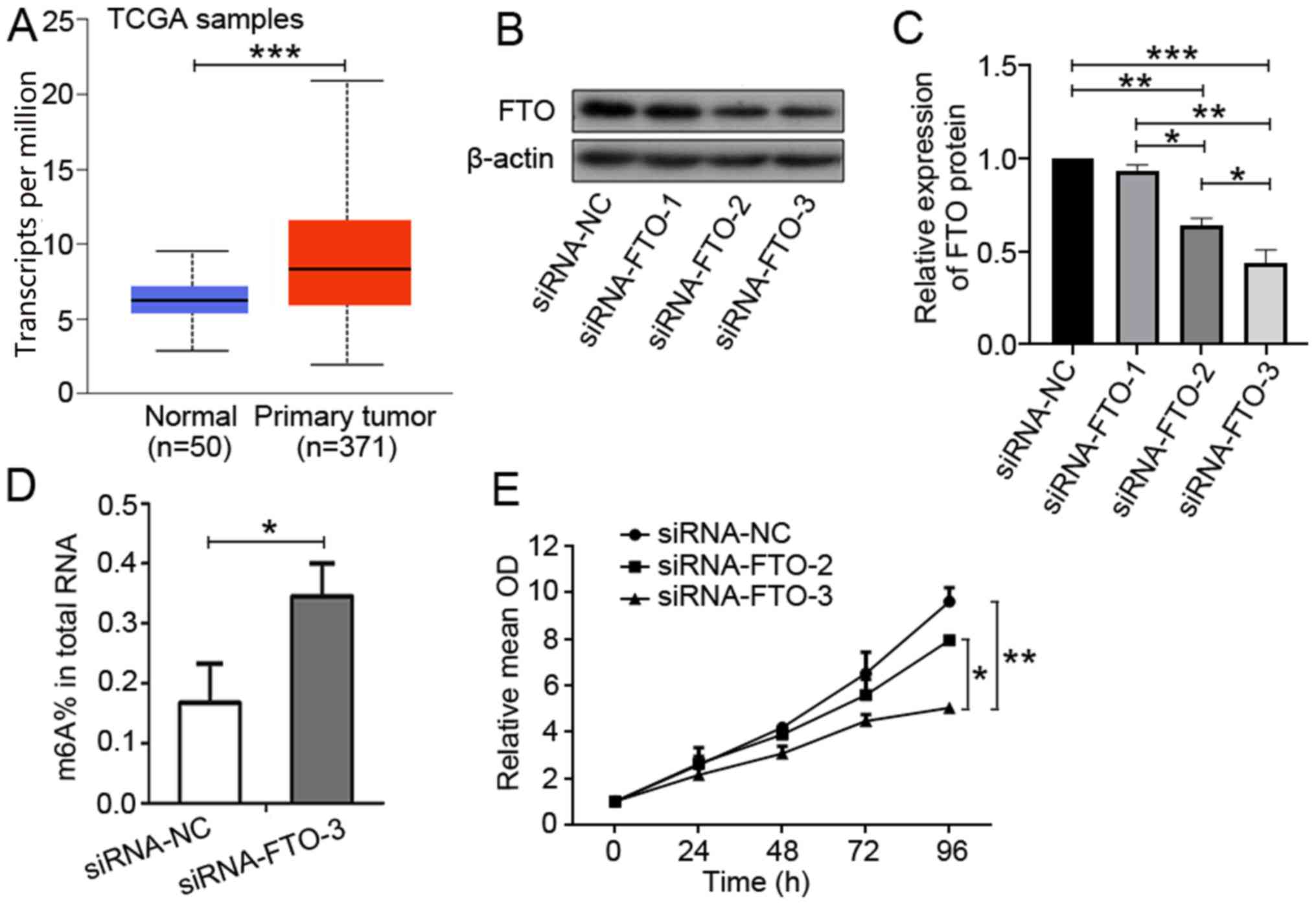
normal liver tissues (P<0.001; Fig.
1A). Overall, these observations suggested that FTO
upregulation may be associated with the development of liver
cancer.
