Introduction
Glioblastoma (GBM) is a malignant and aggressive
primary nervous system tumors worldwide, with a high rate of
recurrence and a poor prognosis compared with other types of
nervous system tumor (1). As
reported in the 2016 World Health Organization classification,
there are four different grades, and two classes, of glioma: Grades
I and II (low-grade gliomas); and grades III and IV (high-grade
gliomas) (2). Despite the advances
in molecular biology and oncogenetics for the treatment of patients
with glioma over the previous decades, the prognosis and overall
survival (OS) time, measured as 12–14 months following surgical
resection, remain poor (3,4). However, the molecular mechanisms of
glioma tumorigenesis remain unclear (5–8).
Therefore, it is essential to identify novel therapeutic and
prognostic targets in glioma.
Long non-coding RNAs (lncRNAs) are a class of newly
identified RNAs with a length ranging from 200 to 1,000
nucleotides, which lack protein-coding ability (9,10).
lncRNAs regulate the expression of genes at epigenetic,
transcriptional and post-transcriptional levels. Previous evidence
has indicated that the abnormal expression of lncRNAs can affect
glioma development and tumorigenesis (11). These lncRNAs may serve as useful
molecular targets for the diagnosis of malignant tumors, including
GBM (11,12). The biological role of lncRNAs in
glioma remains to be investigated.
lncRNA Ewing sarcoma associated transcript 1
(EWSAT1) is a novel cancer-associated lncRNA that serves critical
roles in the occurrence and progression of tumors, including
nasopharyngeal carcinoma (13,14),
osteosarcoma (15), colorectal
(16) and ovarian cancer (17), and Ewing sarcoma (18). However, the biological role and
mechanism of EWSAT1 in gliomas remain to be identified.
Previous studies on the function of microRNA
(miRNA/miR) have demonstrated that miRNAs are involved in the
regulation of gene expression, cell differentiation, metabolism and
invasion (19,20). Previous studies have indicated that
miR-152-3p is closely associated with carcinogenesis, including
glioma (21–23). Therefore, the present study aimed to
explore the potential association between EWSAT1 and miR-152-3p in
glioma.
The present study investigated the expression and
molecular mechanism of EWSAT 1 in glioma. The results suggested
that EWSAT1 was increased in glioma. In addition, EWSAT1 knockdown
suppressed the proliferative and invasive abilities of glioma
cells. The primary aim of the present study was to investigate the
function of lncRNA EWSAT1, and to detect the role of miRNA-152-3p
in the regulatory mechanism of glioma via lncRNA EWSAT1. The
results may provide a theoretical basis for developing new
therapeutic drugs against glioma.
Materials and methods
Cell lines and clinical tissues
A total of 5 glioma cell lines (U251, T98G, LN229,
A172 and SHG44) were obtained from The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences. Normal human
astrocyte (NHA) cells were purchased from The Institute of
Biochemistry and Cell Biology, Chinese Academy of Sciences. All
cells were sub-cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
maintained in a humidified incubator at 37°C with 5%
CO2.
Human GBM samples and adjacent normal tissues were
collected via surgical resection performed at The First Affiliated
Hospital of Jiamusi University. Samples were collected from
patients with GBM (27 females and 15 males; age range, 31–72 years;
median, 51.34 years) between January 2011 and December 2017. None
of the patients received any therapy prior to surgery. All GBM
samples were confirmed by two senior pathologists. The study
protocol was approved by the Ethics Committee of The Institutional
Review Board of Jiamusi University (approval no. JUIRBR-2019-214),
and all procedures were performed in accordance with the principles
outlined in The Declaration of Helsinki (24). Written informed consent was provided
by all patients to participate in the study. The clinical
characteristics of the patients are summarized in Table I.
 | Table I.Clinical characteristics of the
patients with glioblastoma according to long non-coding RNA EWSAT1
level in tissues (n=42). |
Table I.
Clinical characteristics of the
patients with glioblastoma according to long non-coding RNA EWSAT1
level in tissues (n=42).
|
|
| EWSAT1
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | N | Low | High | P-value |
|---|
| Age, years |
|
<60 | 23 | 9 | 14 | 0.382 |
|
≥60 | 19 | 10 | 9 |
|
| Sex |
|
Male | 15 | 9 | 6 | 0.152 |
|
Female | 27 | 10 | 17 |
|
| Karnofsky
Performance Scale score |
|
<60 | 24 | 11 | 13 | 0.929 |
|
≥60 | 18 | 8 | 10 |
|
| Mean tumor
diameter, cm |
|
<5 | 29 | 12 | 17 | 0.453 |
| ≥5 | 13 | 7 | 6 |
|
| Necrosis on
MRI |
|
Yes | 27 | 9 | 18 | 0.038a |
| No | 15 | 10 | 5 |
|
| Seizure |
|
Yes | 14 | 5 | 9 | 0.381 |
| No | 28 | 14 | 14 |
|
Bioinformatic prediction of EWSAT1
expression
The edgeR software package (Bioconductor) in RStudio
3.5.1 (https://www.rstudio.com/; RStudio, Inc.)
was used to detect the differentially expressed lncRNAs in
normalized gene expression profile data from the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/?term=GSE4290) and
The Cancer Genome Atlas (TGCA) GBM database (http://cancergenome.nih.gov) (25,26). For
the normalized gene expression profile data, the edgeR package was
employed to explore significantly abnormally expressed lncRNAs.
Log2 fold-change (FC) >2 and false-discovery rate (FDR) <0.01
were selected as significantly cut-off values, and the aberrantly
expressed candidate lncRNAs were identified. Clinical prognosis
data were acquired from the Gene Expression Profiling Interactive
Analysis (GEPIA) database (http://gepia.cancer-pku.cn/). Gene Ontology (GO) term
enrichment analysis was examined using the Database for Annotation,
Visualization and Integrated Discovery version 6.8 (https://david.ncifcrf.gov/).
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® and chloroform (Invitrogen;
Thermo Fisher Scientific, Inc.) were utilized to extract total
cellular RNA from each group, according to the manufacturer's
protocol. cDNA was synthesized using the PrimeScript™ RT kit
(Takara Biotechnology Co., Ltd.), according to the manufacturer's
protocol. qPCR was performed according to the manufacturer's
instructions of the TransScript Green Two-Step RT-qPCR SuperMix kit
(Beijing Transgen Biotech Co., Ltd.). Thermocycling conditions
consisted of an initial denaturing step at 94°C for 10 min,
followed by 40 cycles of denaturing at 94°C for 5 sec, annealing at
60°C for 30 sec and extending at 72°C for 45 sec. The primers used
were as follows: EWSAT1 forward, 5′-GTGTCTGGCAAGGAACACTA-3′ and
reverse, 5′-GGTGGAGAAGAGGGACAAT-3′; miR-152-3p forward,
5′-GCGCTCAGTGCATGACAGA-3′ and reverse, 5′-GTCGTATCCAGTGCAGGGT-3′;
matrix metalloproteinase (MMP)-2 forward,
5′-CAGGACATTGTCTTTGATGG-3′ and reverse, 5′-TGAAGAAGTAGCTATGACCA-3′;
MMP-9 forward, 5′-AGACCTGGGCAGATTCCAAAC-3′ and reverse
5′-CGGCAAGTCTTCCGAGTAGT-3′; U6 forward,
5′-GGATATTGTTGCCATCAATGACC-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGA-3′; and GAPDH forward
5′-AAGAAGGTGGTGAAGCAGGC-3′ and reverse, 5′-GTCAAAGGTGGAGGAGTGGG-3′.
