Introduction
Numerous guidelines recommend tumor screening via
microsatellite instability (MSI) analysis in all patients with
colorectal cancer (CRC), as MSI status has substantial implications
in CRC diagnosis (1), prognosis
(2) and responses to fluorouracil
(3). Fast progress in cancer
immunotherapy also confirmed the good response of MSI tumors to
checkpoint inhibitor blockade (4,5).
Recently, the US Food and Drug Administration (FDA) approved
pembrolizumab as the first drug for any solid tumor with MSI-high
status (https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm);
the clinical detection of MSI status is in great demand for
patients with CRC or other cancer types.
MSI status is a molecular fingerprint for DNA
mismatch repair (MMR) system deficiency. Dysfunctional MMR genes
(MLH1, MSH2, MSH6 and PMS2) may introduce
length-altering mutations in microsatellites (MS), which are short
tandem repeat sequences dispersed throughout the human genome and
could be used as deficient MMR (dMMR) markers (6,7). Two of
the most common methods for MSI detection have been developed and
are still considered the gold standards. MSI-PCR assesses length
variability at several standardized MS loci, while
immunohistochemical (IHC) staining inspects the protein expression
of MMR genes. High frequency MSI (MSI-H) status is determined by
the fraction of unstable MS markers (MSI-PCR) or the loss of one or
more MMR proteins (IHC). However, considering the various outcomes
of dMMR, these traditional methods may have limitations. The
indirect IHC method cannot detect all abnormalities of MMR genes
and depends heavily on specialized techniques (8). A recent study demonstrated that
5-marker MSI-PCR had inferior sensitivity in 91 prostate tumors
among other methods with more markers (9).
With the drastic development of next-generation
sequencing (NGS) technologies, the clinical management of cancer
patients has been facilitated in a number of ways (10,11). A
prominent advantage of NGS detection is that sufficient genetic
information can be obtained with much lower costs than
low-throughput methods by well-designed tumor sequencing.
Therefore, more MS loci could simultaneously be assessed by NGS
sequencing to provide new possibilities to evaluate MSI status.
Recently, several computational methods for MSI detection, which
can be integrated into existing NGS pipelines, have been designed
and proposed to provide comprehensive information on tumor
genomics. By comparing tumor DNA sequences to normal DNA sequences,
such methods can infer MSI status according to either tumor
mutation burden (12–14) or read count distribution. Two typical
approaches based on read count distribution are mSINGS (15) and MANTIS (16). These methods evaluate each MS locus
within a target marker set and report the MSI status by specifying
certain cut-offs. However, these approaches differ in their
determination of tumor instability. mSINGS compares tumor-only
samples to a pre-constructed baseline-control and evaluates tumor
instability by the fraction of unstable MS loci within a target
set. By contrast, MANTIS compares each target locus in both tumor
and matched healthy samples and calculates tumor instability as the
averaged stepwise difference across all target loci. These two
methods might have disadvantages due to their algorithms. mSINGS
expends more time to analyze one sample for the pileup step and
statistical test for every locus. However, since MANTIS does not
employ a statistical test to calculate the MSI score, this method
may be weak at exploiting informative MS loci and may lead to
biased results.
Herein, a robust and rapid algorithm called
NovoPM-MSI was developed by using a targeted tumor-sequencing panel
to reliably assess the MSI status in CRC patients. The read count
distribution from paired tumor-healthy samples was used to develop
an improved strategy in order to determine tumor instability by
examining target MS loci. We also validated this algorithm against
conventional MSI-PCR in 113 CRC cases. Finally, we demonstrated the
high performance of NovoPM-MSI over mSINGS and MANTIS in MSI status
detection and runtime efficiency.
Materials and methods
Sample source and next-generation
sequencing
To develop the NovoPM-MSI algorithm, retrospective
CRC specimens from 113 patients were utilized. The study was
conducted in accordance with the Declaration of Helsinki and the
protocol was approved by the Ethics Committee of Shengjing Hospital
of China Medical University. All the recruited patients had signed
informed consent for their samples to be used in the study and all
clinical data and specimens were received anonymously. These
specimens were previously analyzed in our CAP-certified laboratory
(Tianjin Novogene Med LAB). All cases were tumor tissue DNA
extracted from formalin-fixed paraffin-embedded (FFPE) specimens
with matched white blood cell (WBC) DNA. These paired cases were
sequenced using a targeted gene panel, NovoPM, between January 2016
and October 2017. This panel covers all coding and 21 non-coding
regions of the 548 genes associated with multiple tumor types. In
the sequencing assay, DNA from tumor and matched WBCs was used to
construct sequencing libraries by a hybrid-capture selection
method. These libraries were sequenced with a highly uniform depth
(targeting >1,000X coverage by non-PCR duplicated read pairs) on
the Illumina Hi-Seq X Ten platform as paired-end reads. The
protocols and reagents have been optimized to ensure uniform
coverage and robust performance for a wide variety of
specimens.