FTO promotes in vitro proliferation
and mobility of the human liver cancer cell line HepG2
FTO may be a modulator of m6A RNA demethylation and
may promote liver cancer oncogenesis (17). siRNAs were used to knock down FTO
expression in the human liver cancer HepG2 cell line. FTO
expression downregulation was confirmed by western blotting
(Fig. 1B and C). In addition, the
m6A RNA level in the total RNA pool was quantified and it was
observed that FTO knockdown was accompanied by a significant
increase in the level of m6A RNA (P=0.032; Fig. 1D), suggesting an m6A demethylation
function for FTO that may serve a role in liver cancer
carcinogenesis. However, the exact mechanism needs to be further
elucidated. Subsequently, a cell proliferation assay was performed
using CCK-8. This analysis indicated that downregulation of FTO
expression suppressed cell proliferation (Fig. 1E).
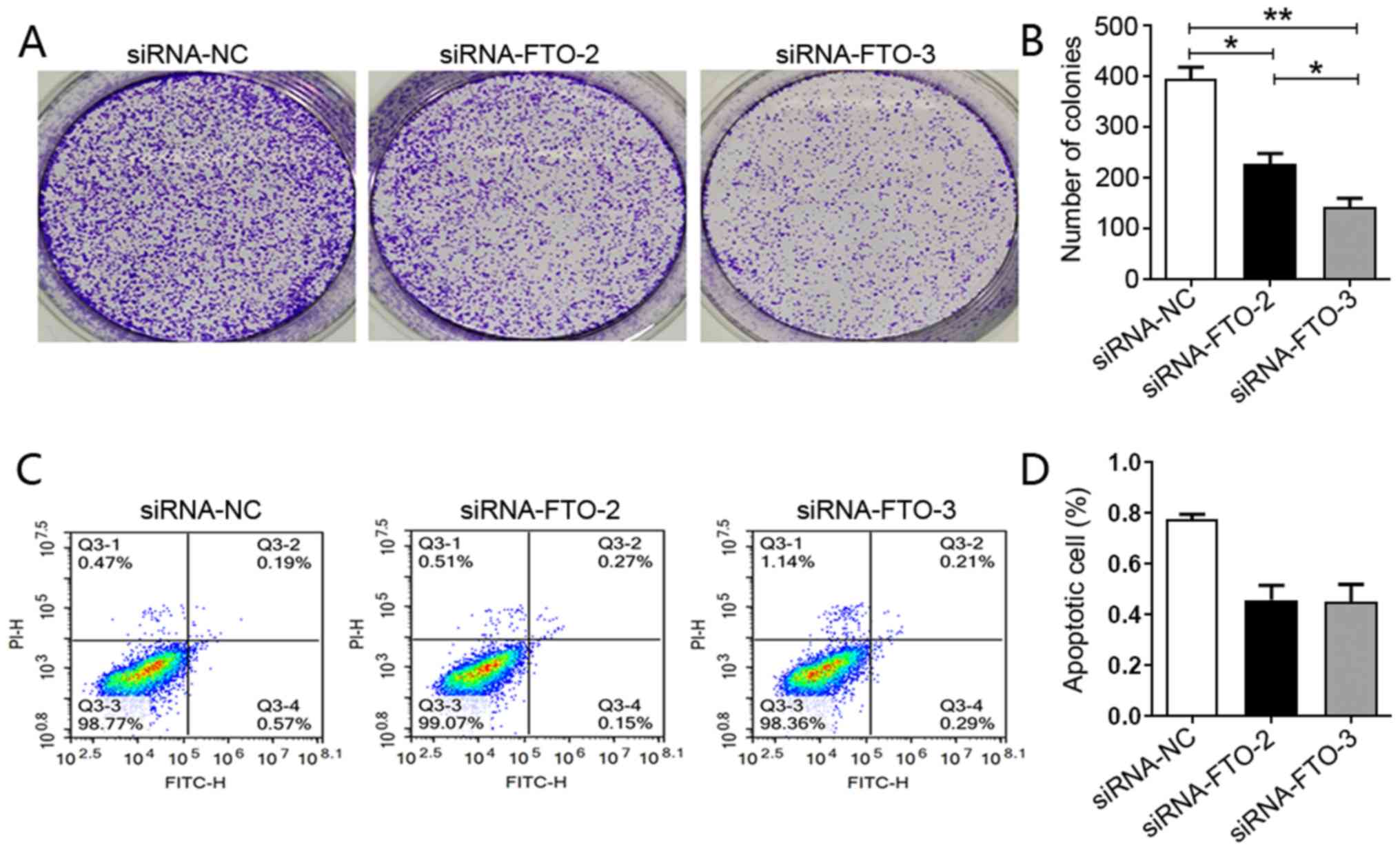