U6 served as the endogenous control. The results were calculated
using the 2−ΔΔCq method (27).
Plasmid transfection
The U251 and T98G cells were maintained in DMEM
containing 10% FBS at 37°C in 5% CO2. When the cell
confluence reached ~80%, the cells were transfected with 20 µM of
each construct (siRNA or miRNA mimics) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol and
incubated at 37°C for 6 h. The culture medium was subsequently
replaced with fresh DMEM supplemented with 10% FBS, and subsequent
experimentation was performed 24 h post-transfection. Small
interfering RNA (siRNA) targeting EWSAT1 and its negative control
(si-NC), miR-152-3p mimic or inhibitor, and their respective
negative control (miR-NC) (20 µM) were obtained from Shanghai
GenePharma Co., Ltd. The sequences were as follows: EWSAT1 siRNA
forward, 5′-UUGGGCUCUCAAUGGUAUCAU-3′ and reverse,
5′-AAGGGAGGGUUACUAACUUUA-3′; si-NC forward,
5′-GGUAAGCAGUGGCUCCUCUAA-3′ and reverse,
5′-ACGUGACACGUUCGGAGAAUU-3′; miR-152-3p mimics forward,
5′-UCAGUGCAACUGACAGAACUUGG-3′ and reverse,
5′-UAGCCACGGUUGUGUAAAGUCUG-3′; miR-152-3p inhibitor forward,
5′-CGCGCUAGCAGCACGUAAAU-3′ and reverse, 5′-GUGCAGGGUCCGAGGUCAUC-3′;
and miR-NC forward, 5′-CAGUACUUUUGUGUAGUACAA-3′ and reverse,
5′-CAGUACUUUUGUGUAGUACAA-3′. Transfection with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was performed according to the manufacturer's
protocol. The cells were then cultured for subsequent experiments
and obtained at 24 h after transfection.
Dual-luciferase reporter assay
To detect the potential target miRNAs of EWSAT1, the
TargetScan (www.targetscan.org/) and StarBase (http://starbase.sysu.edu.cn/) databases were explored.
Among all the statistically relevant miRNAs, miR-152-3p exhibited
the highest score in the two databases and was selected for further
experiments. The wild-type (WT) or mutant (MUT) putative miR-152-3p
binding sites of the 3′-untranslated region (UTR) in EWSAT1 were
ligated into the pMIR-luciferase reporter plasmid vector (Shanghai
GenePharma Co., Ltd.). U251 cells were added in 6-well plates at
1×105 cells/well and the recombinant vectors were
co-transfected with miR-152-3p mimics or miR-NC using
Lipofectamine® 2000. The medium was replaced at 6 h
post-transfection, and the firefly and Renilla luciferase
signals were examined 48 h following transfection using the Dual
Luciferase Reporter Assay kit (Promega Corporation). Firefly
luciferase activity was normalized to that of Renilla
luciferase.
Cell proliferation assays
U251 and T98G cell proliferation was performed by
Cell Counting Kit-8 assay (CCK-8; Beyotime Institute of
Biotechnology) at 24, 48 and 72 h according to the manufacturer's
protocol. The transfected glioma cells were added to 96-well plates
at a density of 3,000 cells/well. Then, a culture solution
containing 10 µl CCK-8 regent was added. Following incubation at
37°C for additional 4 h, cell viability was measured by detecting
the absorbance at a wavelength of 490 nm using a microplate reader
(Bio-Rad Laboratories, Inc.)
Transwell invasion assays
The upper chambers of Transwell plates were
precoated with Matrigel® (Corning Life Sciences) at 37°C
for 30 min. Subsequently, U251 and T98G cells were suspended with
serum-free DMEM (5×104) and seeded in the upper
chambers. DMEM supplemented with 20% FBS was added to the lower
chambers and the plates were incubated in 37°C with 5%
CO2 for 24 h. Cells were fixed with 4% paraformaldehyde
at room temperature for 10 min, and stained with hematoxylin and
eosin (5 min for hematoxylin and 1 min for eosin at room
temperature), using the Staining kit (Beijing Solarbio Science
& Technology Co., Ltd.). The number of migratory cells per
sample was counted in 5 randomly selected fields under a light
microscope (Nikon Corporation) at magnification, ×100.
Western blotting
Proteins were extracted from cell lines (U251 and
T98G) with lysis buffer (BIOSS). The quantity of protein was
detected using the BCA Protein Assay kit (Beyotime Institute of
Biotechnology), and the lysates (20 µg protein) were separated on
10% SDS-PAGE and electrophoretically transferred onto PVDF
membranes (Beyotime Institute of Biotechnology). Following blocking
in 5% skim milk for 1 h at 37°C and incubation with the
corresponding primary antibodies at 4°C overnight [anti-MMP-2
(rabbit polyclonal antibody; 1:1,000; cat. no. 40094s; Cell
Signaling Technology, Inc.), anti-MMP-9 (rabbit polyclonal
antibody; 1:1,000; cat. no. 13667; Cell Signaling Technology, Inc.)
and anti-GAPDH (mouse monoclonal antibody; 1:1,000; SC-47724; Santa
Cruz Biotechnology, Inc.)], the membranes were incubated with
horseradish peroxidase-conjugated anti-mouse or anti-rabbit
secondary antibody (cat. no. ab6721 and ab6728; 1:2,000; Abcam) for
1 h at room temperature. Then, the bands were visualized using ECL
(Beyotime Institute of Biotechnology) and analyzed with a ChemiDoc™
MP Imaging detection system (Bio-Rad Laboratories, Inc.) and Image
Lab software 3.0 (Bio-Rad Laboratories, Inc.).
Immunofluorescence staining
U251 cells (1×105) were collected, and
cell slides were prepared. Glass slides were stained with 0.1%
poly-L-lysine at 4°C overnight. Then, 4% paraformaldehyde with 5%
BSA (cat. no. 9048-46-8; Sigma Aldrich; Merck KGaA) was added for
20 min, and the slides were fixed with 0.1% Triton X-100 for 10 min
at room temperature. The slides were then incubated with rabbit
polyclonal primary antibodies against MMP-2 (cat. no. 40094s;
1:1,000; Cell Signaling Technology, Inc.) and MMP-9 (cat. no.
13667; 1:1,000; Cell Signaling Technology, Inc.) at 4°C for 1 h,
followed by incubation with the corresponding fluorescence-labeled
rabbit secondary antibodies [tetramethylrhodamine
(TRITC)-conjugated goat anti-rabbit IgG (cat. no. SA00007-2; 1:100;
ProteinTech Group, Inc.) and fluorescein isothiocyanate
(FITC)-conjugated goat anti-rabbit IgG (cat. no. SA00003-2;
ProteinTech Group, Inc.)] at room temperature for 1 h. Cell nuclei
were then incubated with DAPI at 4°C (1 µg/ml; cat. no. 4083s; Cell
Signaling Technology, Inc.) for 15 min and observed under a
fluorescence microscope (Nikon Corporation) at magnification,
×400.
