Introduction
Esophageal cancer is one of the most common
malignant tumors. It is associated with high mortality rates and is
the sixth leading cause of cancer-related death in the world
(1,2). China has a high incidence of esophageal
cancer and the highest morbidity and mortality rates in the world.
Approximately one-half of new cases each year are recorded in
China, and the majority is squamous cell carcinoma (3–5). The
incidence of esophageal cancer exhibits significant regional and
ethnic differences (6). Despite
great progress in the diagnosis and treatment of esophageal cancer,
the 5-year survival rate is still poor (7). Therefore, identifying biomarkers
related to the occurrence, development and prognosis of esophageal
cancer is important for early diagnosis and treatment, as well as
for finding new targets and treatment methods.
Tumor protein 53 (TP53) is a recognized tumor
suppressor gene located on chromosome 17q13.1. The TP53 gene
plays an important role in regulating the cell cycle, apoptosis and
DNA damage repair (8,9). Wild-type TP53 can inhibit the
cell cycle and activate apoptosis-related genes that induce
apoptosis and regulate cell proliferation (10). Mutations in TP53 can lead to
the loss of these functions, inducing uncontrolled cell growth and
the promotion of tumor development (11,12).
These TP53 abnormalities may lead to DNA damage and
subsequent aneuploidy.
The TP53 gene mutation is the most common
gene alteration in many tumors, including esophageal cancer. It
also plays an important role in the occurrence and development of
esophageal cancer (13,14). Although the biological significance
of TP53 gene mutation is well characterized, its clinical
significance in esophageal cancer remains controversial, especially
as a prognostic biomarker. Previous studies demonstrated that p53
was highly expressed in esophageal cancer tissue and was associated
with tumor malignancy (15–17). However, other previous studies
reported that the expression of p53 protein had no significant
association with the prognosis of esophageal cancer (18,19). The
detection of mutant p53 protein by immunohistochemistry (IHC) has
some limitations. For example, viral infection, stress and the
regulation of other proteins can also change the aggregation of p53
protein (20). Therefore, the
overexpression of p53 does not always indicate a mutation in the
TP53 gene. Similarly, TP53 mutation does not always
lead to the accumulation of p53 protein in cells. This
characteristic of the TP53 gene limits the use of IHC in
TP53 gene research, which may be the cause of the
conflicting results in the study of TP53 in esophageal
cancer.
Fluorescence in situ hybridization (FISH) is
a molecular pathological method with high sensitivity and
specificity, which uses specific DNA probes to detect chromosomal
aberrations, as well as gene deletion and amplification (21,22). It
has been reported that deletion of the TP53 gene plays an
important role in the occurrence and development of esophageal
cancer (23,24). At present, to the best of the
authors' knowledge, associations between the IHC and FISH methods
in the detection of TP53 in multi-ethnic esophageal cancer
have not been reported.
In the present study, IHC and FISH were used to
analyze the role of TP53 in the occurrence, development and
adverse prognosis of esophageal cancer in different ethnic groups
in Xinjiang (multi-ethnic group). In addition, a tissue microarray
(TMA) group from different regions (including other high-incidence
areas in China, where the etiology of esophageal cancer is
different from Xinjiang province owing to the difference of diet
and living habits) was used to validate the results. The
complementarity, clinical practicability and the accuracy of the
detection of TP53 abnormality in esophageal cancer using
these two methods were evaluated.
Patients and methods
Patients and samples
A total of 369 paraffin-embedded esophageal squamous
cell carcinoma (ESCC) and 187 matched adjacent normal tissue
specimens from patients of Kazakh, Uygur and Han ethnicities, who
underwent radical surgery between January 2012 and December 2014 in
The Thoracic Surgery Department of The First Affiliated Hospital of
Xinjiang Medical University were collected. Glass slides with 4-µm
tissue sections were produced from the paraffin-embedded blocks for
IHC and FISH detection. Among the 369 patients, 257 were male and
112 were female, with an average age of 60.8 years (range, 37–83
years). With regards to differentiation, 77 cases were poorly
differentiated, 196 cases were moderately differentiated and 96
cases were highly differentiated. Lymph node metastasis was
detected in 158 cases, while 211 cases exhibited no lymph node
metastasis. With regards to TNM stage (25), there were 30 cases with stage I, 119
cases with stage II and 220 cases with stage III. None of the
patients had received radiotherapy, chemotherapy or any other
cancer treatment before surgery. Complete follow-up data were
available for 214 patients. The survival time was measured from the
date of surgery to death or the last follow-up date. All
experiments were approved by The Ethics Committee of The First
Affiliated Hospital of Xinjiang Medical University, and written
informed consent was obtained from all participants.
TMA was carried out by Shanghai Core Super
Biotechnology Co., Ltd. There were 117 cases of ESCC with one core
(diameter, 1.5 mm) for each tumor sample, including 87 males and 30
females, with an average age of 64.7 years (range, 39–82 years).
With regards to differentiation, 32 were poorly differentiated, 59
were moderately differentiated and 26 were highly differentiated.
There were 63 cases of lymph node metastasis. Furthermore, there
were 7 cases with stage I, 49 cases with stage II, 57 cases with
stage III and 4 cases with stage IV. The patients received radical
surgery between January 2006 and October 2008, and complete
follow-up data were available for 100 patients. The follow-up
period was terminated in September 2019 and the total follow-up
time was 5.8–7.8 years. In addition, 3 normal esophageal mucosa
tissues, 97 adjacent normal tissues, 13 precancerous lesions and 9
metastatic lesions of ESCC were included in the TMA.
IHC
Tumor samples were fixed with 10% neutral formalin
for 12 h in room temperature, then replaced with new 10% neutral
formalin for 24 h, embedded in paraffin, and sectioned into
4-µm-thick slices. IHC was performed as previously described
(15). In brief, the tissue sections
were incubated at 60°C for 1 h, then dewaxed and hydrated in xylol
and gradient alcohol, and placed in 3% hydrogen peroxide solution
for 10 min to remove endogenous peroxidase. The antigen retrieval
step was performed by heating in a microwave oven at 95°C for 10
min in citrate buffer (pH 7.0). After cooling at room temperature,
the sections were treated with mouse anti-human p53 monoclonal
antibody (1:400; P 5813, Sigma-Aldrich; Merck KGaA) and incubated
at 4°C overnight in a moist chamber. The sections were then treated
with universal secondary antibody (ready to use, PV-6000, Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd) and incubated at 37°C for
25 min. Finally, DAB and hematoxylin were used to visualize
immunoreactivity and for counterstaining, respectively. Both steps
were performed at room temperature for 3 min. Retrospective tissue
sections of confirmed breast cancer were provided by the Department
of Pathology (The First Affiliated Hospital of Xinjiang Medical
University) were used as the positive control. Informed consent was
provided by patients at the time of tissue collection. PBS was used
as the negative control instead of primary antibody.
