Introduction
Gastric cancer (GC) is the second most commonly
diagnosed cancer among men and the third among women, and it was
the second leading cause of cancer-associated death worldwide in
2015 (1). Due to the lack of
specific early symptoms, the majority of patients are not diagnosed
until they have advanced stage GC. The overall prognosis of
patients with GC is poor and the 5-year survival rate is <30%
(2–4). GC is a heterogeneous disease
characterized by different molecular and histological profiles
(5), therefore it is important to
identify novel sensitive and specific biomarkers for early
diagnosis. In addition, a more comprehensive understanding of tumor
suppressor genes may provide novel insight into GC
therapeutics.
MicroRNAs (miRNAs/miRs) are a conserved group of
single-stranded non-coding RNAs that are 17–25 nucleotides long
(6). miRNAs directly bind to the
3′-untranslated region of their target mRNA to regulate gene
expression after transcription, thereby inhibiting translation or
inducing mRNA degradation (7).
miRNAs are involved in carcinogenesis, including tumor initiation
and disease progression (8,9). In cancer, miRNAs can function as
oncogenes or tumor suppressors depending on the function of its
target gene (10,11). Recent studies have demonstrated that
several miRNAs are involved in tumor occurrence and can function as
oncogenes or tumor suppressor genes in GC (12–14). For
example, miR-6852 functions as a tumor suppressor by directly
targeting forkhead box J1 in gastric cancer (13). In addition, by targeting the B cell
lymphoma-2 gene, miR-744 can promote apoptosis in the GC cell line
SGC-7901 (14). Therefore, it is
important to explore the molecular mechanisms underlying miRNA
function in GC to promote the development of targeted
therapies.
The purpose of the present study was to investigate
the expression levels and function of miR-502-5p and its molecular
mechanisms in GC.
Materials and methods
Tissue samples, cells and
reagents
Between July 2017 and February 2018, 32 samples of
GC and adjacent tissues (5 cm away from tumor tissue) were obtained
from patients, including 19 male and 13 female subjects, and the
median age was 57 years (age range, 35–84 years). All patients
underwent gastrectomy at the Changhai Hospital of Naval Medical
University (Shanghai, China). Tissue samples from the patients with
GC were immediately flash frozen in liquid nitrogen following
resection at −196°C. The Changhai Hospital Ethics Committee
(Shanghai, China) approved the present study and all patients
provided written informed consent.
AGS and MKN45 human GC cells and GES human normal
gastric cells were purchased from the American Type Culture
Collection. AGS, MKN45 and GES-1 cells were cultured in RPMI-1640
medium (Hyclone; GE Healthcare Life Sciences) containing 10% fetal
bovine serum (Hyclone; GE Healthcare Life Sciences), 100 IU/ml
penicillin and 100 µg/ml streptomycin.
microRNA and cell transfection
miR-502-5p mimic and miR-502-5p-mimic negative
control (NC) was purchased from Shanghai GenePharma Co., Ltd. The
microRNA was transfected into GC cells using Lipofectamine 2000
reagent (Thermo Fisher Scientific Inc.) according to the
manufacturer's protocol. The time interval between transfection and
subsequent experimentation was 48 h.
RNA and reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from the patient tissue
samples, and AGS, MKN45 and GES cells using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific Inc.). miR-502-5p
expression levels were measured using a TaqMan microRNA assay kit
(Takara Bio. Inc.) according to the manufacturer's instructions,
using U6 as an internal control. Total RNA was then reverse
transcribed into cDNA using a Prime Script RT reagent kit (Takara
Bio. Inc.) according to the manufacturer's instructions. SYBR-Green
(Takara Bio. Inc.) was used to determine SP1 mRNA expression
relative to β-actin. The thermocycling conditions for qPCR were as
follows: 95°C for 5 min followed by 40 cycles of 95°C for 10 sec,
60°C for 30 sec. The primers were designed as follows: miR-502-5p
forward, 5′-CGGGCATCCTTGCTATCTG-3′ and reverse,
5′-CAGCCACAAAAGAGCACAAT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; SP1 forward,
5′-TGGCAGCAGTACCAATGGC-3′ and reverse,
5′-CCAGGTAGTCCTGTCAGAACTT-3′; and β-actin forward,
5′-CCTGGCACCCAGCACAAT-3′ and reverse: 5′-GGGCGGGACTCGTCATAC-3′.