Selection of microsatellite marker
loci
To identify MS repeats across the human genome with
high confidence, a reference FASTA file (hg19/GRCh37) was first
scanned using an in-house pipeline. A qualified MS locus was
defined as follows: i) The base number within a microsatellite
locus should range from 10 to 100; and ii) the minimum length of
each motif should be 5. The total number of MS loci across the
reference genome was 2,946,833. The mononucleotide repeats covered
by the NovoPM panel were extracted from the MS loci in order to
identify MS markers for MSI detection among those repeats. Only
mononucleotide repeats were selected as MS markers, since these
sequences are more sensitive in traditional MSI detection.
Workflow of NovoPM-MSI
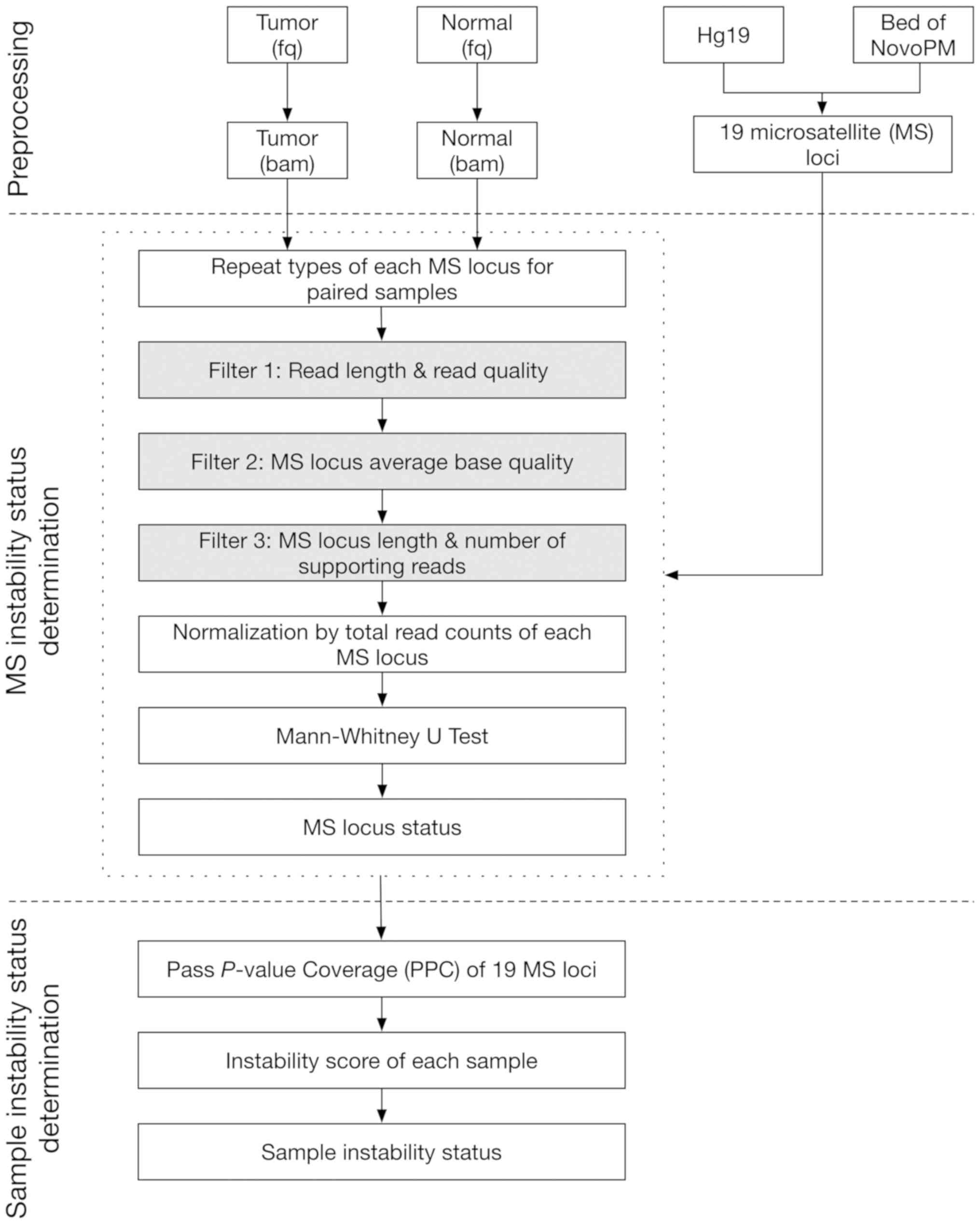
A brief illustration of the NovoPM-MSI detection
workflow is presented in Fig. 1.