The results were independently evaluated by two
pathologists in a blind manner. A total of five high-power fields
of vision (magnification, ×20) were observed under a light
microscope (Leica DM 3000B; Lecia Microsystems GmbH) on each slide
and 100 tumor cells were counted in each field, and the percentage
of positive cells were calculated. Primary lesions with evident
nuclear staining in >10% tumor cells, including the basal cell
layer of the mucosa that had corresponding p53 characteristic
positive staining, was determined as positive. Weak staining
limited to basal cells was considered to be negative (26).
FISH
FISH was performed using a mixture of red-labeled
TP53 and green-labeled chromosome 17 centromere control
probe (Cytocell Ltd.). The amount of p53 probe is 40–50 ng/test and
159 kb in length covers the whole TP53 gene, and amount of
control probe is 60–75 ng/test, respectively. Briefly, tissue
samples were incubated at 60°C for 2 h, then dewaxed with xylol,
hydrated with gradient ethanol solution, and washed with ultra-pure
water. The slides were then incubated at 80°C in Vysis pretreatment
solution (Vysis, Inc.; Abbot Pharmaceutical, Co., Ltd.) for 15 min.
After washing with ultra-pure water, the slides were incubated in
Vysis protease I solution (Vysis, Inc.; Abbot Pharmaceutical, Co.,
Ltd.) at 37°C for 10 min, and then dehydrated in a series of cold
ethanol solutions (70, 80, 90 and 100%, each for 2 min) after
washing with ultra-pure water. Following this, 10 µl of the hybrid
mixture was added and the glass slides were covered with rubber
cement. Slides were then denatured at 75°C for 5 min and hybridized
at 37°C overnight, using a hybridization instrument. The next day,
the slides were washed with 2X SSC (ID Labs™ Inc. Biotechnology,
http://www.lookbio.com/) solution for 2 min at
72°C, then at room temperature for 1 min. Subsequently, 5 µl DAPI
was applied to each spot and covered with a cover slip. A confocal
laser scanning microscope (Leica Microsystems GmbH, magnification,
×100) was used for imaging and analysis. The length of stimulated
luminescence were: Blue (DAPI) 405 nm, green (FITC) 488nm, red
(Texas Red) 561 nm, respectively.
For analysis, ≥100 non-overlapping and complete
nuclei with fluorescent signals were counted, and the results were
calculated according to the percentage of nuclei with signal
changes. There are two red and two green signals in normal nuclei.
Samples with >30% of tumor nuclei with either none or only one
red signal per probe were determined to indicate TP53
deletion.
Statistical analysis
Data were analyzed using SPSS 20.0 software (IBM
Corp.). The χ2 test and Fisher's exact test were
conducted. Survival analysis was performed using Kaplan-Meier and
log-rank tests. Univariate and multivariate analyses were conducted
by Cox regression. P<0.05 was considered to indicate a
statistically significant difference.
Results
Association of p53 protein expression
with clinicopathological factors
The positive expression rates of p53 protein in
cancer tissue and adjacent normal tissue were 54.5% (201/369) and
1.1% (2/187), respectively, and this difference was statistically
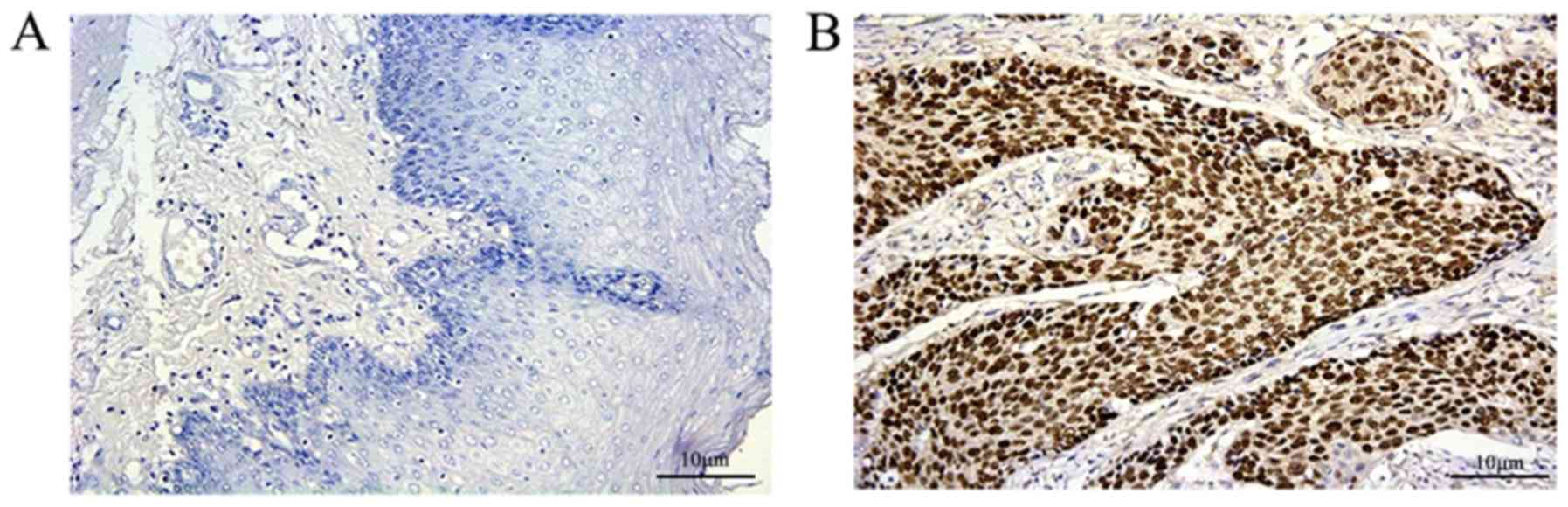
significant (P<0.001; Fig. 1;
Table I). The expression of p53
protein was significantly associated with sex (P=0.026) and tumor
differentiation (P=0.032). There were no significant associations
between p53 expression and age, TNM stage, lymph node metastasis (N
classification), depth of invasion (T classification) or vascular
invasion. In addition, the expression rates of p53 in ESCC of the
Kazakh, Uygur and Han ethnic groups were 56.3, 53.6 and 53.7%,
respectively. There was no significant difference in the expression
of p53 protein among the different ethnicities (P>0.05; data not
shown).
 | Table I.Association between p53 expression
and clinicopathological characteristics of patients with ESCC. |
Table I.