Each sample was analyzed in triplicate and levels were quantified
using the 2−ΔΔCq method (15).
MTT cell proliferation assay
AGS and MKN45 cells were transfected with miR-502-5p
mimics or NC for 24 h as aforementioned. AGS and MKN45 cells were
collected and 5×103 cells per well were seeded into
96-well plates in triplicate. Following 1–4 days in the incubator
at 37°C with an atmosphere of 5% CO2, 10 µl MTT assay
solution was added to each well for 4 h at 37°C. Next, 100 µl DMSO
was added to each well for 30 min to dissolve the purple formazan,
and optical density was measured at 490 nm with a microplate reader
(Bio-Rad Laboratories, Inc.).
Migration and invasion Transwell
assays
In the migration assay, 1×105 cells were
plated in 200 µl serum-free medium in the top Transwell chamber. In
the invasive assay, 1×105 cells were plated in 200 µl
serum-free medium in the top Transwell chamber with a
Matrigel-coated membrane. The matrigel was pre-coated at 37°C for
30 min. In both the migration and invasion experiments, 500 µl
medium containing 10% FBS was added into the lower chamber as a
chemoattractant. After 24 h, the cells on the top surface of the
Transwell chamber were removed using cotton swabs. The cells on the
bottom surface were fixed at room temperature with 100% methanol
for 30 min, and then stained with 0.05% crystal violet for 30 min
at room temperature. Five visual fields were randomly selected to
photograph with an Olympus IX51 light microscope (Olympus
Corporation; magnification, ×20).
Western blot analysis
Transfected AGS and MKN45 cells were lysed with RIPA
buffer (Cell Signaling Technology, Inc.) containing complete
protease inhibitor cocktail (Roche Diagnostics), phosphatase
inhibitors (Roche Diagnostics), 5 mM dithiothreitol (DTT,
Sigma-Aldrich; Merck KGaA) and 1 mM phenyl methyl sulfonyl fluoride
(Sigma-Aldrich; Merck KGaA). The supernatant of the cell lysate was
collected and protein concentrations were determined using the
bicinchoninic protein assay kit (Thermo Fisher Scientific Inc.)
according to the manufacturer's instructions. Then 20 µg protein
was loaded onto a 10% gel, resolved using SDS-PAGE and transferred
onto PVDF membranes. Membranes were then blocked with 5% fat-free
milk for 2 h at room temperature. Subsequently, membranes were
incubated with primary antibodies against SP1 (1:1,000; cat. no.
WL02251; Wanleibio Co., Ltd.) and β-actin (1:1,000; cat. no.
P30002M; Abmart Pharmaceutical Technology Co., Ltd.) at 4°C
overnight, washed three times with TBST (0.05% Tween-20) and
incubated with anti-mouse horseradish peroxidase-conjugated
secondary antibody (1:2,000; cat. no. 7054S; Cell Signaling
Technology, Inc.) at room temperature for 2 h. Following three
washes with TBST, immunoreactive bands were visualized using ECL
working fluid (Biochannel, Nanjing, China; http:
//www.biochannel.cn/page19.html?product_id=299). This
experiment was repeated three times.
Identification of miR-502-5p target
genes
Target Scan version.7.2 (http: //www.targetscan.org/), miRandaversion.2010
(http: //www.microrna.org/microrna/getGeneForm.do)
and miRBase version.22.1 (http:
//www.mirbase.org/) were used to predict the candidate target
genes of miR-502-5p.
Statistical analysis
All data are expressed as the mean ± standard
deviation, and statistical analyses were performed using SPSS
version 17.0 (SPSS, Inc.). Student's paired t-tests were used for
comparisons between two groups and one-way ANOVA followed by
Tukey's post hoc test was used for multiple comparisons. P<0.05
was considered to indicate a statistically significant
difference.