During the data pre-processing stage, the sequencing reads obtained
after the in-house QC pipeline assessment were aligned using the
Burrows-Wheeler Aligner (version 0.7.8) (17) against the reference genome
(hg19/GRCh37) and indexed by SAMtools (version 0.1.19) (18). Duplicate reads were removed using
Picard Tools (version 1.96) (http://broadinstitute.github.io/picard/) from BAM
files. A reference genome and target loci were required as well as
paired tumor-normal BAM files in NovoPM-MSI.
Low-quality reads were removed step by step if they
met the following conditions: i) reads from paired tumor-normal BAM
files that were either too short (<35 bp) or had low average
base quality (<25.0); ii) reads covering the MS locus that had
delimited length and lower average base quality (<30.0); and
iii) reads with no clipped parts that were shorter than a second
length threshold (<35). At each MS locus, minimum coverage was
required in both tumor and normal samples (>30X) and each repeat
type for an MS locus should have at least three qualified reads to
support that type. A repeat length that was too long (>3× the
standard deviation from mean) was defined as an outlier and was not
considered in the subsequent steps.
Qualified supporting reads were then normalized by
the average sequencing depth for the tumor and normal samples,
prior to determining the instability for each MS locus. In
NovoPM-MSI, the length distribution at each MS locus was
characterized by the normalized supporting reads in tumor and
normal samples. For example, for one MS locus with X number of
repeat types, two vectors were obtained by counting the supporting
reads for each repeat type in the normal and tumor samples, given
as Ni and Ti (i=1, 2, 3, …, X), respectively. This locus was
determined as unstable if the two vectors were significantly
different by using the non-parametric Mann-Whitney U test
(P<0.05).
Finally, an instability score was calculated by the
fraction of unstable loci within the target MS markers. An
empirical cut-off was set as 0.2 based on the guidelines for
defining MSI positivity (15,19).
MSI status determination by PCR
All the paired tumor-normal CRC samples were tested
by the MSI-PCR method to obtain the standard MSI status. The PCR
panel included six mononucleotide repeat markers (NR21, BAT26,
NR27, BAT25, NR24 and MONO27) for MSI testing and three other
markers (Penta C, Penta D and Amelogenin) for the sample
contamination control. A fluorescence profile of amplified
microsatellite DNA from paired samples was produced by capillary
fluorescence electrophoresis (ABI 3730×l Genetic Analyzer, Applied
Biosystems). If fluorescence peaks were present at any marker in
the tumor sample but absent in the normal sample, then that marker
was determined as unstable. MSI-H samples had two or more unstable
markers, while MSI-L and MSS samples had one or zero unstable
markers. Thus, the cut-off for the MSI-PCR method was 33.3%
(2/6).
MSI detection by mSINGS and
MANTIS
For mSINGS, a baseline control was first constructed
by using 26 WBC samples from randomly selected MSI-negative cases.
All the tumor samples were then tested by mSINGS, with defaults.
The software determined an unstable locus by using the Z-test if
the number of the types of repeat lengths was larger than
(reference mean + 3*SD), where the reference mean and SD were from
the baseline-control. The software reported the percentage of
unstable loci, according to which MSI-H samples were determined if
the percentage was higher than 20% (15).
Additionally, all the paired samples were tested by
MANTIS, which required paired tumor-healthy samples and a target
marker set. The input files for MANTIS were the same as those
required for NovoPM-MSI. MANTIS calculates the absolute value of
the stepwise difference between tumor and healthy samples at each
target MS locus using supporting read counts. Then, an average
distance value across all the targeted loci was obtained to present
the tumor instability. The samples were considered MSI-H when the
average distance value was >0.4 (16).
Performance comparison among three
NGS-based tools
Three computational tools, NovoPM-MSI, MANTIS and
mSINGS, were tested, with all samples having a standard MSI status
assigned by MSI-PCR. Four performance indices, sensitivity (SN),
specificity (SP), accuracy (ACC) and Mathew's correlation
coefficient (MCC), were used to evaluate the detection performance
among these tools. All the indices were calculated by
convention.
Runtime efficiency was also tested by evaluating the
processing time for each sample. Runtime data were compared by
tools using pair-wise comparisons and the significance was adjusted
by Bonferroni's correction (Welch's t-test, adjusted α=0.017).
Results
Target MS marker of NovoPM-MSI
A total of 19 MS loci of mononucleotide repeats
located in non-coding regions were finally selected as the target
marker set. These sensitive loci were sequenced after targeted gene
enrichment during the NovoPM assay. Detailed information regarding
these targets is listed in Table
I.
 | Table I.Microsatellite markers used in NovoMSI
detection. |
Table I.