Association between p53 expression
and clinicopathological characteristics of patients with ESCC.
|
|
| p53 expression |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | n | Positive, n
(%) | Negative, n
(%) | χ2 | P-value |
|---|
| Specimen |
|
|
| 152.682 | <0.001 |
|
Adjacent normal tissue | 187 | 2 (1.1) | 185 (98.9) |
|
|
|
ESCC | 369 | 201 (54.5) | 168 (45.5) |
|
|
| Sex |
|
|
| 4.179 | 0.026 |
|
Female | 112 | 70 (62.5) | 42 (37.5) |
|
|
|
Male | 257 | 131 (51.0) | 126 (49.0) |
|
|
| Age, years |
|
|
| 1.658 | 0.119 |
|
>65 | 145 | 85 (58.6) | 60 (41.4) |
|
|
|
≤65 | 224 | 116 (51.8) | 108 (48.2) |
|
|
| TNM stage |
|
|
| 0.615 | 0.450 |
|
I–II | 232 | 130 (56.0) | 102 (44.0) |
|
|
|
III | 137 | 71 (51.8) | 66 (48.2) |
|
|
| T
classification |
|
|
| 0.739 | 0.226 |
|
T1-T2 | 145 | 83 (57.2) | 62 (42.8) |
|
|
|
T3-T4 | 224 | 118 (52.7) | 106 (47.3) |
|
|
| N
classification |
|
|
| 1.642 | 0.120 |
| No | 211 | 121 (57.3) | 90 (42.7) |
|
|
|
Yes | 158 | 80 (50.6) | 78 (49.4) |
|
|
|
Differentiation |
|
|
| 3.904 | 0.032 |
|
Good | 96 | 44 (45.8) | 52 (54.2) |
|
|
|
Moderate-poor | 273 | 157 (57.5) | 116 (42.2) |
|
|
| Tumor size |
|
|
| 3.484 | 0.074 |
|
<4 | 204 | 120 (58.8) | 84 (41.2) |
|
|
| ≥4 | 165 | 81 (49.1) | 84 (50.9) |
|
|
| Vascular
invasion |
|
|
| 0.049 | 0.893 |
|
Negative | 294 | 161 (54.8) | 133 (45.2) |
|
|
|
Positive | 75 | 40 (53.3) | 35 (45.7) |
|
|
In the TMA analysis, the positive expression rate of
p53 protein was 58.1% (68/117) in cancer tissues and 11.3% (11/97)
in adjacent normal tissues. This difference was statistically
significant (P<0.001). There was a significant association
between p53 protein expression and the depth of invasion (P=0.011;
Table II), but there was no
significant difference in p53 protein expression between other
clinicopathological factors, such as sex, age, TNM stage and lymph
node metastasis (Table II).
 | Table II.Association between p53 expression
and clinicopathological characteristics of patients with ESCC
(tissue microarray). |
Table II.
Association between p53 expression
and clinicopathological characteristics of patients with ESCC
(tissue microarray).
|
|
| p53 expression,
IHC |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | N | Positive, n
(%) | Negative, n
(%) | χ2 | P-value |
|---|
| Specimen |
|
|
| 49.833 | <0.001 |
|
Adjacent normal tissue | 97 | 11 (11.3) | 86 (88.7) |
|
|
|
ESCC | 117 | 68 (58.1) | 49 (41.9) |
|
|
| Sex |
|
|
| 0.035 | 0.509 |
|
Female | 30 | 17 (56.7) | 13 (43.3) |
|
|
|
Male | 87 | 51 (58.6) | 36 (41.4) |
|
|
| Age, years |
|
|
| 0.106 | 0.447 |
|
>60 | 76 | 45 (59.2) | 31 (40.8) |
|
|
|
≤60 | 41 | 23 (56.1) | 18 (43.9) |
|
|
| TNM stage |
|
|
| 0.337 | 0.347 |
|
I–II | 56 | 31 (55.4) | 25 (44.6) |
|
|
|
III–IV | 61 | 37 (60.7) | 24 (39.3) |
|
|
| T
classification |
|
|
| 6.405 | 0.011 |
|
T1-T2 | 23 | 8 (34.8) | 15 (65.2) |
|
|
|
T3-T4 | 94 | 60 (63.8) | 34 (36.2) |
|
|
| N
classification |
|
|
| 0.054 | 0.483 |
| No | 54 | 32 (59.3) | 22 (40.7) |
|
|
|
Yes | 63 | 36 (57.1) | 27 (42.6) |
|
|
|
Differentiation |
|
|
| 0.96 | 0.567 |
|
Good | 26 | 15 (57.7) | 11 (42.3) |
|
|
|
Moderate-poor | 91 | 53 (58.2) | 38 (41.8) |
|
|
TP53 gene deletion and its
associations with clinicopathological factors
FISH was performed in 214 of the 369 patients with
complete follow-up data. The deletion rates of the TP53 gene
were 31.8% (68/214) in cancer tissue and 8.3% (2/24) in adjacent
normal tissue, and the difference was statistically significant
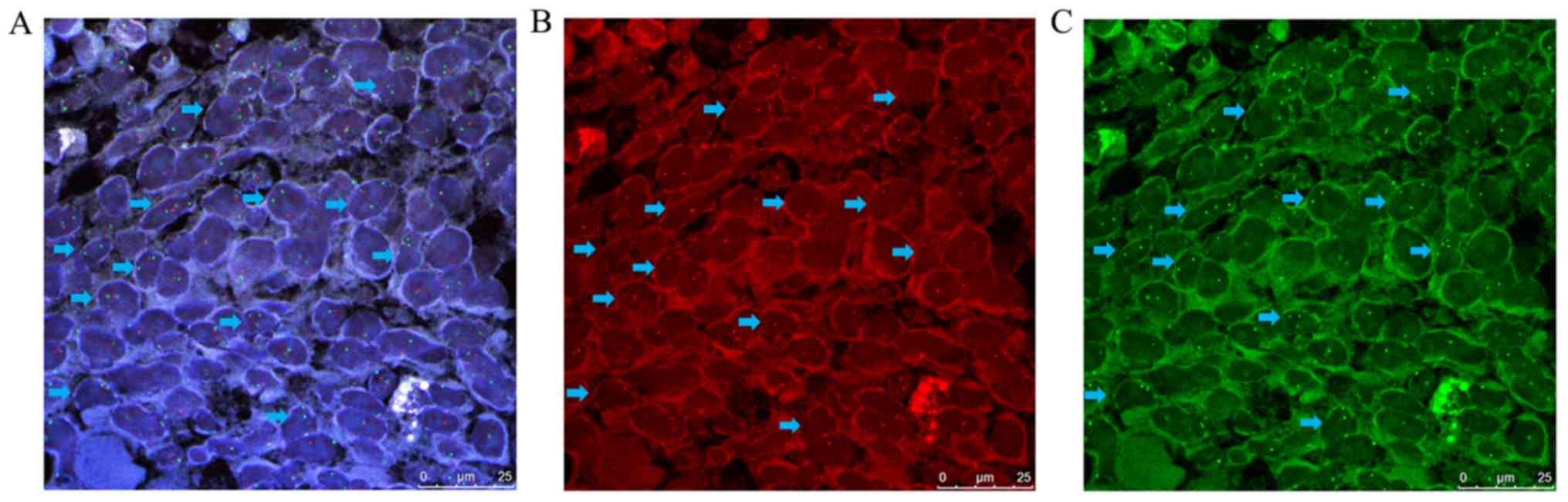
(P=0.01; Fig. 2; Table III). Among these, the TP53
deletion rates in Kazakh, Uygur and Han ethic groups were 31.0,
36.1 and 28.2%, respectively; there was no significant difference
when comparing the different ethnic groups (P>0.05, data not
shown). TP53 gene deletion was significantly associated with
tumor differentiation (P=0.002), lymph node metastasis (P=0.005)
and vascular invasion (P<0.001; Table III). There were no significant
associations between TP53 gene deletion and other
clinicopathological factors (P>0.05).