To further explore the role of this gene in liver
cancer carcinogenesis, a colony formation assay was performed and
it was observed that downregulation of FTO expression impaired the
ability of HepG2 cells to form colonies (Fig. 2A and B). To establish the mechanism
by which loss of FTO expression impaired these processes, an
apoptosis assay was performed. Although transfection with siRNA-FTO
appeared to decrease apoptosis, there was no significant difference
in apoptosis between siRNA-NC and siRNA-FTO HepG2 cells, suggesting
that FTO knockdown did not affect the apoptosis of HepG2 cells
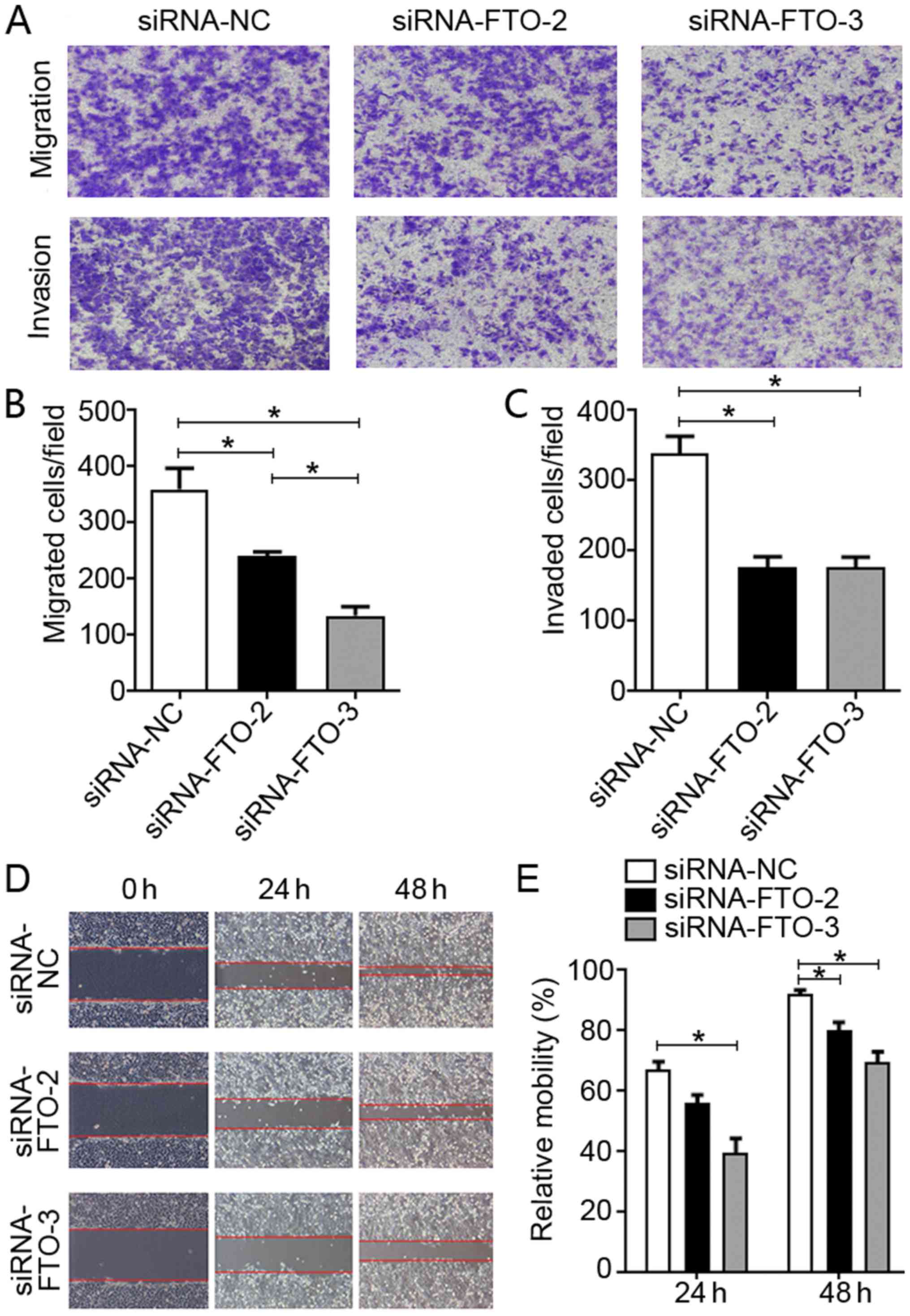
(Fig. 2C and D). Cell migration and
wound healing assays suggested that FTO promoted the invasion and
migration of liver cancer cells, as this was suppressed with FTO
knockdown (Fig. 3). Overall, these
findings indicated that FTO may promote the proliferation and
migration of liver cancer cells in vitro.