Microsatellite markers used in NovoMSI
detection.
| Chr | Start | End | Function | Gene | Type |
|---|
| 1 | 161309452 | 161309480 | Intronic | SDHC | (T)28 |
| 3 | 12649448 | 12649472 | Intronic | RAF1 | (A)24 |
| 3 | 37067099 | 37067120 | Intronic | MLH1 | (T)21 |
| 3 | 185802071 | 185802094 | Intronic | ETV5 | (A)23 |
| 4 | 55598211 | 55598236 | Intronic | KIT | (T)25 |
| 6 | 135527343 | 135527365 | Intronic | MYB | (A)22 |
| 7 | 140498359 | 140498380 | Intronic | BRAF | (T)21 |
| 7 | 140508151 | 140508177 | Intronic | BRAF | (A)26 |
| 8 | 38303723 | 38303746 | Intronic | FGFR1 | (T)23 |
| 8 | 48732074 | 48732095 | Intronic | PRKDC | (A)21 |
| 9 | 5062500 | 5062531 | Intronic | JAK2 | (A)31 |
| 9 | 87357704 | 87357731 | Intronic | NTRK2 | (T)27 |
| 9 | 87479651 | 87479673 | Intronic | NTRK2 | (T)22 |
| 12 | 11962746 | 11962772 | ncRNA_intronic | ETV6 | (T)26 |
| 13 | 49039094 | 49039118 | Intronic | RB1 | (T)24 |
| 15 | 99192754 | 99192778 | ncRNA_exonic |
IRAIN/IGF1R | (T)24 |
| 17 | 29559061 | 29559087 | Intronic | NF1 | (T)26 |
| 21 | 42863078 | 42863102 | Intronic | TMPRSS2 | (T)24 |
| X | 123195593 | 123195618 | Intronic | STAG2 | (T)25 |
Validation of three computational
methods against conventional MSI-PCR
Among the 113 paired CRC tumor-normal cases, nine
samples were assigned MSI-H and 104 samples were assigned MSS
according to the gold standard of MSI-PCR. The fraction of MSI-H
cases in the present study collection was 7.9%, which was slightly
lower than the number of MSI-H cases in the larger CRC population
(15-20%) (20,21).
The initial cut-offs to determine the MSI-H status
for NovoPM-MSI, MANTIS and mSINGS were set at 0.2, 0.2 and 0.4,
respectively. The validation of these methods against the MSI-PCR
method is listed in Table II. All
three computational methods correctly detected eight positive cases
and one false negative case, which were not same for each tool,
thus reaching the same sensitivity of 88.9%. These analyses
differed in specificity with descending order of mSINGS (103/104,
99%), NovoPM-MSI (101/104, 97.1%) and MANTIS (90/104, 86.5%).
According to Matthew's correlation coefficient, a more balanced
measure, the performance of NovoPM-MSI (0.786) was slightly