 | Table III.Association between p53 gene deletion
and clinicopathological characteristics of patients with ESCC. |
Table III.
Association between p53 gene deletion
and clinicopathological characteristics of patients with ESCC.
|
| p53 FISH |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Positive, n
(%) | Negative, n
(%) | χ2 | P-value |
|---|
| Specimen |
|
| 5.712 | 0.01 |
|
Adjacent normal tissue | 2 (8.3) | 22 (91.7) |
|
|
|
ESCC | 68 (31.8) | 146 (68.2) |
|
|
| Sex |
|
| 0.051 | 0.872 |
|
Female | 19 (30.6) | 43 (69.4) |
|
|
|
Male | 49 (32.2) | 103 (67.8) |
|
|
| Age, years |
|
| 1.653 | 0.224 |
|
>60 | 29 (37.2) | 49 (62.8) |
|
|
|
≤60 | 39 (28.7) | 97 (71.3) |
|
|
| TNM stage |
|
| 3.23 | 0.089 |
|
I–II | 39 (27.7) | 102 (72.3) |
|
|
|
III | 29 (39.7) | 44 (60.3) |
|
|
| T
classification |
|
| 0.043 | 0.881 |
|
T1-T2 | 26 (31.0) | 58 (69.0) |
|
|
|
T3-T4 | 42 (32.3) | 88 (67.7) |
|
|
| N
classification |
|
| 7.278 | 0.005 |
| No | 30 (24.4) | 93 (75.6) |
|
|
|
Yes | 38 (41.8) | 53 (58.2) |
|
|
|
Differentiation |
|
| 8.963 | 0.002 |
|
Good | 11 (17.2) | 53 (82.8) |
|
|
|
Moderate-Poor | 57 (38.0) | 93 (62.0) |
|
|
| Tumor size |
|
| 0.273 | 0.66 |
|
<4 | 38 (33.3) | 76 (66.7) |
|
|
| ≥4 | 30 (30.0) | 70 (70.0) |
|
|
| Vascular
invasion |
|
| 16.274 | <0.001 |
|
Negative | 45 (25.7) | 130 (74.3) |
|
|
|
Positive | 23 (59.0) | 16 (41.0) |
|
|
In the TMA analysis, the deletion rates of the
TP53 gene were 47.9% (56/117) in cancer tissues and 2.1%
(2/97) in adjacent normal tissues (P<0.001). TP53 gene
deletion was significantly associated with lymph node metastasis
(P=0.003) and TNM stage (P=0.01). However, there was no significant
difference in TP53 gene deletion between other
clinicopathological factors (P>0.05; Table IV).
 | Table IV.Association between p53 gene deletion
and clinicopathological characteristics of patients with ESCC
(tissue microarray). |
Table IV.
Association between p53 gene deletion
and clinicopathological characteristics of patients with ESCC
(tissue microarray).
|
|
| p53 FISH |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | N | Positive (%) | Negative (%) | χ2 | P-value |
|---|
| Specimen |
|
|
| 56.31 | <0.001 |
|
Adjacent normal tissue | 97 | 2 (2.1) | 95 (97.9) |
|
|
|
ESCC | 117 | 56 (47.9) | 61 (52.1) |
|
|
| Sex |
|
|
| 0.484 | 0.487 |
|
Male | 87 | 40 (46) | 47 (54) |
|
|
|
Female | 30 | 16 (53.3) | 14 (46.7) |
|
|
| Age, years |
|
|
| 1.506 | 0.22 |
|
≤60 | 40 | 16 (40) | 24 (60) |
|
|
|
>60 | 77 | 40 (51.9) | 37 (48.1) |
|
|
| N
classification |
|
|
| 8.484 | 0.003 |
| No | 54 | 18 (33.3) | 36 (66.7) |
|
|
|
Yes | 63 | 38 (60.3) | 25 (39.7) |
|
|
| T
classification |
|
|
| 0.008 | 0.557 |
|
T1-T2 | 23 | 11 (47.8) | 12 (52.2) |
|
|
|
T3-T4 | 94 | 45 (47.9) | 49 (52.1) |
|
|
| TNM stage |
|
|
| 6.353 | 0.01 |
|
I–II | 56 | 20 (35.7) | 36 (64.3) |
|
|
|
III–IV | 61 | 36 (41.0) | 25 (59.0) |
|
|
|
Differentiation |
|
|
| 1.184 | 0.194 |
|
Good | 26 | 10 (38.5) | 16 (61.5) |
|
|
|
Moderate-poor | 91 | 46 (50.5) | 45 (49.5) |
|
|
Association between TP53 gene
abnormalities and prognosis
Kaplan-Meier survival analysis was used to further
analyze the relationship between TP53 gene abnormalities and
ESCC prognosis. There were no significant associations between p53
protein expression and the overall survival of patients with ESCC
(P>0.05, data not shown). However, TP53 gene deletion was
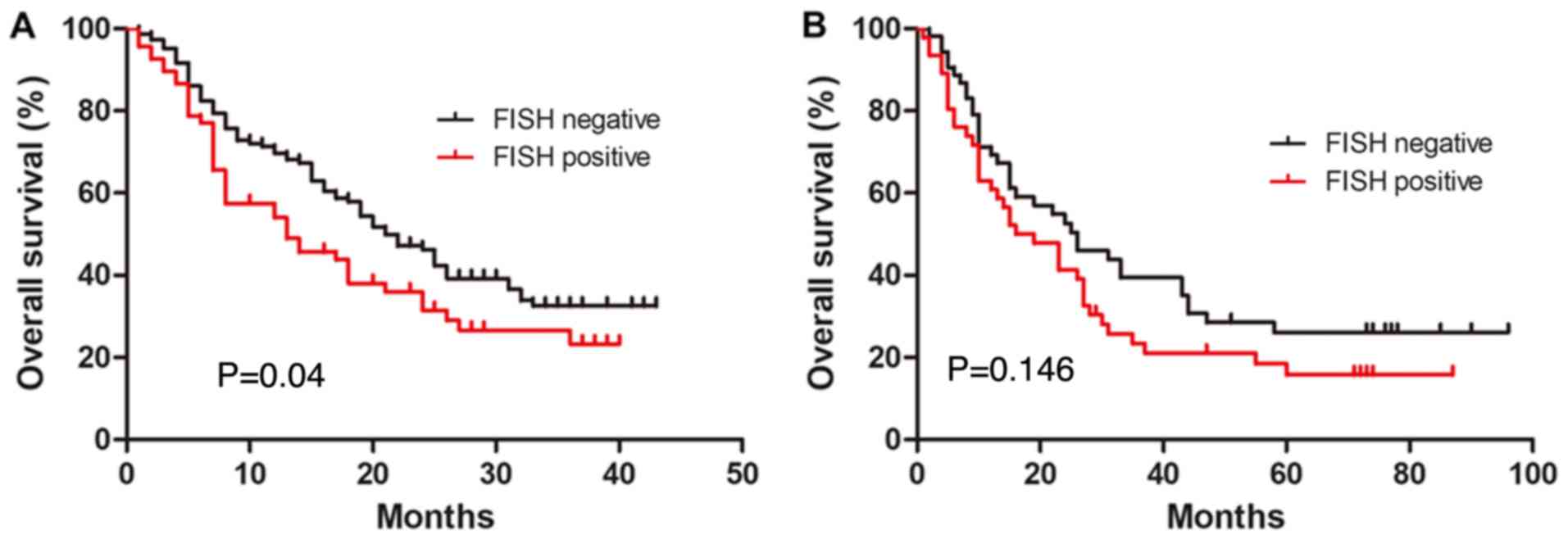
negatively associated with overall survival. Patients with
TP53 gene deletion had a worse prognosis (P<0.05;
Fig. 3A). In addition, univariate
Cox regression analysis demonstrated that TP53 gene