Prognostic value of FTO in patients
with liver cancer
The potential clinical prognostic value of FTO was
investigated. The expression levels of FTO in patients with liver
cancer at Zhejiang Provincial People's Hospital were analyzed using
IHC and the association between FTO expression and 5-year OS was
explored. The results indicated a significantly higher FTO
expression in liver cancer tissues compared with adjacent normal
tissues (Fig. 4A; data not shown).
Additionally, a significant positive association was identified
between FTO expression and Edmonson Grade (4) (χ2=10.523; P=0.001; Table I), which was an established
prognostic indicator for liver cancer in multivariate analysis
(coefficient, 0.990; P=0.006; Table
II). The Kaplan-Meier analysis suggested that reduced FTO
expression was indicative of a good prognosis. Although not
statistically significant, a 68% 5-year OS was associated with low
FTO expression compared with a 48% 5-year OS for high FTO
expression (P=0.077; Fig. 4B).
 | Table I.FTO expression in liver cancer
tissues. |
Table I.
FTO expression in liver cancer
tissues.
|
|
| FTO expression,
n |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical
parameters | Total no. | Low | High | χ2 | P-value |
|---|
| Age, years |
|
|
| 0.190 | 0.663 |
|
<55 | 128 | 81 | 47 |
|
|
| ≥55 | 202 | 123 | 79 |
|
|
| Sex |
|
|
| 0.456 | 0.500 |
| Male | 268 | 168 | 100 |
|
|
|
Female | 62 | 36 | 26 |
|
|
| Size, cm |
|
|
| 1.328 | 0.249 |
| ≤5 | 192 | 123 | 69 |
|
|
|
>5 | 130 | 75 | 55 |
|
|
| NA | 8 |
|
|
|
|
| Tumour number |
|
|
| 0.730 | 0.393 |
|
Single | 270 | 164 | 106 |
|
|
|
Multiple | 60 | 40 | 20 |
|
|
| Edmondson
grade |
|
|
| 10.523 | 0.001a |
|
I+II | 205 | 139 | 66 |
|
|
|
III | 119 | 59 | 60 |
|
|
| NA | 6 |
|
|
|
|
| Metastasis |
|
|
| 0.525 | 0.469 |
| M0 | 297 | 186 | 111 |
|
|
| M1 | 27 | 15 | 12 |
|
|
| NA | 6 |
|
|
|
|
| Microvascular
invasion |
|
|
| 3.449 | 0.063 |
|
Absence | 124 | 83 | 41 |
|
|
|
Presence | 121 | 67 | 54 |
|
|
| NA | 85 |
|
|
|
|
| HBs antigen |
|
|
| 1.135 | 0.287 |
|
Negtive | 63 | 35 | 28 |
|
|
|
Positive | 261 | 164 | 97 |
|
|
| NA | 6 |
|
|
|
|
| Cirrhosis |
|
|
| 0.231 | 0.631 |
|
Negative | 110 | 66 | 44 |
|
|
|
Positive | 220 | 138 | 82 |
|
|
| AFP, µg/l |
|
|
| 2.056 | 0.152 |
|
<50 | 145 | 92 | 53 |
|
|
|
≥50 | 124 | 68 | 56 |
|
|
| NA | 61 |
|
|
|
|
 | Table II.Univariate and multivariate Cox
regression analyses for the clinicopathological parameters in
patients with liver cancer. |
Table II.