inferior to that of mSINGS (0.879), with MANTIS (0.516) showing the
worst performance.
 | Table II.Performance comparison among NovoMSI,
MANTIS and mSINGS, using microsatellite instability-PCR results as
the gold standard. |
Table II.
Performance comparison among NovoMSI,
MANTIS and mSINGS, using microsatellite instability-PCR results as
the gold standard.
| Method | TP | TN | FP | FN | SN (%) | SP (%) | ACC (%) | MCC |
|---|
| NovoPM-MSI | 8 | 101 | 3 | 1 | 88.90 | 97.10 | 96.50 | 0.786 |
| MANTIS | 8 | 90 | 14 | 1 | 88.90 | 86.50 | 86.70 | 0.516 |
| mSINGS | 8 | 103 | 1 | 1 | 88.90 | 99.00 | 98.20 | 0.879 |
Detection comparison among NovoPM-MSI,
MANTIS and mSINGS
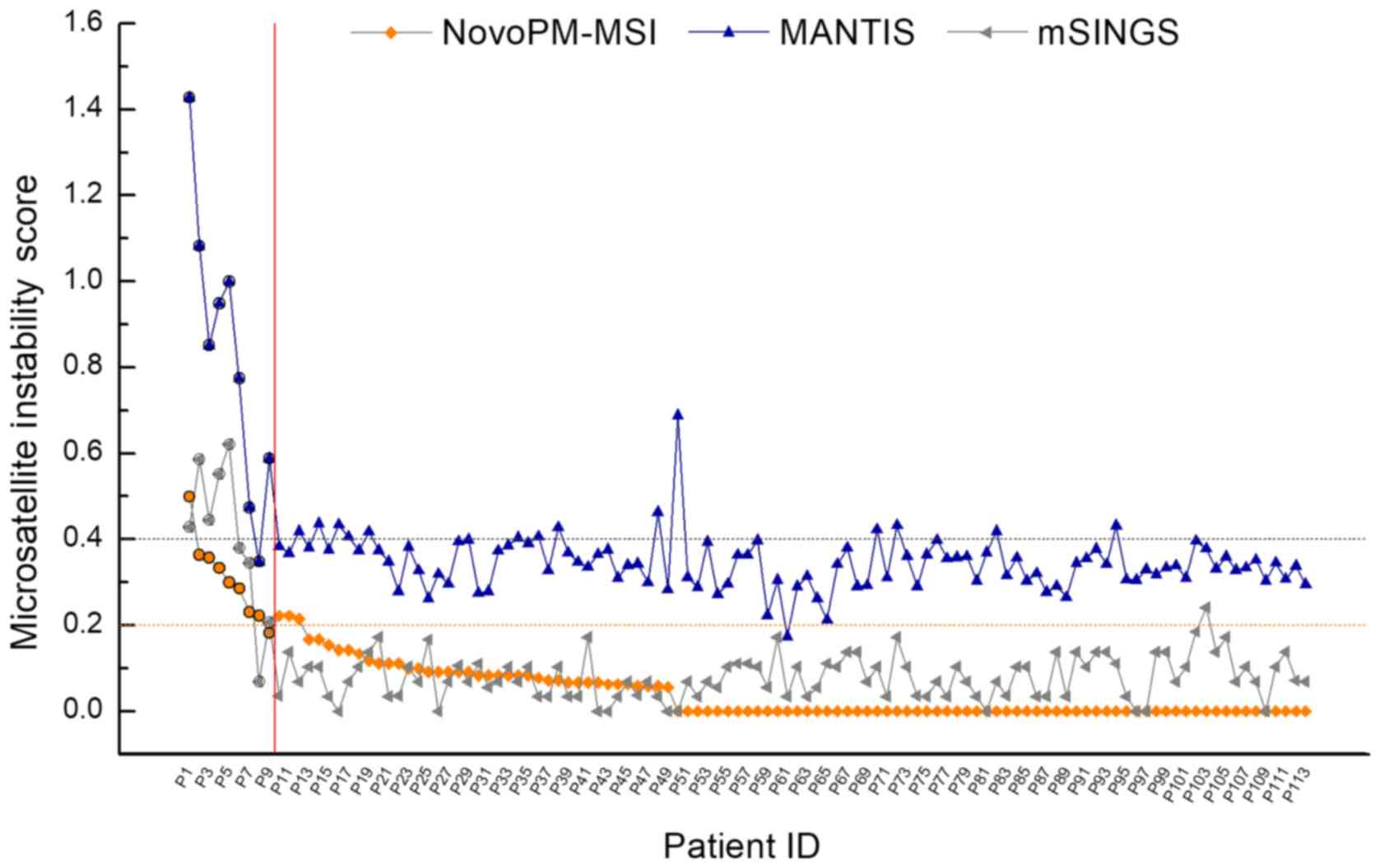
By plotting the instability score against samples,
the detection performance among these computational tools was
further assessed. NovoPM-MSI showed the least fluctuations within
either the MSI-H (Fig. 2, left side
of vertical line) or MSS category (Fig.
2, right side of vertical line). Although mSINGS had the
highest MCC (0.879, Table II), the
two misclassified samples were far from the default cut-off
(0.2).
MANTIS showed the worst performance, with an
accuracy of 86.7% (98/113) for the 14 false-positive samples that
were reported (default cut-off: 0.4) and an acute fluctuate
instability score produced.
In addition, the steady decrease in the instability
score reported by NovoPM-MSI across MSI-H and MSS samples suggested
an intermediate status for the MSI phenotype. We then modified a
calling strategy of NovoPM-MSI by setting an ambiguous interval for
the instability score. The small range was optimized by current
observations set from 0.17 to 0.23 to include four samples with
‘false’ result. The samples were explicitly assigned MSI-H or MSS
if the instability scores reported by NovoPM-MSI were >0.23 or
<0.17. The samples were assigned MSI-ambiguous if the
instability score fell into the ambiguous interval and these
samples were recommended for further MSI-PCR testing. By setting
the ambiguous interval, NovoPM-MSI reached 100% sensitivity and
specificity. However, such intervals could not be properly utilized
to improve the sensitivity and specificity of mSINGS.