deletion, lymph node metastasis and vascular invasion were
significantly associated with the overall survival of patients with
ESCC (P<0.05; Table V).
Multivariate Cox analysis demonstrated that lymph node metastasis
and vascular invasion were independent prognostic factors for
ESCC.
 | Table V.Univariate and multivariate analysis
of overall survival in patients with esophageal squamous cell
carcinoma. |
Table V.
Univariate and multivariate analysis
of overall survival in patients with esophageal squamous cell
carcinoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex, female vs.
male | 1.271
(0.843–1.918) | 0.252 |
|
|
| Age, ≤60 vs. >60
years | 1.365
(0.955–1.952) | 0.087 |
|
|
| T classification,
T1-T2 vs. T3-T4 | 1.335
(0.926–1.925) | 0.121 |
|
|
| N classification,
negative vs. positive | 2.336
(1.630–3.348) | <0.001 | 0.500
(0.341–0.733) | <0.001 |
| Differentiation,
good vs. moderate-poor | 1.229
(0.964–1.569) | 0.097 |
|
|
| Tumor size, <4
cm vs. ≥4 cm | 1.116
(0.785–1.587) | 0.54 |
|
|
| Vascular invasion,
negative vs. positive | 2.386
(1.585–3.593) | <0.001 | 0.563
(0.363–0.874) | 0.01 |
| p53 FISH, negative
vs. positive | 1.457
(1.009–2.105) | 0.04 | 0.863
(0.589–1.265) | 0.45 |
| p54 IHC, negative
vs. positive | 1.122
(0.785–1.603) | 0.527 |
|
|
In the TMA group, Kaplan-Meier analysis demonstrated
that both p53 protein expression and TP53 gene deletion were
not associated with the overall survival of patients with ESCC.
However, patients with the TP53 gene deletion had a worse
prognosis than those without the deletion (Fig. 3B). In addition, univariate Cox
regression analysis showed that the depth of invasion, lymph node
metastasis and tumor differentiation were significantly associated
with overall survival (P<0.05; Table
VI). Furthermore, multivariate Cox analysis suggested that the
depth of invasion and tumor differentiation were independent
prognostic factors, patients with deeper invasion and lower
differentiation had a worse prognosis.
 | Table VI.Univariate and multivariate analysis
of overall survival in patients with esophageal squamous cell
carcinoma (tissue microarray). |
Table VI.
Univariate and multivariate analysis
of overall survival in patients with esophageal squamous cell
carcinoma (tissue microarray).
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex, female vs.
male | 1.842
(1.043–3.254) | 0.035 | 0.728
(0.446–1.187) | 0.203 |
| Age, ≤60 vs. >60
years | 0.990
(0.615–1.594) | 0.967 |
|
|
| T classification,
T1-T2 vs. T3-T4 | 0.339
(0.155–0.745) | 0.007 | 1.925
(1.073–3.451) | 0.028 |
| N classification,
negative vs. positive | 0.544
(0.340–0.871) | 0.011 | 0.449
(0.201–1.007) | 0.052 |
| Differentiation,
good vs. moderate-poor | 0.295
(0.150–0.580) | <0.001 | 0.324
(0.162–0.646) | 0.001 |
| p53 FISH, negative
vs. positive | 0.701
(0.445–1.105) | 0.126 |
|
|
| p53 IHC, negative
vs. positive | 1.117
(0.705–1.770) | 0.638 |
|
|
Concordance between the results of IHC
and FISH
The χ2 test suggested that there was a
significant association between the results of IHC and FISH
(P=0.007). The positive concordance rate between IHC and FISH was
41.1% (39/95), the negative concordance rate was 75.6% (90/119),
and the total concordance rate was 60.3% (129/214). There was also
a significant association between IHC and FISH in the TMA analysis
(P=0.001). The positive concordance rate of IHC and FISH was 60.3%
(41/68), while the negative concordance rate was 69.4% (34/49), and
the total concordance rate was 64.1% (75/117; Table VII).
 | Table VII.Concordance between the results of
IHC and FISH in esophageal squamous cell carcinoma. |
Table VII.