Statistical analysis
Data were expressed as the mean ± standard deviation
of 3 independent experiments. SPSS 21.0 (IBM Corp.) was applied for
data analysis excluding the Kaplan-Meier. Student's t-test or
one-way ANOVA with Tukey's post hoc test were performed to
determine differences between groups. The associations between
EWSAT1 level and the clinicopathological characteristics of the
patients were analyzed using the χ2 or Fisher's exact
tests. Pearson's correlation analysis was performed to determine
the correlation between EWSAT1 and miR-152-3p expression levels.
GraphPad Prism software 5.0 (GraphPad Software, Inc.) was used to
detect the Kaplan-Meier curve and a log-rank test was performed to
assess the survival percentage. The aberrantly expressed lncRNAs
were assessed based on the Benjamini-Hochberg method (28). P<0.05 was considered to indicate a
statistically significant difference.
Results
Upregulation of EWSAT1 in GBM
Using the GEO dataset GSE4290, the top 20 aberrantly
expressed lncRNAs were identified with cut-off values of log2 FC
>2 and FDR <0.01 (Fig. 1A).
EWSAT1 was the most significantly differentially expressed lncRNA.
EWSAT1 was identified to be upregulated in glioma tissues, and the
level increased according to glioma grade (Fig. 1A and B). The clinical survival data
collected from the GEPIA database (http://gepia.cancer-pku.cn/) demonstrated that
increased EWSAT1 levels in the GBM samples were associated with a
poorer OS, according to the median survival time of patients
(P=0.023; n=158; Fig. 1C). To detect
the potential role of EWSAT1 in glioma progression, the associated
gene expression profiles were explored by using the (GO) database
(https://david.ncifcrf.gov/). It was
observed that the most significant GO biological processes mainly
included cell invasion and adhesion (Fig. 1D). GBM is the most malignant and
aggressive primary nervous system tumor worldwide (1,2), thus it
is essential to detect the clinical characteristics of patients
with GBM (Grade IV in glioma), according to EWSAT1 expression in
tissues (GBM, n=42). In the clinical GBM samples, the results of
the present study demonstrated that the expression level of EWSAT1
exhibited a significant association with patient necrosis on
magnetic resonance imaging scans (grade IV; Table I; P=0.038). In addition, a higher
level of EWSAT1 was observed in patients with GBM (grade IV)
compared with that observed in adjacent normal brain tissues
(Fig. 1E). The Kaplan-Meier curves
also exhibited a worse prognosis in patients with glioma expressing
a high level of plasmacytoma variant translocation 1 (P=0.0014;
Fig. 1F). Thus, EWSAT1 was
identified to be involved in the progression of glioma.
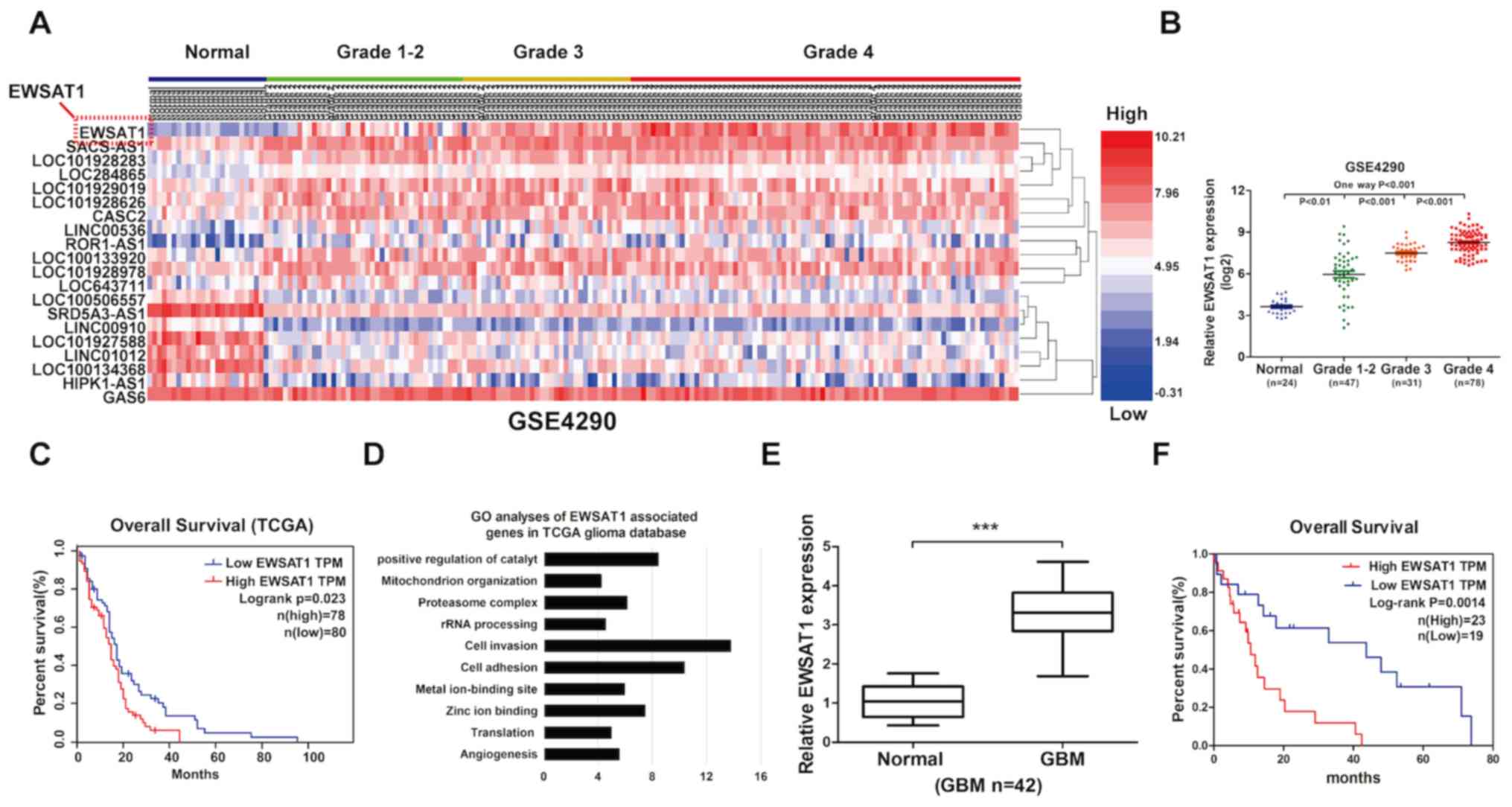 | Figure 1.lncRNA EWSAT1 is upregulated in human
GBM samples and is correlated with worse survival. (A) A heatmap
demonstrated the aberrant expression of lncRNAs from the GEO
database. Red bar represents high expression, while blue bar
represents low expression. (B) Gene expression levels of EWSAT1 in
the GEO cohort compared with the normal cohort. (C) Kaplan-Meier
curve of overall survival in the GEO database, demonstrating that a
high level of EWSAT1 is associated with poor prognosis (n=158). (D)
Gene function of EWSAT1-associated genes were identified using GO
analysis. (E) An increased gene expression level of EWSAT1 was
present in clinical tissues of patients with GBM (n=42) compared
with matched adjacent normal tissue, as determined by reverse
transcription-quantitative PCR analysis. (F) Kaplan-Meier curve of
overall survival in patients with glioma indicated that a high
level of EWSAT1 was correlated with worse prognosis (n=42). Each
experiment was performed in triplicate. ***P<0.001. lncRNA, long
non-coding RNA; EWSAT1, Ewing sarcoma associated transcript 1; GBM,
glioblastoma; GO, Gene Ontology; TCGA, The Cancer Genome Atlas;
GEO, Gene Expression Omnibus; TPM, Trans Per Kilobase of exon model
per Million. |
Knockdown of EWSAT1 suppresses GBM
cell viability and invasion
As aforementioned, upregulated EWSAT1 levels were
demonstrated in glioma cells. Therefore, it was hypothesized that
EWSAT1 serves as a regulator of glioma development. To confirm
this, the level of EWSAT1 was detected in glioma cell lines (U251,
LN229, T98G, A172 and SHG44) and compared with those in NHA cells
by RT-qPCR. The results demonstrated that, compared with the NHA
cells, the most significant increases in EWSAT1 levels were
observed in the U251 and T98G cells (Fig. 2A). Then, an siRNA was used to silence
EWSAT1 expression, and significantly decreased expression levels of
EWSAT1 were observed compared with those induced by si-NC in both
U251 and T98G cells (Fig. 2B). The
result of the CCK-8 assay revealed inhibited cell viability in
EWSAT1-silenced U251 and T98G cells compared with that of cells
transfected with the si-NC following 24–72 h transfection (Fig. 2C). As demonstrated by the results of
the Transwell assay, EWSAT1 silencing decreased the number of
invasive cells (Fig. 2D). These
results revealed that silencing EWSAT1 repressed cell proliferative
and invasive abilities in vitro.