Results
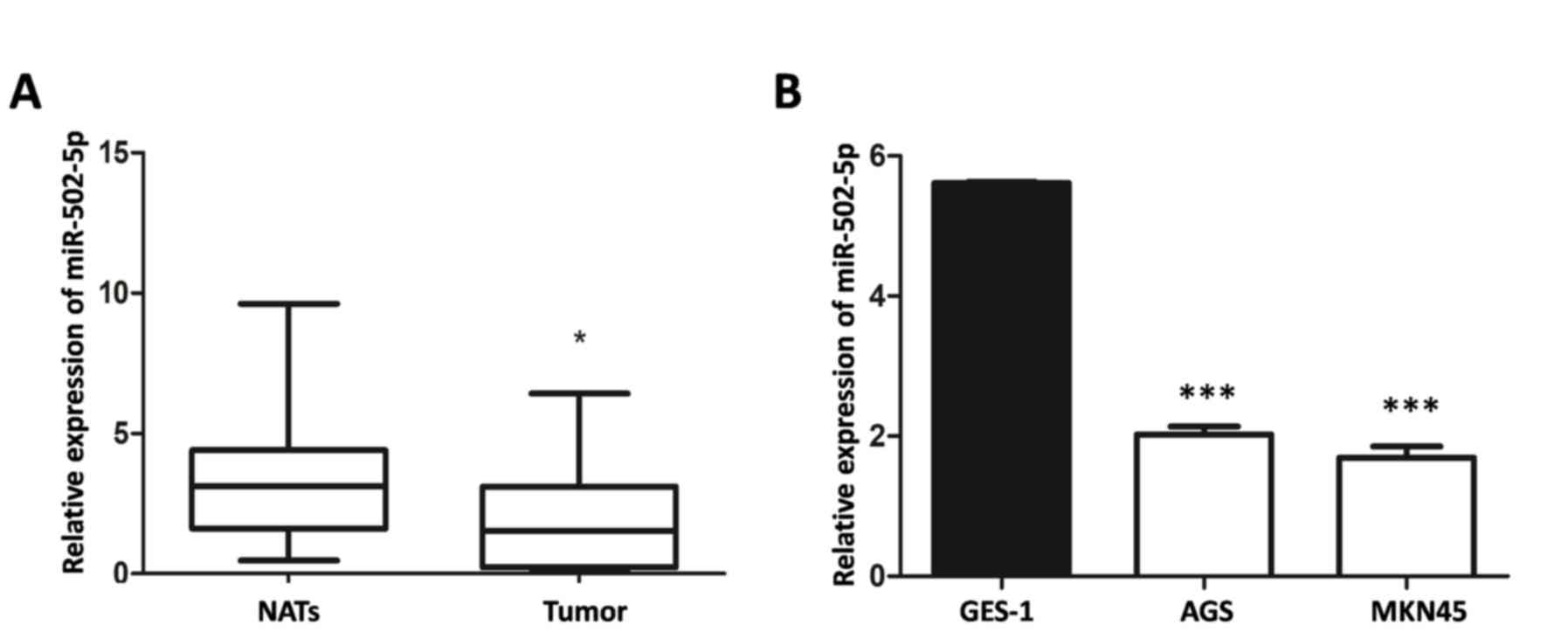
Expression of miR-502-5p in GC tissues
and cells
Expression levels of miR-502-5p in 32 GC tissues and
two GC cell lines were analyzed using RT-qPCR. The expression level
of miR-502-5p in GC tissues was significantly lower compared with
those in matched normal adjacent tissues (P<0.05; Fig. 1A). Compared with normal GES-1 gastric
epithelial cells, the two GC cell lines exhibited significantly
lower miR-502-5p expression (P<0.001; Fig. 1B).