Concordance between the results of
IHC and FISH in esophageal squamous cell carcinoma.
| A, Multi ethnic
group |
|---|
|
|---|
|
| p53 FISH |
|
|---|
|
|
|
|
|---|
| p53 IHC | Positive, % | Negative, % | Concordance rate,
% |
|---|
| Negative | 29 | 90 | 75.6 |
| Positive | 39 | 56 | 41.1 |
| Total | 68 | 146 | 60.3 |
|
| B, Tissue
microarray |
|
| p53
FISH |
|
|
|
|
|
| p53 IHC | Positive,
% | Negative,
% | Concordance
rate, % |
|
| Negative | 15 | 34 | 69.4 |
| Positive | 41 | 27 | 60.3 |
| Total | 56 | 61 | 64.1 |
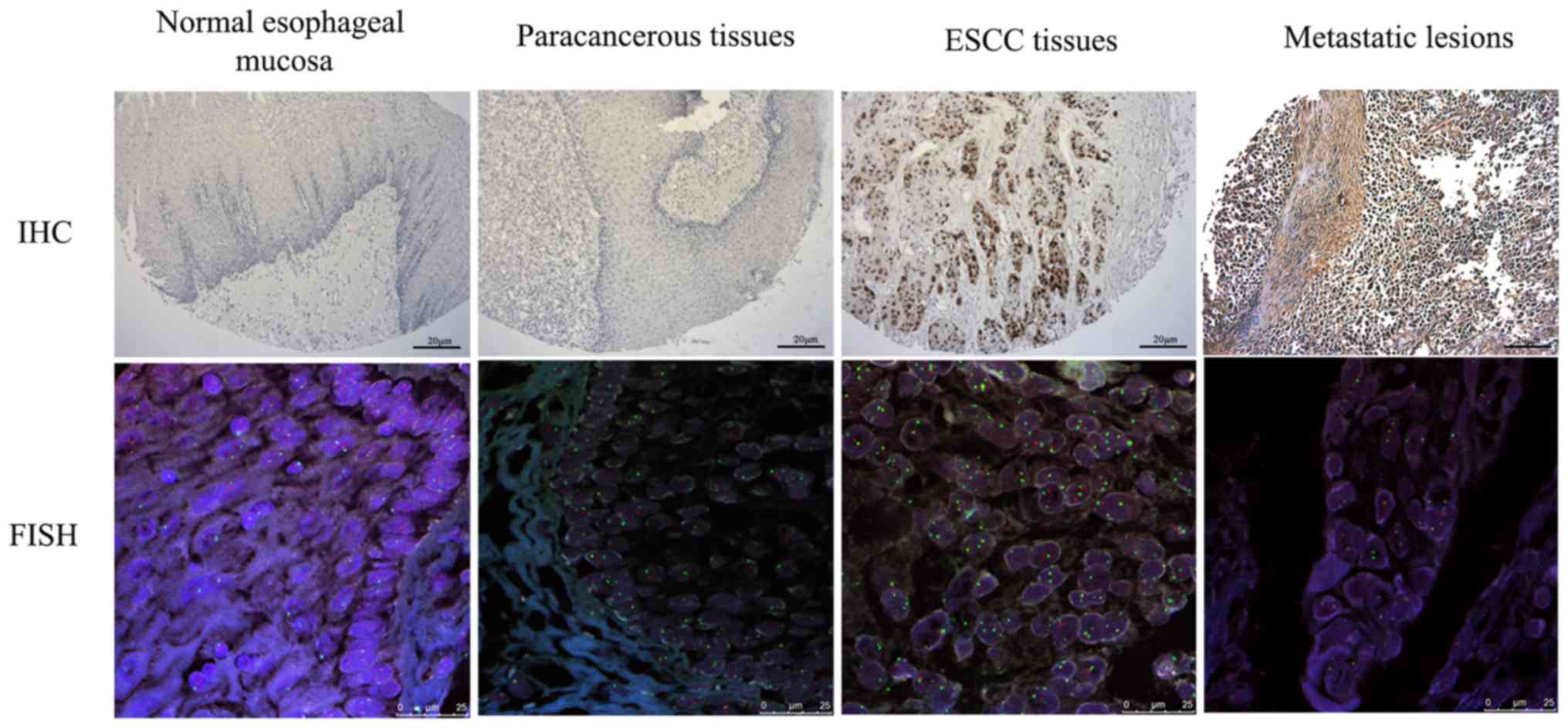
The present study also analyzed TP53 gene
deletion and protein expression in different stages of esophageal
cancer (Fig. 4). The positive rates
of TP53 gene deletion in normal esophageal mucosa, adjacent
normal tissues, precancerous lesions, ESCC tissues and metastatic
lesions of esophageal cancer were 0% (0/3), 2.1% (2/97), 30.8%
(4/13), 47.9% (56/117) and 88.9% (8/9), respectively. The positive
expression rates of p53 protein in these tissues were 0% (0/3),
11.3% (11/97), 46.2% (6/13), 58.1% (68/117) and 77.8% (7/9),
respectively. There were significant differences in TP53
gene deletion and protein expression when comparing the different
histological stages (P<0.05; Table
VIII).
 | Table VIII.p53 abnormalities detected by FISH
and IHC at different histological stages. |
Table VIII.
p53 abnormalities detected by FISH
and IHC at different histological stages.
| A, FISH |
|
|
|
| p53 |
|
|---|
|
|
|
|
|
|---|
| Histology | n | Positive (%) | Negative | P-value |
|---|
| Normal esophageal
mucosa | 3 | 0 | 3 |
|
| Adjacent normal
tissue | 97 | 2 (2.1) | 95 |
|
| Precancerous
lesion | 13 | 4 (30.8) | 9 | <0.001 |
| ESCC tissue | 117 | 56 (47.9) | 61 |
|
| Metastatic
lesion | 9 | 8 (88.9) | 1 |
|
|
| B, IHC |
|
|
|
| p53 |
|
|
|
|
|
|
|
Histology | n | Positive
(%) |
Negative | P-value |
|
| Normal esophageal
mucosa | 3 | 0 | 3 |
|
| Adjacent normal
tissue | 97 | 11 (11.3) | 86 |
|
| Precancerous
lesion | 13 | 6 (46.2) | 7 | <0.001 |
| ESCC tissue | 117 | 68 (58.1) | 49 |
|
| Metastatic
lesion | 9 | 7 (77.8) | 2 |
|
|
| C, FISH and
IHC |
|
|
|
| p53 |
|
|
|
Histology | n | Positive
(%) |
Negative | P-value |
|
| Normal esophageal
mucosa | 3 | 0 | 3 |
|
| Adjacent normal
tissue | 97 | 13 (13.4) | 84 |
|
| Precancerous
lesion | 13 | 9 (69.2) | 4 | <0.001 |
| ESCC tissue | 117 | 83 (70.9) | 34 |
|
| Metastatic
lesion | 9 | 8 (88.9) | 1 |
|
Discussion
Mutations in p53 protein are carcinogenic and can
promote the invasion, metastasis, proliferation and survival of
tumors (27). The accumulation of
p53 protein may indicate TP53 gene mutation and detection of
p53 protein accumulation by IHC can indirectly evaluate TP53
gene mutation. However, IHC may miss TP53 gene deletion
(20). DNA FISH can directly detect
the loss of the TP53 gene locus, which is one of the main
molecular features of the tumor suppressor gene variation and an
important marker for the transformation of some normal cells.