Univariate and multivariate Cox
regression analyses for the clinicopathological parameters in
patients with liver cancer.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Parameters | No. | Coefficient | HR | 95% CI | P-value | Coefficient | HR | 95% CI | P-value |
|---|
| Age (<55/≥55
years) | 128/202 | −0.427 | 0.653 | 0.412–1.034 | 0.069 | −0.275 | 0.759 | 0.381–1.514 | 0.435 |
| Sex
(male/female) | 268/62 | 0.414 | 1.512 | 0.888–2.577 | 0.128 | −0.194 | 0.823 | 0.384–1.766 | 0.618 |
| Tumor size
(≤50/>50 mm) | 192/130 | 0.714 | 2.042 | 1.283–3.248 | 0.003a | 0.448 | 1.565 | 0.729–3.356 | 0.250 |
| Tumor number
(single/multiple) | 270/60 | 0.160 | 1.173 | 0.644–2.138 | 0.601 | 0.860 | 2.364 | 0.982–5.692 | 0.055 |
| Edmondson grade
(I+II/III) | 205/119 | 1.018 | 2.769 | 1.733–4.423 | 0.000a | 0.990 | 2.691 | 1.319–5.490 | 0.006a |
| Metastasis
(M0/M1) | 297/27 | 1.402 | 4.063 | 2.173–7.596 | 0.000a | 1.424 | 4.153 | 1.481–9.625 | 0.007a |
| Microvascular
invasion (−/+) | 124/121 | 0.637 | 1.891 | 1.136–3.148 | 0.041a | −0.170 | 0.844 | 0.382–1.862 | 0.674 |
| HBs antigen
(−/+) | 63/261 | 0.125 | 1.133 | 0.633–2.030 | 0.674 | −0.173 | 0.841 | 0.267–2.652 | 0.768 |
| Cirrhosis
(−/+) | 110/220 | 0.167 | 1.182 | 0.717–1.948 | 0.513 | 0.781 | 2.183 | 0.881–5.406 | 0.092 |
| AFP (<50/≥50
µg/l) | 145/124 | 0.837 | 2.310 | 1.315–4.058 | 0.004a | 0.504 | 1.655 | 0.801–3.418 | 0.174 |
| FTO (−/+) | 185/124 | 0.412 | 1.150 | 0.950–2.399 | 0.081 | 0.451 | 1.570 | 0785–3.141 | 0.202 |
Discussion
The present study reports a critical role of FTO in
liver cancer tumorigenesis. The present analyses indicated that
upregulation of FTO expression was frequently observed in liver
cancer tissues. Knockdown of FTO significantly suppressed
proliferation and migration of cultured liver cancer cells in
vitro. Additionally, FTO knockdown led to a significant
elevation of m6A methylation, suggesting that FTO-mediated m6A
demethylation may contribute to liver cancer. However, the
mechanism of this process remains unclear and merits further
investigation. The IHC analysis revealed that FTO expression was
markedly elevated in liver cancer tissues compared with adjacent
normal tissues. Furthermore, a positive association between FTO
expression and Edmonson Grade was observed, suggesting that FTO may
be a potential prognostic indicator for liver cancer. The
Kaplan-Meier analysis suggested that low FTO expression was
indicative of good prognosis.
RNA methylation has recently emerged as an important
modulator of tumorigenesis. m6A methylation is one of the most
common modifications found in eukaryotic mRNA and is a critical
event in RNA metabolism (26). It is
becoming increasingly evident that the ‘writers’, ‘erasers’ and
‘readers’ of m6A serve critical roles in tumorigenesis (27). FTO has been identified as a m6A
‘eraser’ and is associated not only with increased body mass and
obesity (28), but also with
carcinogenesis (29). The
association between FTO single nucleotide polymorphisms and
tumorigenesis has been previously investigated in prostate,
pancreatic, breast and colorectal cancer (29,30).