Runtime comparison among NovoPM-MSI,
MANTIS and mSINGS
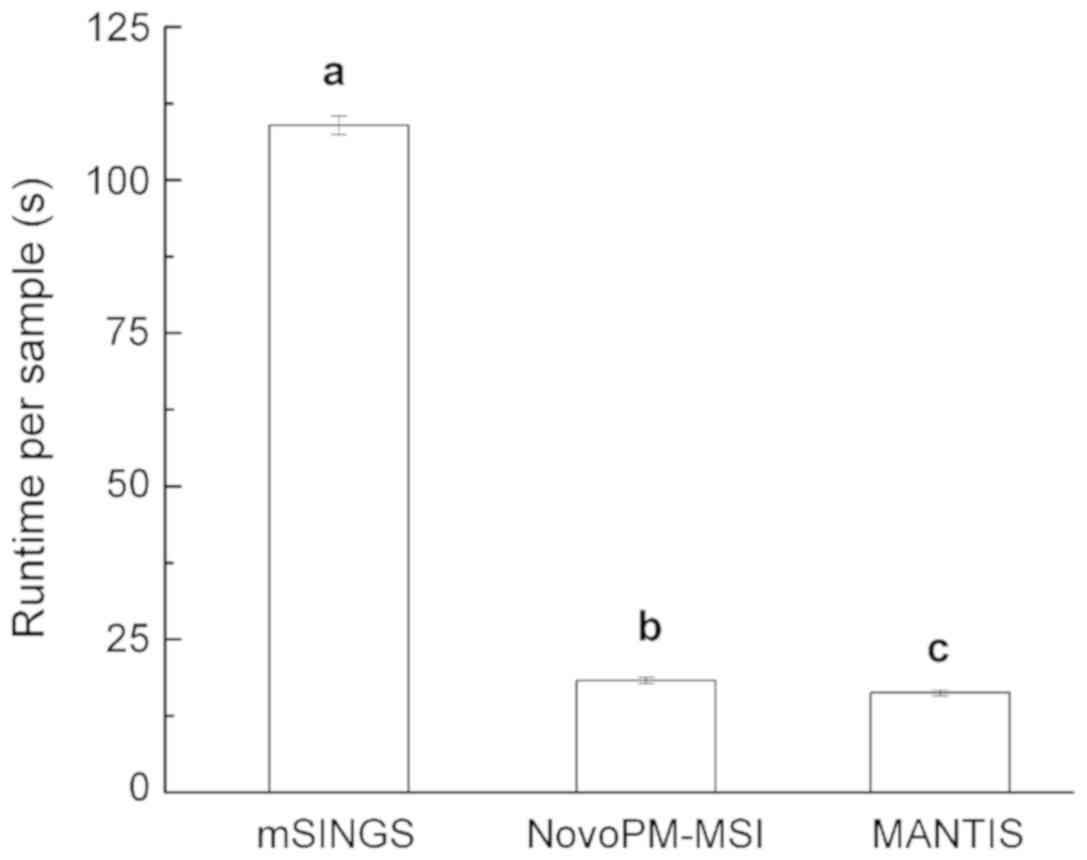
In addition to the accuracy of detection, the
runtime for each sample is also important when evaluating tool
performance. The ranking of runtime efficiency was MANTS (16.3
sec)>NovoPM-MSI (18.3 sec)>mSINGS (109.0 sec). All the
pair-wise comparisons were highly significant among the three tools
(Welch's t-test, P<0.01, Fig.
3).
Discussion
In the present study, we described an NGS-based
strategy, NovoPM-MSI, for MSI detection in paired tumor-healthy
samples. We selected 19 mononucleotide MS repeats as target loci
(Table I) since several studies have
shown that mononucleotide repeats are the most sensitive in MSI
detection (22–24). This new method evaluates each of the
target MS loci by comparing the length distribution between tumor
and normal samples and then calculates the percentage of unstable
MS loci and reports the instability score as well as the MSI status
according to empirical cut-offs. NovoPM-MSI has comparable
reliability to the gold standard MSI-PCR, with robust performance
(Table II).
We also compared NovoPM-MSI to two other NGS-based
MSI detection tools, mSINGS (15)
and MANTIS (16). All three tools
exploit information from the read count distribution but differ in
the instability evaluation. MANTIS assesses step-wise differences
between tumor and matched normal samples at the same MS locus and
calculates an average difference across all the MS loci to reflect
the general instability of a sample but not a single locus.
Notably, the aggregate averaged distance may be biased when using
relatively fewer markers. One advantage of MANTIS is that this
approach is less susceptible to sequencing errors or poorly
performing loci, but this advantage becomes increasingly apparent
when using more target loci, which may be limited in most gene
panels. In the present study, MANTIS produced the most
false-positive calls (14 in 104) and showed the most fluctuations
in the instability score (Table II
and Fig. 2). mSINGS utilizes
incidentally sequenced MS loci and evaluates the instability of
each MS locus by comparison with a reference distribution generated
from pooled normal samples. This approach assesses single locus
instability using the Z-score test and reports the percentage of
unstable loci as an instability score. mSINGS performed well for
the 113 cases in the present study, with both high sensitivity
(88.9%) and specificity (99%). However, its disadvantage in
runtime, which was nearly six times longer than that of MANTIS, was
also evident (Fig. 3). Additionally,
the two misclassified samples could not be saved through setting an
ambiguous interval. By contrast, NovoPM-MSI combines the advantages
of the above tools. First, this approach examined an intermediate
marker set (Table I), which was
customized for most gene panels. Second, NovoPM-MSI runs much
faster than mSINGS (18.3 vs. 109.0 sec) and has a runtime
comparable to that of MANTIS (16.3 sec). Finally, by setting an
ambiguous interval (0.17-0.23) from empirical observations, the
detection performance of NovoPM-MSI was greatly improved compared
with that of the other tools. In short, NovoPM-MSI efficiently
detected MSI status in a robust and rapid manner.