The present study demonstrated that the positive
rates of p53 protein expression in ESCC of in two groups, a
multi-ethnic group and a TMA group, were 54.5 and 58.1%,
respectively. It was also demonstrated that p53 protein expression
was associated with tumor differentiation and depth of invasion,
which indicated that the TP53 mutation was not only related
to the occurrence of ESCC, but also to prognostic factors such as
malignant phenotype, invasion and metastasis, which were indirectly
related to adverse prognosis. However, there was no significant
association between p53 protein expression and other clinical
factors, such as lymph node metastasis, TNM stage and overall
survival, which was consistent with previous studies by Taghavi
et al (19) and Yen et
al (28). By contrast, previous
studies have reported that mutated p53 protein could promote the
invasion and metastasis of tumors and affect the prognosis of
patients with ESCC (15,29,30).
These conflicting results could be due to different patient
selection processes, differences in the types of antibodies used
and cut-ff values for p53 expression used in the different studies
(18,31).
The present study also detected p53 protein
expression in Han, Uygur and Kazakh ethnic groups from Xinjiang,
and identified that there were no significant differences in p53
protein expression among the three ethnicities, indicating that p53
protein expression was similar in patients with ESCC from different
ethnic groups. In addition, the present results demonstrated that
the expression of p53 protein was significantly associated with sex
in the multi-ethnic group. However, the majority of previous
studies on p53 suggested that there were no associations with sex
(15,19,28). It
was hypothesized that the present results could be an incidental
anomaly due to the small sample size. The possible association
between p53 and sex may be further confirmed by a larger sample
size in future studies.
Moreover, the present study also detected TP53 gene
deletion. TP53 gene deletion rates were 31.8 and 47.9% in
the tumor tissues of the multi-ethnic and the TMA groups,
respectively. In addition, TP53 gene deletion was also
related to prognostic factors such as tumor differentiation and
lymph node metastasis. In a previous study of p53 IHC and loss of
heterozygosity (LOH) analysis with similar experimental conditions,
including 94 resected ESCC samples, the TP53 LOH rate was
67.5%, but it was not associated with the malignant phenotype of
ESCC (22). In our previous study,
FISH was used to detect TP53 gene deletion in patients with
ESCC of Kazakh background, which suggested that TP53 gene
deletion was related to tumor differentiation and lymph node
metastasis (32). In the present
study, additional ethnicities and samples were included, and
TP53 gene deletion was demonstrated to be more common in
patients with poor differentiation, lymph node metastasis and
vascular invasion at advanced stages, which was consistent with our
previous study, suggesting that the altered TP53 may play an
important role in predicting the prognosis of ESCC. In addition,
there was a significant association between TP53 gene
deletion and overall survival in multi-ethnic groups. Although
there was no significant association between TP53 gene
deletion and overall survival in the TMA group, patients with
TP53 gene deletion displayed a tendency towards worse
prognosis, compared with those without the deletion, which was
consistent with the result of multi-ethnic groups. Overall, the
present results suggested that TP53 gene deletion was
associated with poor prognosis and may be a potential and effective
prognostic indicator of ESCC.
The expression rates of p53 protein were 54.5 and
58.1%, and the deletion rates of the TP53 gene were 31.8 and
47.9% in the two groups, respectively. The total concordance rates
of FISH and IHC for detecting TP53 gene abnormality were
60.3 and 64.1%, respectively. When both methods were used
simultaneously, the detection rate of TP53 gene
abnormalities was significantly increased. A previous study by
Graesslin et al (33)
reported that there was no significant association between
TP53 gene deletion and p53 protein expression in endometrial
cancer, and there was no significant association between
TP53 gene deletion and clinical prognostic factors.
Therefore, FISH was not recommended for the routine detection of
the TP53 gene in endometrial cancer (33). However, the present results support
the combination of FISH and IHC in assessing TP53
abnormalities in ESCC. Although FISH allows objective
quantification of results by counting intranuclear signals, there
are some disadvantages to this method, such as the influence of
non-tumor cells on the results. However, the present study
suggested that IHC combined with FISH could overcome this
limitation.
In addition, the present study also detected
TP53 abnormalities in different stages of ESCC and found
that TP53 gene deletion and protein expression increased
with the progression of ESCC. In a study of Barrett's esophagus,
Davelaar et al (34) reported
that the positive rates of TP53 FISH and IHC increased with
the progression of the disease, and the combined use of FISH and
IHC technology could significantly improve the detection rate of
TP53 gene abnormalities at different stages of the disease,
which is similar to the present results in ESCC. These results also
indicated that TP53 gene abnormalities are not only an early
event of esophageal cancer, but also play an important role in
tumor progression.
However, the sample size was relatively small and
only one core of tumor tissue for each patient was included in TMA,
which may account for the difference in the results observed
between the TMA and the multi-ethnic group. Therefore, verification
of the results with TMA, including larger sample size and more
tumor cores is required in subsequent studies.
In conclusion, the present results demonstrated that
the TP53 gene mutation played an important role in the
occurrence and development of ESCC. The factors affecting p53
overexpression are complex, and TP53 gene deletion is one of
the most important. These results suggested that FISH could be used
as an additional method in the diagnosis of ESCC, especially when
IHC results are negative. The application of FISH could increase
the accuracy of chromosome karyotype determination, improve the
detection efficiency of TP53 gene abnormalities in ESCC, and
help accurately evaluate the malignancy and prognosis of esophageal
cancer.
Acknowledgements
Not applicable.