However, the m6A demethylase role of FTO in cancer development has
only recently emerged. It has been previously demonstrated that FTO
expression is enriched in the tumor area and that it promotes
proliferation, migration and lymph node metastasis in gastric
cancer (19). In addition, Li et
al (31) revealed that FTO
expression is associated with an overall decrease in cancer
survival. It has also been reported that FTO significantly enhances
proliferation while inhibiting apoptosis in lung cancer cell lines
(32).
Mechanistically, FTO has been demonstrated to
suppress the m6A methylation of ubiquitin specific peptidase 7
while increasing mRNA stability via mRNA demethylation (33). These results indicate that FTO may
promote oncogenesis and that suppressing its expression may offer a
beneficial therapeutic strategy for various types of cancer. The
results of the present study revealed that FTO may promote
proliferation and migration of the liver cancer HepG2 cell line
in vitro. However, the wound healing assay was inconclusive
as it could not exclude the possibility that the suppressed
migration observed upon FTO knockdown was not caused by a reduction
in cell proliferation. Results from the Transwell assay supported
the aforementioned observation that reduced FTO expression may
inhibit invasion and migration of HepG2 cells. Although FTO seemed
to promote proliferation of HepG2 cells, it was also observed that
this does not significantly affect apoptosis. Huang et al
(33) demonstrated that FTO
depletion decreases the expression levels of the mitotic checkpoint
complex and G2/M regulators in mouse GC-1 cells. Wu et al
(34) observed that FTO knockdown
markedly decreases the expression levels of cyclin A2 and CDK2,
both crucial cell cycle regulators, leading to delayed entry of
methylene diphenyl diisocyanate-induced cells into G2 phase.
However, whether FTO-knockdown affects the cell cycle of HepG2
cells is unclear, and the mechanism of FTO promoting proliferation
of HepG2 cells is likely more complex than just through cell cycle
arrest. Therefore, these mechanisms need to be further clarified.
By suggesting a potential role of FTO in liver cancer, the results
of the present study strengthened earlier reports of a role of FTO
in oncogenesis. However, further research is required to elucidate
the mechanism by which FTO-mediated modulation of m6A demethylation
affects liver cancer oncogenesis.
The role served by m6A methylation in the
development of liver cancer has been previously explored. It has
been reported that the methyltransferase METTL3 promotes
tumorigenicity and metastasis of liver cancer in vitro and
in vivo by suppressing the expression of suppressor of
cytokine signaling 2 via an m6A-YTHDF2-dependent mechanism
(22). The m6A methyltransferase
WTAP has also been reported to serve a crucial role in the
development of liver cancer via the HuR-ETS1-p21/p27 axis (23). Although more evidence is required to
support a role of FTO in liver cancer, it has been reported that
FTO suppresses the proliferation and migration of intrahepatic
cholangiocarcinoma cells by impairing the mRNA stability of the TEA
domain transcription factor 2 oncogene (35). However, data from the present study
revealed that FTO could promote the progression of liver cancer
while downregulating m6A RNA methylation levels.
In conclusion, the present study revealed that FTO
may contribute to liver cancer oncogenesis via the downregulation
of m6A RNA methylation levels. Further investigations are required
to better understand the association between the development of
liver cancer and m6A methylation. The present study reported FTO as
a potential prognostic indicator for liver cancer and a potential
novel therapeutic target for the management of liver cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Project of
Application on Public Welfare Technology in Zhejiang Province
(grant no. LGF18H160022), the Joint Fund of Zhejiang Provincial
Natural Science Foundation (grant no. LYY18H310002) and the
Zhejiang Medical Technology Plan Project (grant no. 2017ZD003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZY, SW and ZX designed the study. ZY, SW, WC, XZ,
JC, JJ, MW and LZ performed the data collection and collation. All
the authors were involved in the analysis and interpretation of
data. ZY wrote the paper, with the help of the co-authors. SW, WC
and XZ reviewed and revised the manuscript. All authors agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Zhejiang Provincial People's Hospital (Hangzhou,
China). Written informed consent was obtained from all
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI
|
|
2
|
Akinyemiju T, Abera S, Ahmed M, Alam N,
Alemayohu MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y,
Artaman A, et al Global Burden of Disease Liver Cancer
Collaboration, : The burden of primary liver cancer and underlying
etiologies from 1990 to 2015 at the global, regional, and national
level: Results from the global burden of Disease Study 2015. JAMA
Oncol. 3:1683–1691. 2017.PubMed/NCBI
|
|
3
|
Njei B, Rotman Y, Ditah I and Lim JK:
Emerging trends in hepatocellular carcinoma incidence and
mortality. Hepatology. 61:191–199. 2015.PubMed/NCBI
|
|
4
|
Wang H, Lu Z and Zhao X: Tumorigenesis,
diagnosis, and therapeutic potential of exosomes in liver cancer. J
Hematol Oncol. 12:1332019.PubMed/NCBI
|
|
5
|
Chok KS, Ng KK, Poon RT, Lo CM and Fan ST:
Impact of postoperative complications on long-term outcome of
curative resection for hepatocellular carcinoma. Br J Surg.