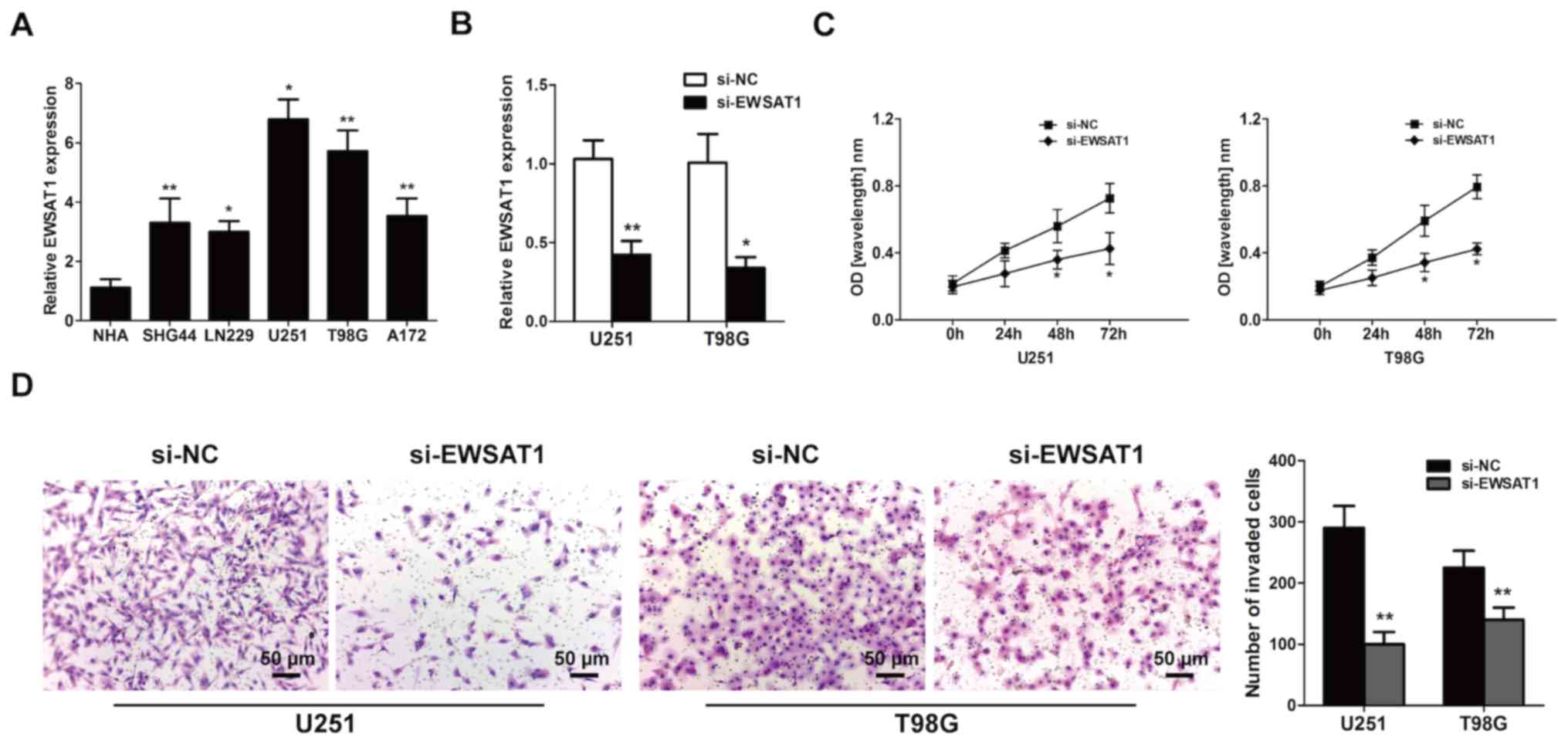 | Figure 2.Silencing of the long non-coding RNA
EWSAT1 inhibits the proliferative and invasive abilities of
glioblastoma cell lines in vitro. (A) The EWSAT1 gene
expression levels in the GBM U251, LN229, T98G, A172 and SHG44 cell
lines compared with those in normal human astrocyte cells were
assessed by RT-qPCR analysis. (B) EWSAT1 gene expression was
efficiently knocked down by siRNA in U251 and T98G cells compared
with the si-NC group, as detected by RT-qPCR assay. (C) GBM cell
growth was measured using the Cell Counting Kit-8 assay (si-EWSAT1
vs. si-NC). (D) Transwell invasion assay was used to detect the
invasive ability. Data are presented as the mean ± standard
deviation of 3 independent experiments. Magnification, ×100; scale
bar, 50 µm. *P<0.05 and **P<0.01. EWSAT1, Ewing sarcoma
associated transcript 1; GBM, glioblastoma; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control; siRNA, small
interfering RNA. |
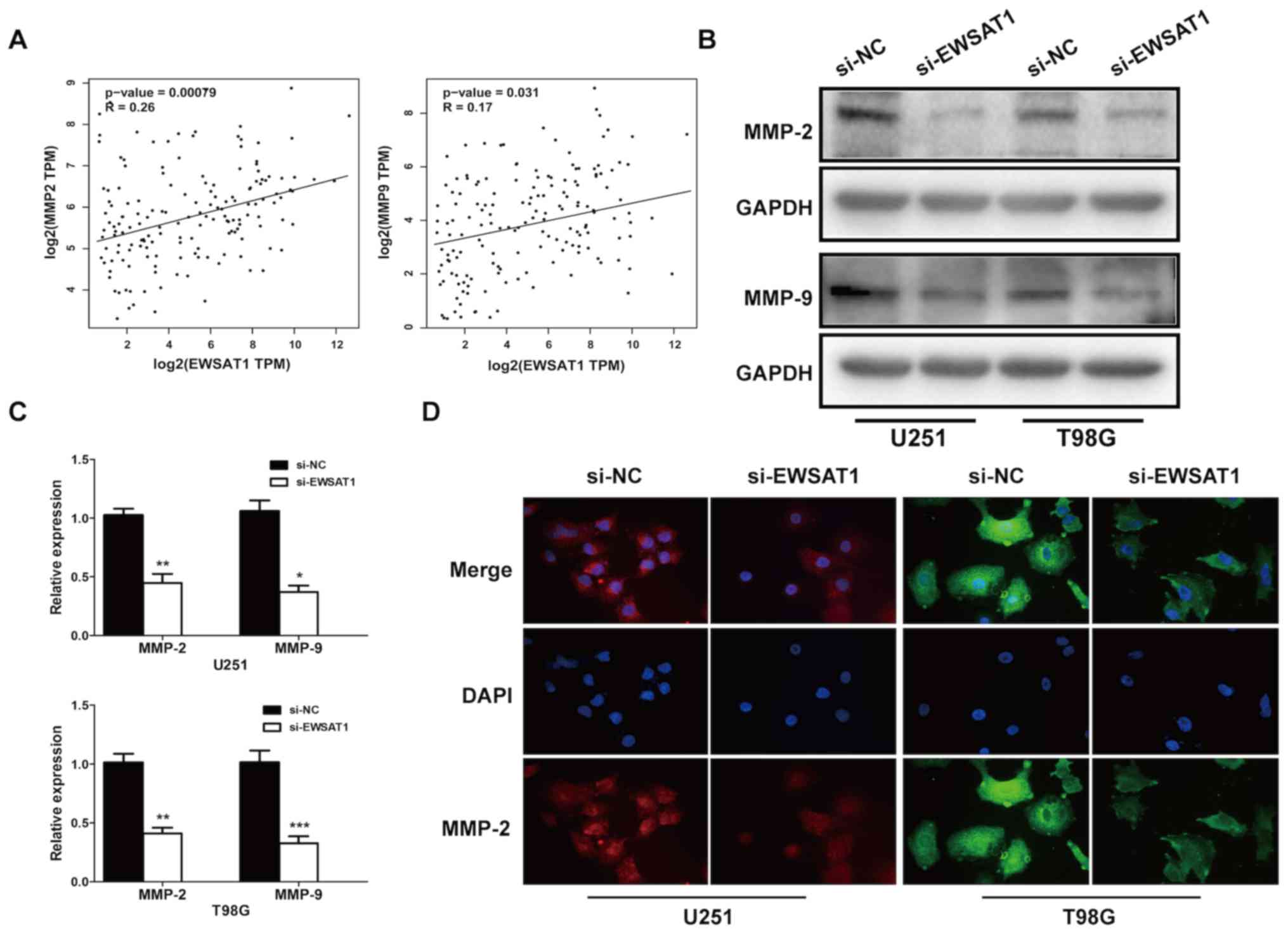
To further investigate the underlying mechanism by
which EWSAT1 affected the invasive capacity of GBM, TCGA database
was explored, and a significant positive correlation was identified
between EWSAT1 expression and the invasion-associated markers MMP-2
(r=0.26; P=0.00079) and MMP-9 (r=0.17; P=0.031) (Fig. 3A). Then, the mRNA, protein levels and
cellular location of MMP-2 and MMP-9 were determined. Knockdown of
EWSAT1 decreased the levels of MMP-2 and MMP-9 in glioma (Fig. 3B and C), and suppressed the