Funding
The present study was funded by The National Natural
Science Foundation (grant no. 81360305).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MN and EA was mainly responsible for designing and
performing the experiments. JA and AA was mainly responsible for
analyzing and interpreting the data, and performed statistical
analysis. LZ and IS provided the study materials and revised the
manuscript critically, and also guided the research design and
experiment. AT was mainly responsible for drafting the manuscript
and collecting clinical data, including clinicopathological data
and patient follow-up data. RC and ZH helped with performing the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were approved by The Ethics
Committee of The First Affiliated Hospital of Xinjiang Medical
University, and written informed consent was obtained from all
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zeng H, Zheng R, Zhang S, Zuo T, Xia C,
Zou X and Chen W: Esophageal cancer statistics in China, 2011:
Estimates based on 177 cancer registries. Thorac Cancer. 7:232–237.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y,
Ueda J, Wei W, Inoue M and Tanaka H: Epidemiology of esophageal
cancer in Japan and China. J Epidemiol. 23:233–242. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Soussi T: The p53 pathway and human
cancer. Brit J Surg. 92:1331–1332. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vousden KH and Lane DP: p53 in health and
disease. Nat Rev Mol Cell Biol. 8:275–283. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
da Silva GN, Evangelista AF, Magalhães DA,
Macedo C, Búfalo MC, Sakamoto-Hojo ET, Passos GA and Salvadori DM:
Expression of genes related to apoptosis, cell cycle and signaling
pathways are independent of TP53 status in urinary bladder cancer
cells. Mol Biol Rep. 38:4159–4170. 2011. View Article : Google Scholar
|
|
11
|
Cardin R, Piciocchi M, Tieppo C, Maddalo
G, Zaninotto G, Mescoli C, Rugge M and Farinati F: Oxidative DNA
damage in Barrett mucosa: Correlation with telomeric dysfunction
and p53 mutation. Ann Surg Oncol. 20 (Suppl 3):S583–S589. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Di Agostino S, Strano S and Blandino G:
Gender, mutant p53 and PML: A growing ‘affaire’ in tumor
suppression and oncogenesis. Cell Cycle. 12:1824–1825. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Greenblatt MS, Bennett WP, Hollstein M and
Harris CC: Mutations in the p53 tumor suppressor gene: Clues to
cancer etiology and molecular pathogenesis. Cancer Res.
54:4855–4878. 1994.PubMed/NCBI
|
|
14
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang K, Chen L, Zhang J, Wu Z, Lan L,
Wang L, Lu B and Liu Y: Elevated p53 expression levels correlate
with tumor progression and poor prognosis in patients exhibiting
esophageal squamous cell carcinoma. Oncol Lett. 8:1441–1446. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Okumura H, Kita Y, Yokomakura N, Uchikado
Y, Setoyama T, Sakurai H, Omoto I, Matsumoto M, Owaki T, Ishigami S
and Natsugoe S: Nuclear expression of 14-3-3 sigma is related to
prognosis in patients with esophageal squamous cell carcinoma.
Anticancer Res. 30:5175–5179. 2010.PubMed/NCBI
|
|
17
|
Zhao Z, Wang P, Gao Y and He J: The high
expression instead of mutation of p53 is predictive of overall
survival in patients with esophageal squamous-cell carcinoma: A
meta-analysis. Cancer Med. 6:54–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Murata A, Baba Y, Watanabe M, Shigaki H,
Miyake K, Karashima R, Imamura Y, Ida S, Ishimoto T, Iwagami S, et
al: p53 immunohistochemical expression and patient prognosis in
esophageal squamous cell carcinoma. Med Oncol. 30:7282013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Taghavi N, Biramijamal F, Sotoudeh M,
Moaven O, Khademi H, Abbaszadegan MR and Malekzadeh R: Association
of p53/p21 expression with cigarette smoking and prognosis in
esophageal squamous cell carcinoma patients. World J Gastroenterol.
16:4958–4967. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nenutil R, Smardova J, Pavlova S,
Hanzelkova Z, Muller P, Fabian P, Hrstka R, Janotova P, Radina M,
Lane DP, et al: Discriminating functional and non-functional p53 in
human tumours by p53 and MDM2 immunohistochemistry. J Pathol.
207:251–259. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Halling KC and Kipp BR: Fluorescence in
situ hybridization in diagnostic cytology. Hum Pathol.
38:1137–1144. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tafe LJ, Allen SF, Steinmetz HB, Dokus BA,
Cook LJ, Marotti JD and Tsongalis GJ: Automated processing of
fluorescence in-situ hybridization slides for HER2 testing in
breast and gastro-esophageal carcinomas. Exp Mol Pathol.
97:116–119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Egashira A, Morita M, Yoshida R, Saeki H,
Oki E, Sadanaga N, Kakeji Y, Tsujitani S and Maehara Y: Loss of p53
in esophageal squamous cell carcinoma and the correlation with
survival: Analyses of gene mutations, protein expression, and loss
of heterozygosity in Japanese patients. J Surg Oncol. 104:169–175.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bellini MF, Cadamuro AC, Succi M, Proenca
MA and Silva AE: Alterations of the TP53 gene in gastric and
esophageal carcinogenesis. J Biomed Biotechnol. 2012:8919612012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fagundes RB, Melo CR, Pütten AC, Moreira
LF and de Barros SG: p53 immunoexpression: An aid to conventional
methods in the screening of precursor lesions of squamous
esophageal cancer in patients at high-risk? Cancer Detect Prev.
29:227–232. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Muller PA and Vousden KH: p53 mutations in
cancer. Nat Cell Biol. 15:2–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yen CC, Tsao YP, Chen PC, Wu YC, Liu JH,
Pan CC, Liu CY, Tzeng CH, Chen PM, Chen YJ, et al: PML protein as a
prognostic molecular marker for patients with esophageal squamous
cell carcinomas receiving primary surgery. J Surg Oncol.
103:761–767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang L, Yu X, Li J, Zhang Z, Hou J and Li
F: Prognostic significance of p53 expression in patients with
esophageal cancer: A meta-analysis. BMC Cancer. 16:3732016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Melling N, Norrenbrock S, Kluth M, Simon
R, Hube-Magg C, Steurer S, Hinsch A, Burandt E, Jacobsen F, Wilczak
W, et al: p53 overexpression is a prognosticator of poor outcome in
esophageal cancer. Oncol Lett. 17:3826–3834. 2019.PubMed/NCBI
|
|
31
|
Yamamoto S, Yashima K, Kawata S, Hosoda K,
Tamoto A, Ikebuchi Y, Matsumoto K, Kawaguchi K, Harada K, Murawaki
Y and Isomoto H: Frequent aberrant p53 and Fhit expression in
endoscopically resected superficial hypopharyngeal cancer and
esophageal cancer. Oncol Lett. 14:587–592. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Niyaz M, Turghun A, Ping ZH, Zhu Z,
Sheyhedin I, Ren C and Awut I: TP53 gene deletion in esophageal
cancer tissues of patients and its clinical significance. Mol Med
Rep. 7:122–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Graesslin O, Chantot-Bastaraud S,
Lorenzato M, Birembaut P, Quéreux C and Daraï E: Fluorescence in
situ hybridization and immunohistochemical analysis of p53
expression in endometrial cancer: Prognostic value and relation to
ploidy. Ann Surg Oncol. 15:484–492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Davelaar AL, Calpe S, Lau L, Timmer MR,
Visser M, Ten Kate FJ, Parikh KB, Meijer SL, Bergman JJ, Fockens P
and Krishnadath KK: Aberrant TP53 detected by combining
immunohistochemistry and DNA-FISH improves Barrett's esophagus
progression prediction: A prospective follow-up study. Genes
Chromosomes Cancer. 54:82–90. 2015. View Article : Google Scholar : PubMed/NCBI
|