96:81–87. 2009.PubMed/NCBI
|
|
6
|
Cha CH, Ruo L, Fong Y, Jarnagin WR, Shia
J, Blumgart LH and DeMatteo RP: Resection of hepatocellular
carcinoma in patients otherwise eligible for transplantation. Ann
Surg. 238:315–321, discussion 321–323. 2003.PubMed/NCBI
|
|
7
|
Riaz A, Miller FH, Kulik LM, Nikolaidis P,
Yaghmai V, Lewandowski RJ, Mulcahy MF, Ryu RK, Sato KT, Gupta R, et
al: Imaging response in the primary index lesion and clinical
outcomes following transarterial locoregional therapy for
hepatocellular carcinoma. JAMA. 303:1062–1069. 2010.PubMed/NCBI
|
|
8
|
Riaz A, Ryu RK, Kulik LM, Mulcahy MF,
Lewandowski RJ, Minocha J, Ibrahim SM, Sato KT, Baker T, Miller FH,
et al: Alpha-fetoprotein response after locoregional therapy for
hepatocellular carcinoma: Oncologic marker of radiologic response,
progression, and survival. J Clin Oncol. 27:5734–5742.
2009.PubMed/NCBI
|
|
9
|
Hawkins MA and Dawson LA: Radiation
therapy for hepatocellular carcinoma: From palliation to cure.
Cancer. 106:1653–1663. 2006.PubMed/NCBI
|
|
10
|
Palmer DH: Sorafenib in advanced
hepatocellular carcinoma. N Engl J Med. 359:2498, author reply
2498–2499. 2008.
|
|
11
|
El-Khoueiry AB, Sangro B, Yau T, Crocenzi
TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH III, et
al: Nivolumab in patients with advanced hepatocellular carcinoma
(CheckMate 040): An open-label, non-comparative, phase 1/2 dose
escalation and expansion trial. Lancet. 389:2492–2502.
2017.PubMed/NCBI
|
|
12
|
Voutsadakis IA: PD-1 inhibitors
monotherapy in hepatocellular carcinoma: Meta-analysis and
systematic review. Hepatobiliary Pancreat Dis Int. 18:505–510.
2019.PubMed/NCBI
|
|
13
|
Machnicka MA, Milanowska K, Osman Oglou O,
Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S,
Dunin-Horkawicz S, Rother KM, et al: MODOMICS: A database of RNA
modification pathways - 2013 update. Nucleic Acids Res.
41:D262–D267. 2013.PubMed/NCBI
|
|
14
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012.PubMed/NCBI
|
|
15
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014.PubMed/NCBI
|
|
16
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG, et al: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011.PubMed/NCBI
|
|
17
|
Bartosovic M, Molares HC, Gregorova P,
Hrossova D, Kudla G and Vanacova S: N6-methyladenosine demethylase
FTO targets pre-mRNAs and regulates alternative splicing and 3-end
processing. Nucleic Acids Res. 45:11356–11370. 2017.PubMed/NCBI
|
|
18
|
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C,
Huang H, Nachtergaele S, Dong L, Hu C, et al: FTO Plays an
Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine
RNA Demethylase. Cancer Cell. 31:127–141. 2017.PubMed/NCBI
|
|
19
|
Xu D, Shao W, Jiang Y, Wang X, Liu Y and
Liu X: FTO expression is associated with the occurrence of gastric
cancer and prognosis. Oncol Rep. 38:2285–2292. 2017.PubMed/NCBI
|
|
20
|
Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R,
Wang YY and Zhe H: FTO regulates the chemo-radiotherapy resistance
of cervical squamous cell carcinoma (CSCC) by targeting β-catenin
through mRNA demethylation. Mol Carcinog. 57:590–597.