expression of the invasion-associated markers MMP-2 and MMP-9 in
glioma cells (Fig. 3D). Therefore,
these data suggested that EWSAT1 knockdown suppressed both
proliferation and invasion in GBM.
Correlation between EWSAT1 and
miR-152-3p
StarBase v2.0 (http://starbase.sysu.edu.cn) was employed to predict
potential lncRNA-miRNA interactions. A cohort of potential miRNAs
that could interact with EWSAT1 was predicted. Pearson's
correlation analysis suggested that there was an inverse
correlation between EWSAT1 and miR-152-3p level (n=158; r=−0.25;
P=0.0011; Fig. 4A). Then, miR-152-3p
expression was detected in 42 GBM and adjacent normal brain samples
by RT-qPCR, and a significantly decrease was identified in GBM
tissues (P<0.001; Fig. 4B). In
addition, the expression of miR-152-3p was significantly
upregulated in the EWSAT1-silenced group compared with the si-NC
group (Fig. 4C). A luciferase
reporter assay was subsequently used to confirm the putative
miR-152-3p target site (Fig. 4D).
Next, miR-152-3p mimics or inhibitor mimics were used to examine
transfection efficiency in the U251 and T98G cell lines (Fig. 4E). In addition, luciferase reporters
carrying either the predicted miR-152-3p WT (EWSAT1-WT) or its
mutated fragment (EWSAT1-MUT) were constructed. The results from
luciferase reporter assays showed that miR-152-3p significantly
suppressed the luciferase activity in the cells transfected with
the EWSAT1-WT but not mutant EWSAT1-MUT 3′-UTR (Fig. 4F). These data suggested that EWSAT1
may interact with miR-152-3p in glioma cells.