The advantages of NGS-based methods for MSI
detection over traditional methods are obvious. MSI status can be
simultaneously available with other genomic alterations, including
SNV, gene fusion and indels, by NGS pipelines, without dedicated
MSI detection. In addition, NGS-based methods enable the
examination of more MS loci than that of the MSI-PCR method,
providing a comprehensive assessment of MSI status. Although
considered the gold standard, MSI-PCR reportedly shows 97%
sensitivity and 95% specificity (25). In the present study, the three
NGS-based methods showed performances comparable to those of
conventional PCR (Table II). Recent
studies have suggested that the MSI-H phenotype should be
subdivided to guide important clinical applications (15,26–28). The
rough binary classification of MSI only provides qualitative
results and cannot meet this requirement. Alternatively, NGS-based
methods can fill the gap by reporting both qualitative status and
quantitative instability score. The NovoPM-MSI method reported in
the present study showed reasonable classification (Fig. 2).
The present study may have some limitations. The
NovoPM-MSI method may be limited by the small number of MSI-H cases
(9/113), primarily due to the low frequency of MSI-H phenotypes in
CRCs driven by MMR deficiency (~15%) (20,21). The
accuracy of NovoPM-MSI would be increased if more MSI-H cases were
accumulated, regardless of the one or two cut-offs used. We are now
endeavoring to collect more CRC cases with different MSI status to
build large validation cohort for NovoPM-MSI. In addition, using
white blood cells as a normal control is relatively easy, but in
most clinical settings, only tumor samples are available (e.g.,
retrospective FFPE without matched normal specimen). Under such
conditions, tumor-only methods, including mSINGS, are required. In
addition, MSI status is most closely studied in CRCs; it is found
to be present in endometrial, ovarian, cervical, gastric and
prostate cancers (27). The current
NovoPM-MSI method has been developed for CRCs, but its
applicability to other cancers may be tested in the future if cases
from more cancer types are accumulated.
In conclusion, findings of the present study show
that NovoPM-MSI can rapidly detect MSI status with high accuracy
and robustness compared to conventional MSI-PCR and other NGS
methods based on read count distributions. As the genetic
characteristics of tumors become increasingly more critical in the
clinical management of patients, we hope that the NovoPM-MSI method
will provide more valuable information in clinical service when
conducting tumor sequencing.
Acknowledgements
The authors would like to thank Ms Wanchun Zang, an
experienced analyst for clinical data (Beijing Novogene
Bioinformatics Technology Co. Ltd.), for her helpful suggestions on
the manuscript.
Funding
This work was supported by grants from the research
program of Novogene Bioinformatics Technology Co., Ltd. (grant no.
P101ZY17010010).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ, GS, LL, GC and XZ designed the study. LZ and GS
improved the algorithm, analyzed and interpreted all the data. LZ,
GS, LL and GC wrote the manuscript. YY contributed to sample
collection and project coordination. GC and XZ provided the
research materials, supervised the study and edited the manuscript.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shengjing Hospital of China Medical University.
Written informed consent was obtained from all the patients.
Patient consent for publication
Not applicable.
Competing interests
LZ, GS, LL, YY and GC are employees of Beijing
Novogene Bioinformatics Technology Co., Ltd. The authors declare
that they have no competing interests.
References
|
1
|
Zeinalian M, Hashemzadeh-Chaleshtori M,
Salehi R and Emami MH: Clinical aspects of microsatellite
instability testing in colorectal cancer. Adv Biomed Res. 7:282018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Popat S, Hubner R and Houlston RS:
Systematic review of microsatellite instability and colorectal
cancer prognosis. J Clin Oncol. 23:609–618. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ribic CM, Sargent DJ, Moore MJ, Thibodeau
SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R,
Shepherd LE, et al: Tumor microsatellite-instability status as a
predictor of benefit from fluorouracil-based adjuvant chemotherapy
for colon cancer. N Engl J Med. 349:247–257. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Weinberg BA, Xiu J, Hwang JJ, Shields AF,
Salem ME and Marshall JL: Immuno-oncology biomarkers for gastric
and gastroesophageal junction adenocarcinoma: Why PD-L1 testing may
not be enough. Oncologist. 23:1171–1177. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boland CR and Goel A: Microsatellite
instability in colorectal cancer. Gastroenterology.
138:2073–2087.e2073. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kelkar YD, Strubczewski N, Hile SE,
Chiaromonte F, Eckert KA and Makova KD: What is a microsatellite: A
computational and experimental definition based upon repeat
mutational behavior at A/T and GT/AC repeats. Genome Biol Evol.