2018.PubMed/NCBI
|
|
21
|
Liu Y, Wang R, Zhang L, Li J, Lou K and
Shi B: The lipid metabolism gene FTO influences breast cancer cell
energy metabolism via the PI3K/AKT signaling pathway. Oncol Lett.
13:4685–4690. 2017.PubMed/NCBI
|
|
22
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018.PubMed/NCBI
|
|
23
|
Chen Y, Peng C, Chen J, Chen D, Yang B, He
B, Hu W, Zhang Y, Liu H, Dai L, et al: WTAP facilitates progression
of hepatocellular carcinoma via m6A-HuR-dependent epigenetic
silencing of ETS1. Mol Cancer. 18:1272019.PubMed/NCBI
|
|
24
|
Wang W, Liu L, Zhou Y, Ye Q, Yang X, Jiang
J, Ye Z, Gao F, Tan X, Zhang G, et al: Hydroxychloroquine enhances
the antitumor effects of BC001 in gastric cancer. Int J Oncol.
55:405–414. 2019.PubMed/NCBI
|
|
25
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017.PubMed/NCBI
|
|
26
|
Vu LP, Cheng Y and Kharas MG: The Biology
of m6A RNA Methylation in Normal and Malignant Hematopoiesis.
Cancer Discov. 9:25–33. 2019.PubMed/NCBI
|
|
27
|
Pan Y, Ma P, Liu Y, Li W and Shu Y:
Multiple functions of m6A RNA methylation in cancer. J Hematol
Oncol. 11:482018.PubMed/NCBI
|
|
28
|
Loos RJ and Bouchard C: FTO: The first
gene contributing to common forms of human obesity. Obes Rev.
9:246–250. 2008.PubMed/NCBI
|
|
29
|
Nock NL, Plummer SJ, Thompson CL, Casey G
and Li L: FTO polymorphisms are associated with adult body mass
index (BMI) and colorectal adenomas in African-Americans.
Carcinogenesis. 32:748–756. 2011.PubMed/NCBI
|
|
30
|
Jafari Nedooshan J, Kargar S, Neamatzadeh
H, Haghighi F, Dehghani Mohammad Abadi R and Seddighi N: Lack of
association of the fat mass and obesity associated (FTO) gene
rs9939609 polymorphism with breast cancer risk: A systematic review
and meta-analysis based on case - control studies. Asian Pac J
Cancer Prev. 18:1031–1037. 2017.PubMed/NCBI
|
|
31
|
Li Y, Zheng D, Wang F, Xu Y, Yu H and
Zhang H: Expression of demethylase genes, FTO and ALKBH1, is
associated with prognosis of gastric cancer. Dig Dis Sci.
64:1503–1513. 2019.PubMed/NCBI
|
|
32
|
Li J, Han Y, Zhang H, Qian Z, Jia W, Gao
Y, Zheng H and Li B: The m6A demethylase FTO promotes the growth of
lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem
Biophys Res Commun. 512:479–485. 2019.PubMed/NCBI
|
|
33
|
Huang T, Gao Q, Feng T, Zheng Y, Guo J and
Zeng W: FTO knockout causes chromosome instability and G2/M arrest
in mouse GC-1 cells. Front Genet. 9:7322019.PubMed/NCBI
|
|
34
|
Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q,
Liu Q, Cai M, Wang F, Wang Y, et al: FTO regulates adipogenesis by
controlling cell cycle progression via m6A-YTHDF2 dependent
mechanism. Biochim Biophys Acta Mol Cell Biol Lipids.
1863:1323–1330. 2018.PubMed/NCBI
|
|
35
|
Rong ZX, Li Z, He JJ, Liu LY, Ren XX, Gao
J, Mu Y, Guan YD, Duan YM, Zhang XP, et al: Downregulation of fat
mass and obesity associated (FTO) promotes the progression of
intrahepatic cholangiocarcinoma. Front Oncol. 9:3692019.PubMed/NCBI
|