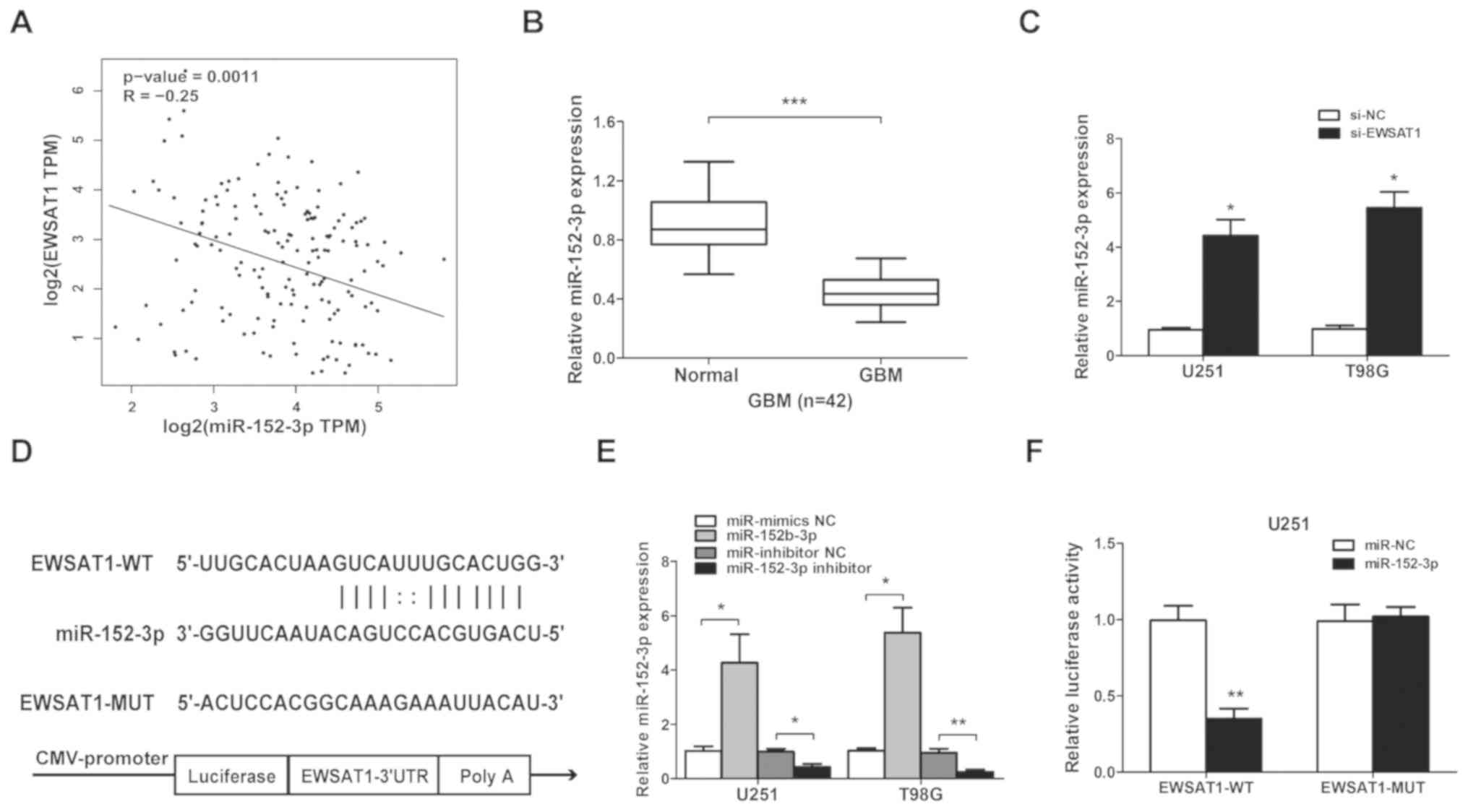 | Figure 4.Long non-coding RNA EWSAT1 is
targeted by miR-152-3p at the 3′-UTR. (A) Pearson's correlation
coefficient analysis between EWSAT1 and miR-152-3p expression
levels. (B) The levels of miR-152-3p in GBM compared with adjacent
normal tissues were detected using RT-qPCR analysis. ***P<0.001
(C) miR-152-3p expression was examined by RT-qPCR following
transfection with si-EWSAT1 or si-NC in GBM cell lines. (D) Target
site of miR-152-3p in the 3′-UTR of EWSAT1. (E) miR-152-3p
expression is upregulated by miR-152-3p mimics compared with
miR-mimics NC (*P<0.05), or inhibited by miR-152-3p inhibitor
compared with the miR-inhibitor NC (*P<0.05 and **P<0.01).
(F) The relative luciferase activity was identified following
co-transfection of miR-152-3p mimics compared with the miR-NC in
EWSAT1-WT- or EWSAT1-MUT-transfected U251 cells using the
dual-luciferase reporter assay. **P<0.01. The experiments were
repeated 3 times. EWSAT1, Ewing sarcoma associated transcript 1;
miR, microRNA; UTR, untranslated region; GBM, glioblastoma;
RT-qPCR, reverse transcription-quantitative PCR; NC, negative
control; si, small interfering; WT, wild-type; MUT, mutant. |
miR-152-3p inhibits the effects of
EWSAT1 in GBM cells
The Kaplan-Meier survival analysis demonstrated that
low miR-152-3p expression levels were associated with significantly
poorer survival in the GEPIA database (Fig. 5A). The level of miR-152-3p in glioma
cells was evaluated, and it was identified that miR-152-3p was
significantly inhibited in these cells compared with NHA cells
(Fig. 5B). Then, rescue experiments
were performed using U251 cells. Transfection of the miR-152-3p
inhibitor in the si-EWSAT1-transfected group led to a decreased
level of miR-152-3p compared with the miR-inhibitor NC (Fig. 5C). The level of miR-152-3p increased
when the cells were transfected with si-EWSAT1. However, following
treatment with the miR-152-3p inhibitor, the inhibition effect on
cell invasion induced by EWSAT1 knockdown was also impeded in the
U251 cells (Fig. 5D). These data
indicated that miR-152-3p was a vital mediator of EWSAT1-regulated
proliferation and invasion processes.
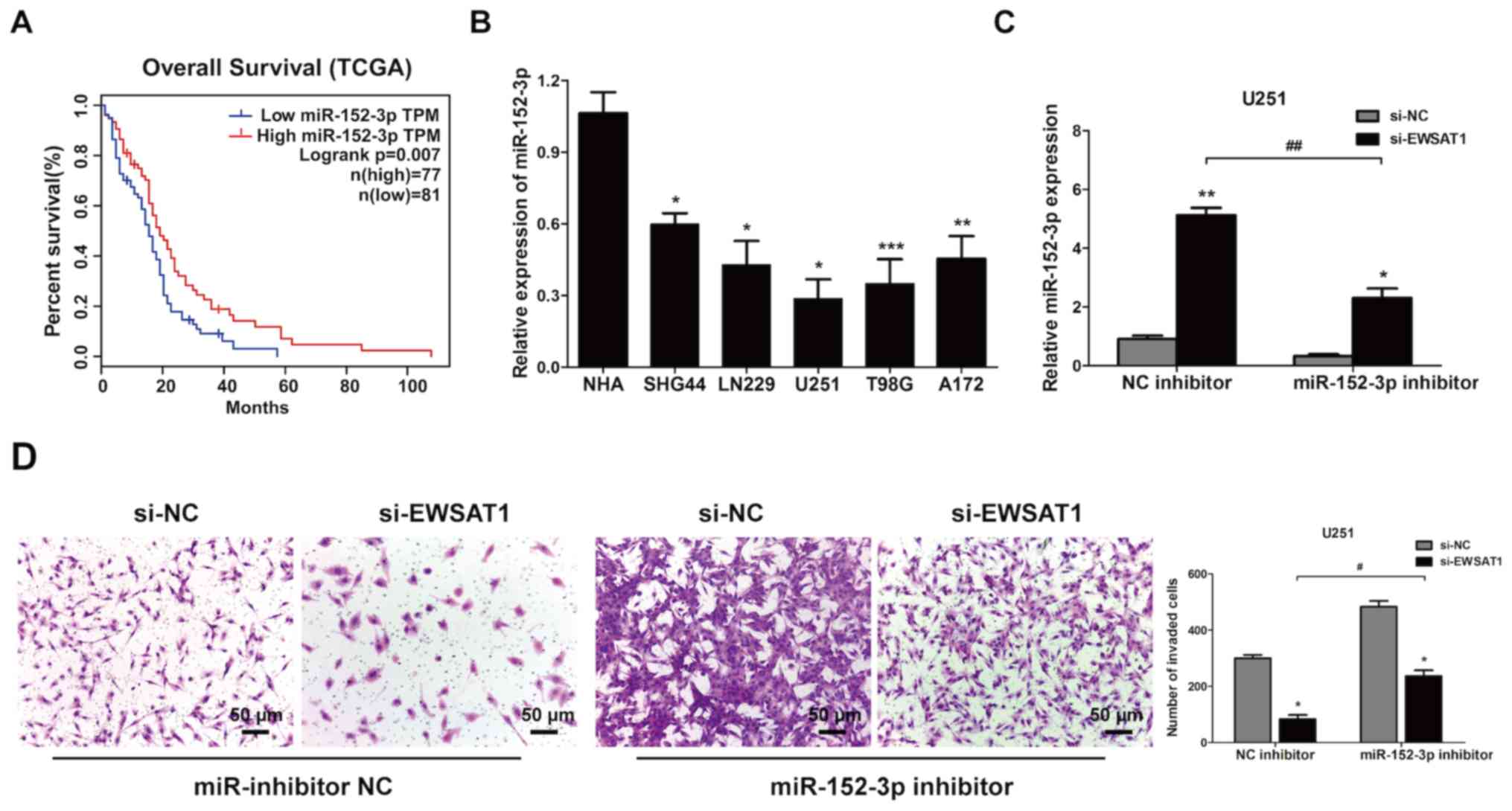 | Figure 5.miR-152-3p inhibits the effect of
long non-coding RNA EWSAT1 in GBM cells. (A) Kaplan-Meier survival
curve analysis of patient data from The Cancer Genome Atlas
database with high or low levels of miR-152-3p (n=158). (B)
miR-152-3p levels in GBM cell lines compared with normal human
astrocyte cells were explored by reverse transcription-quantitative
PCR. *P<0.05, **P<0.01 and ***P<0.001 vs. NHA cells. (C)
The gene expression levels of miR-152-3p were determined in U251
cells co-transfected with si-EWSAT1 + miR-152-3p inhibitor, and
with si-EWSAT1 + miR-inhibitor NC (*P<0.05 and **P<0.01 vs.