2:620–635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Müller A, Giuffre G, Edmonston TB, Mathiak
M, Roggendorf B, Heinmöller E, Brodegger T, Tuccari G, Mangold E,
Buettner R, et al: Challenges and pitfalls in HNPCC screening by
microsatellite analysis and immunohistochemistry. J Mol Diagn.
6:308–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hempelmann JA, Lockwood CM, Konnick EQ,
Schweizer MT, Antonarakis ES, Lotan TL, Montgomery B, Nelson PS,
Klemfuss N, Salipante SJ and Pritchard CC: Microsatellite
instability in prostate cancer by PCR or next-generation
sequencing. J Immunother Cancer. 6:292018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roychowdhury S, Iyer MK, Robinson DR,
Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA,
Quist MJ, et al: Personalized oncology through integrative
high-throughput sequencing: A pilot study. Sci Transl Med.
3:111ra1212011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Uzilov AV, Ding W, Fink MY, Antipin Y,
Brohl AS, Davis C, Lau CY, Pandya C, Shah H, Kasai Y, et al:
Development and clinical application of an integrative genomic
approach to personalized cancer therapy. Genome Med. 8:622016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang MN, McPherson JR, Cutcutache I, Teh
BT, Tan P and Rozen SG: MSIseq: Software for assessing
microsatellite instability from catalogs of somatic mutations. Sci
Rep. 5:133212015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nowak JA, Yurgelun MB, Bruce JL,
Rojas-Rudilla V, Hall DL, Shivdasani P, Garcia EP, Agoston AT,
Srivastava A, Ogino S, et al: Detection of mismatch repair
deficiency and microsatellite instability in colorectal
adenocarcinoma by targeted next-generation sequencing. J Mol Diagn.
19:84–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stadler ZK, Battaglin F, Middha S,
Hechtman JF, Tran C, Cercek A, Yaeger R, Segal NH, Varghese AM,
Reidy-Lagunes DL, et al: Reliable detection of mismatch repair
deficiency in colorectal cancers using mutational load in
next-generation sequencing panels. J Clin Oncol. 34:2141–2147.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salipante SJ, Scroggins SM, Hampel HL,
Turner EH and Pritchard CC: Microsatellite instability detection by
next generation sequencing. Clin Chem. 60:1192–1199. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kautto EA, Bonneville R, Miya J, Yu L,
Krook MA, Reeser JW and Roychowdhury S: Performance evaluation for
rapid detection of pan-cancer microsatellite instability with
MANTIS. Oncotarget. 8:7452–7463. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R; 1000 Genome
Project Data Processing Subgroup, : The sequence alignment/map
format and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boland CR, Thibodeau SN, Hamilton SR,
Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA,
Fodde R, Ranzani GN and Srivastava S: A National Cancer Institute
Workshop on Microsatellite Instability for cancer detection and
familial predisposition: Development of international criteria for
the determination of microsatellite instability in colorectal
cancer. Cancer Res. 58:5248–5257. 1998.PubMed/NCBI
|
|
20
|
Grady WM and Carethers JM: Genomic and
epigenetic instability in colorectal cancer pathogenesis.
Gastroenterology. 135:1079–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vilar E and Gruber SB: Microsatellite
instability in colorectal cancer-the stable evidence. Nat Rev Clin
Oncol. 7:153–162. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bacher JW, Flanagan LA, Smalley RL, Nassif
NA, Burgart LJ, Halberg RB, Megid WM and Thibodeau SN: Development
of a fluorescent multiplex assay for detection of MSI-high tumors.
Dis Markers. 20:237–250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Middha S, Zhang L, Nafa K, Jayakumaran G,
Wong D, Kim HR, Sadowska J, Berger MF, Delair DF, Shia J, et al:
Reliable pan-cancer microsatellite instability assessment by using
targeted next-generation sequencing data. JCO Precis Oncol.
2017:2017.PubMed/NCBI
|
|
24
|
Umar A, Boland CR, Terdiman JP, Syngal S,
de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ,
Hamelin R, et al: Revised Bethesda guidelines for hereditary
nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite
instability. J Natl Cancer Inst. 96:261–268. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hempelmann JA, Scroggins SM, Pritchard CC
and Salipante SJ: MSIplus for integrated colorectal cancer
molecular testing by next-generation sequencing. J Mol Diagn.
17:705–714. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de la Chapelle A and Hampel H: Clinical
relevance of microsatellite instability in colorectal cancer. J
Clin Oncol. 28:3380–3387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hause RJ, Pritchard CC, Shendure J and
Salipante SJ: Classification and characterization of microsatellite
instability across 18 cancer types. Nat Med. 22:1342–1350. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pawlik TM, Raut CP and Rodriguez-Bigas MA:
Colorectal carcinogenesis: MSI-H versus MSI-L. Dis Markers.
20:199–206. 2004. View Article : Google Scholar : PubMed/NCBI
|