NC), and in cells co-transfected with si-EWSAT1 + miR-152-3p
inhibitor compared with si-EWSAT1 + miR-inhibitor NC
(##P<0.01). (D) The invasive abilities of the cells
were verified using Matrigel assays following transfection with
si-EWSAT1 and miR-152-3p inhibitors. Magnification, ×100; scale
bar, 50 µm. *P<0.05 vs. NC. #P<0.05. The
experiment was repeated 3 times. miR, microRNA; EWSAT1, Ewing
sarcoma associated transcript 1; GBM, glioblastoma; NC, negative
control; si, small interfering. |
Discussion
GBM is the most common and aggressive type of
central nervous system tumor. Due to its infiltrative growth
pattern, anti-apoptotic nature and resistance to chemotherapy,
total surgical resection is difficult to perform for this malignant
brain tumor. Therefore, it is essential to explore its basic
biological mechanisms, and to develop novel targets for recurrence
prevention and therapy. It has been previously demonstrated that
lncRNAs are involved in a variety of human carcinomas; therefore,
an in-depth analysis of these molecules may provide an
understanding of the diagnosis, prognosis and therapeutic methods
of human cancer (29–32). Moreover, the mechanism by which
EWSAT1 serves as an oncogene in GBM requires further study.
In the present study, the level of lncRNA EWSAT1,
which is a novel tumor suppressor located on chromosome 15q23, was
significantly increased in glioma samples. EWSAT1 overexpression
was markedly associated with a poorer prognosis. Moreover, it was
demonstrated that EWSAT1 promoted cell proliferation and invasion
using gain and loss-of-function assays in vitro. The level
of miR-152-3p was decreased in GBM tissues. The expression of
miR-152-3p was negatively correlated with that of EWSAT1. By using
bioinformatics analysis, miR-152-3p was identified as a promising
target of EWSAT1. Together, these results indicated that EWSAT1
exerted a oncogene role in GBM progression, suggesting a potential
therapeutic method for GBM.
Previous studies have reported that lncRNAs are
potential miRNA sponges (33,34).
Silencing of the lncRNA SCAMP1 inhibits malignant biological
behavior in glioma through the miR-499a-5p/LIM homeobox
transcription factor 1 alpha/NLR family CARD domain containing 5
signaling pathway (35). HOX
transcript antisense RNA directly inhibited WIF-1 level by
regulating the promoter region methylation of its histone H3K27,
and then activating the Wnt/β-catenin signaling pathway (36). In hepatocellular carcinoma, Zhang
et al (37) demonstrated that
FLVCR1-AS1 sponges miR-513c to regulate tumor biological
functions.
Multiple previous studies have revealed that the
abnormal expression of miRNAs serves a vital role in the occurrence
and progression of cancer (38).
Feng et al (39) showed that
miR-152-3p serves as direct regulator of KLF4 in prostate cancer.
It was reported that increased EWSAT1 expression was associated
with poor outcomes in patients with osteosarcoma (15). Zhang et al (16) suggested that the downregulation of
the lncRNA EWSAT1 suppressed cell proliferation and invasion of
CRC, which indicated that EWSAT1 may be a potential target of CRC
treatment. The lncRNA EWSAT1 promotes ovarian cancer progression
through regulating the expression of miR-330-5p (17). Thus, these data appear to indicate an
association between the lncRNA EWSAT1 and miR-152-3p in GBM.
In the present study, the putative binding site
between EWSAT1 and miR-152-3p was identified using a luciferase
reporter assay system; the results confirmed EWSAT1 as a potential
tumor suppressor that regulates miR-152-3p, subsequently inhibiting
the growth and invasion of glioma. The suppressive effect of
si-EWSAT1 was reversed when co-transfected with miR-152-3p
inhibitor. Collectively, these results on the role of EWSAT1, and
the mechanisms underlying the proliferation and invasion of glioma
cells, provide essential information on its role in
tumorigenesis.
In conclusion, EWSAT1 acts as an oncogenic lncRNA
that facilitates the development and progression of GBM through
miR-152-3p, indicating that EWSAT1 may function as a potential
biomarker and therapeutic target for GBM.
Acknowledgements
Not applicable.
Funding
The present study was supported by Heilongjiang
Health and Health Committee Scientific Research (grant no.
2018377).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HY and WC designed the study, performed experiments,
analyzed the data and wrote the manuscript. GJ performed the in
vitro experiments. JY and WW analyzed the data and drafted the
manuscript. HL designed and supervised the study, and edited the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Institutional Review Board of Jiamusi University
(Jiamusi, China; approval no. JUIRBR-2019-214) and all procedures
were performed in accordance with the principles outlined in The
Declaration of Helsinki. Written informed consent was provided by
all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lai NS, Wu DG, Fang XG, Lin YC, Chen SS,
Li ZB and Xu SS: Serum microRNA-210 as a potential noninvasive
biomarker for the diagnosis and prognosis of glioma. Br J Cancer.
112:1241–1246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maher EA, Furnari FB, Bachoo RM, Rowitch
DH, Louis DN, Cavenee WK and DePinho RA: Malignant glioma: Genetics
and biology of a grave matter. Genes Dev. 15:1311–1333. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Davies E, Clarke C and Hopkins A:
Malignant cerebral glioma-i: Survival, disability, and morbidity
after radiotherapy. BMJ. 313:1507–1512. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sathornsumetee S, Reardon DA, Desjardins
A, Quinn JA, Vredenburgh JJ and Rich JN: Molecularly targeted
therapy for malignant glioma. Cancer. 110:13–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li C, Jing H, Ma G and Liang P: Allicin
induces apoptosis through activation of both intrinsic and
extrinsic pathways in glioma cells. Mol Med Rep. 17:5976–5981.
2018.PubMed/NCBI
|
|
6
|
Chen CC, Taniguchi T and D'Andrea A: The
Fanconi anemia (FA) pathway confers glioma resistance to DNA
alkylating agents. J Mol Med (Berl). 85:497–509. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tamura R, Tanaka T, Miyake K, Yoshida K
and Sasaki H: Bevacizumab for malignant gliomas: Current
indications, mechanisms of action and resistance, and markers of
response. Brain Tumor Pathol. 34:62–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li C, Zheng H, Hou W, Bao H, Xiong J, Che
W, Gu Y, Sun H and Liang P: Long non-coding RNA linc00645 promotes
TGF-β-induced epithelial-mesenchymal transition by regulating
miR-205-3p-ZEB1 axis in glioma. Cell Death Dis. 10:7172019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Q, Sun S, Yu W, Jiang J, Zhou F, Qiu
G, Xu S and Jiang X: Altered expression of long non-coding RNAs
during genotoxic stress-induced cell death in human glioma cells. J
Neurooncol. 112:283–292. 2015. View Article : Google Scholar
|
|
12
|
Vital AL, Tabernero MD, Castrillo A,
Rebelo O, Tão H, Gomes F, Nieto AB, Resende Oliveira C, Lopes MC
and Orfao A: Gene expression profiles of human glioblastomas are
associated with both tumor cytogenetics and histopathology. Neuro
Oncol. 12:991–1003. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kong L, Li X, Wang H, He G and Tang A:
Calycosin inhibits nasopharyngeal carcinoma cells by influencing
EWSAT1 expression to regulate the TRAF6-related pathways. Biomed
Pharmacother. 106:342–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song P and Yin SC: Long non-coding RNA
EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro
by targeting miR-326/-330-5p. Aging (Albany NY). 8:2948–2960. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang GY, Zhang JF, Hu XM, Luo ZP and Ma
YZ: Clinical significance of long non-coding RNA EWSAT1 as a novel
prognostic biomarker in osteosarcoma. Eur Rev Med Pharmacol Sci.
21:5337–5341. 2017.PubMed/NCBI
|
|
16
|
Zhang R, Li JB, Yan XF, Jin K, Li WY, Xu
J, Zhao J, Bai JH and Chen YZ: Increased EWSAT1 expression promotes
cell proliferation, invasion and epithelial-mesenchymal transition
in colorectal cancer. Eur Rev Med Pharmacol Sci. 22:6801–6808.
2018.PubMed/NCBI
|
|
17
|
Fu X, Zhang L, Dan L, Wang K and Xu Y:
LncRNA EWSAT1 promotes ovarian cancer progression through targeting
miR-330-5p expression. Am J Transl Res. 9:4094–4103.
2017.PubMed/NCBI
|
|
18
|
Marques Howarth M, Simpson D, Ngok SP,
Nieves B, Chen R, Siprashvili Z, Vaka D, Breese MR, Crompton BD,
Alexe G, et al: Long noncoding RNA EWSAT1-mediated gene repression
facilitates Ewing sarcoma oncogenesis. J Clin Invest.
124:5275–5290. 2014. View
Article : Google Scholar
|
|
19
|
Sempere LF, Freemantle S, Pitha-Rowe I,
Moss E, Dmitrovsky E and Ambros V: Expression profiling of
mammalian microRNAs uncovers a subset of brain-expressed microRNAs
with possible roles in murine and human neuronal differentiation.
Genome Biol. 5:R132004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Malzkorn B, Wolter M, Liesenberg F,
Grzendowski M, Stühler K, Meyer HE and Reifenberger G:
Identification and functional characterization of microRNAs
involved in the malignant progression of gliomas. Brain Pathol.
20:539–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun J, Tian X, Zhang J, Huang Y, Lin X,
Chen L and Zhang S: Regulation of human glioma cell apoptosis and
invasion by miR-152-3p through targeting DNMT1 and regulating NF2:
MiR-152-3p regulate glioma cell apoptosis and invasion. J Exp Clin
Cancer Res. 36:1002017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ge S, Wang D, Kong Q, Gao W and Sun J:
Function of miR-152 as a tumor suppressor in human breast cancer by
targeting PIK3CA. Oncol Res. 25:1363–1371. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu X, Li J, Qin F and Dai S: MiR-152 as a
tumor suppressor microRNA: Target recognition and regulation in
cancer. Oncol Lett. 11:3911–3916. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
General Assembly of the World Medical
Association, . World Medical Association Declaration of Helsinki:
Ethical principles for medical research involving human subjects. J
Am Coll Dent. 81:14–18. 2014.PubMed/NCBI
|
|
25
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:(Database Issue).
D991–D995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kong B, Yang T, Chen L, Kuang YQ, Gu JW,
Xia X, Cheng L and Zhang JH: Protein-protein interaction network
analysis and gene set enrichment analysis in epilepsy patients with
brain cancer. J Clin Neurosci. 21:316–319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
29
|
Xu T, Lin CM, Cheng SQ, Min J, Li L, Meng
XM, Huang C, Zhang L, Deng ZY and Li J: Pathological bases and
clinical impact of long noncoding RNAs in prostate cancer: A new
budding star. Mol Cancer. 17:1032018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lalevee S and Feil R: Long noncoding RNAs
in human disease: Emerging mechanisms and therapeutic strategies.
Epigenomics. 7:877–879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bhan A and Mandal SS: Long noncoding RNAs:
Emerging stars in gene regulation, epigenetics and human disease.
ChemMedChem. 9:1932–1956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yarmishyn AA and Kurochkin IV: Long
noncoding RNAs: A potential novel class of cancer biomarkers. Front
Genet. 6:1452015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu Y, Deng C, Zhang H, Zhang J, Peng B and
Hu C: Long non-coding RNA XIST promotes cell growth and metastasis
through regulating miR-139-5p mediated Wnt/β-catenin signaling
pathway in bladder cancer. Oncotarget. 8:94554–94568. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li C, Wan L, Liu Z, Xu G, Wang S, Su Z,
Zhang Y, Zhang C, Liu X, Lei Z and Zhang HT: Long non-coding RNA
XIST promotes TGF-β-induced epithelial-mesenchymal transition by
regulating miR-367/141-ZEB2 axis in non-small cell lung cancer.
Cancer Lett. 418:185–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zong Z, Song Y, Xue Y, Ruan X, Liu X, Yang
C, Zheng J, Cao S, Li Z and Liu Y: Knockdown of LncRNA SCAMP1
suppressed malignant biological behaviours of glioma cells via
modulating miR-499a-5p/LMX1A/NLRC5 pathway. J Cell Mol Med.
23:5048–5062. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY,
Xue WQ, Chen YB, Zhang Y and Jia WH: HOTAIR, a prognostic factor in
esophageal squamous cell carcinoma, inhibits WIF-1 expression and
activates Wnt pathway. Cancer Sci. 104:1675–1682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang K, Zhao Z, Yu J, Chen W, Xu Q and
Chen L: LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate
cancer cell proliferation, migration, and invasion in
hepatocellular carcinoma. J Cell Biochem. 119:6045–6056. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Feng F, Liu H, Chen A, Xia Q, Zhao Y, Jin
X and Huang J: miR-148-3p and miR-152-3p synergistically regulate
prostate cancer progression via repressing KLF4. J Cell Biochem.
120:17228–17239. 2019. View Article : Google Scholar : PubMed/NCBI
|