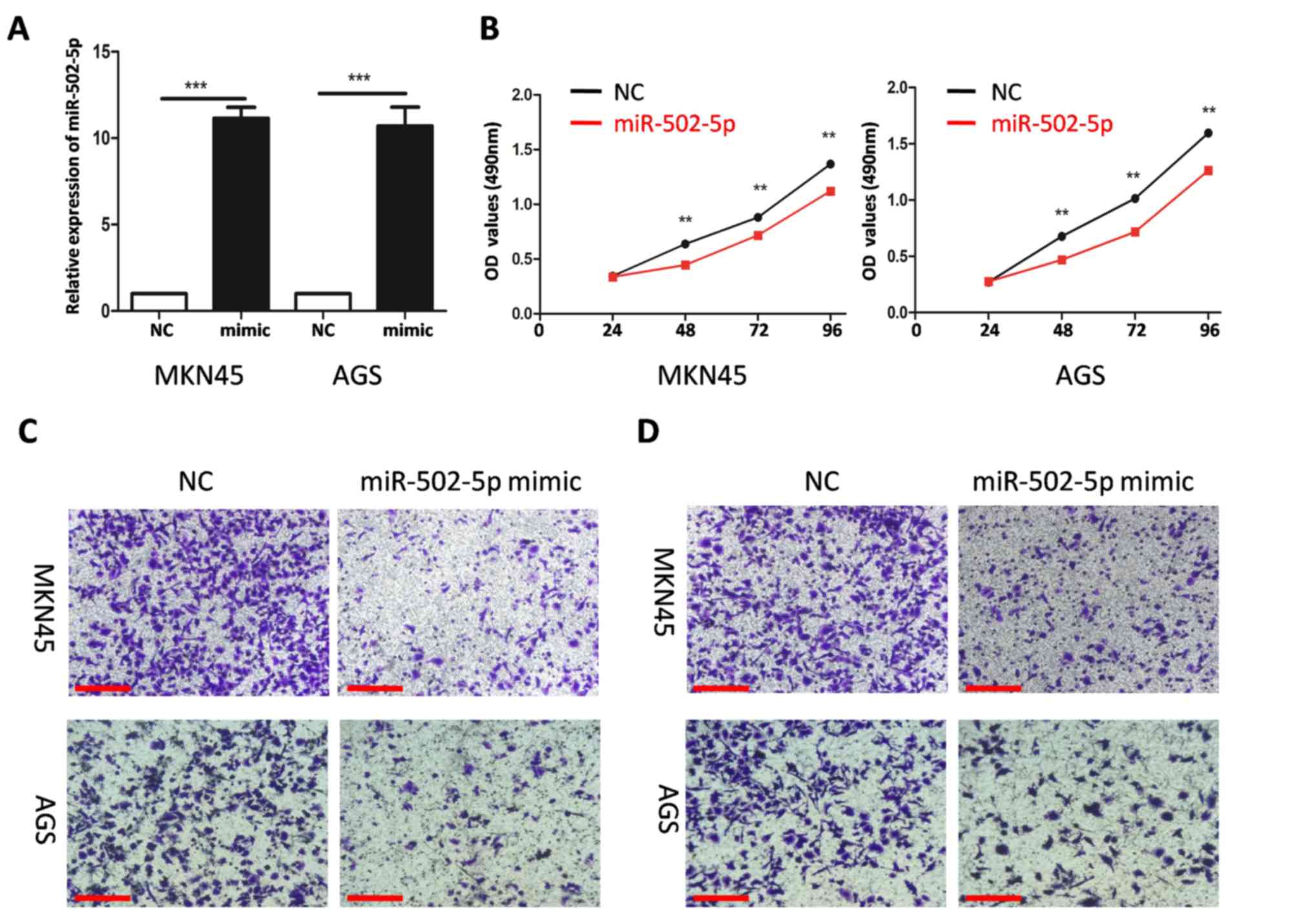
miR-502-5p inhibits the proliferation,
migration and invasion of GC cells
To study the effect of miR-502-5p on GC cells, NC or
mimics of miR-502-5p were transfected into MKN45 and AGSGC cells.
The transfection efficiency of miR-502-5p was evaluated using
RT-qPCR. The level of miR-502-5p in MKN45 and AGS cells transfected
with miR-502-5p mimics was significantly higher compared with those
transfected with NC (both P<0.001; Fig. 2A). Overexpression of miR-502-5p
significantly decreased proliferation at all time points compared
with the NC group (all P<0.01; Fig.
2B) and reduced the cellular migration and invasion capacities
of MKN45 and AGSGC cells (Fig. 2C and
D).
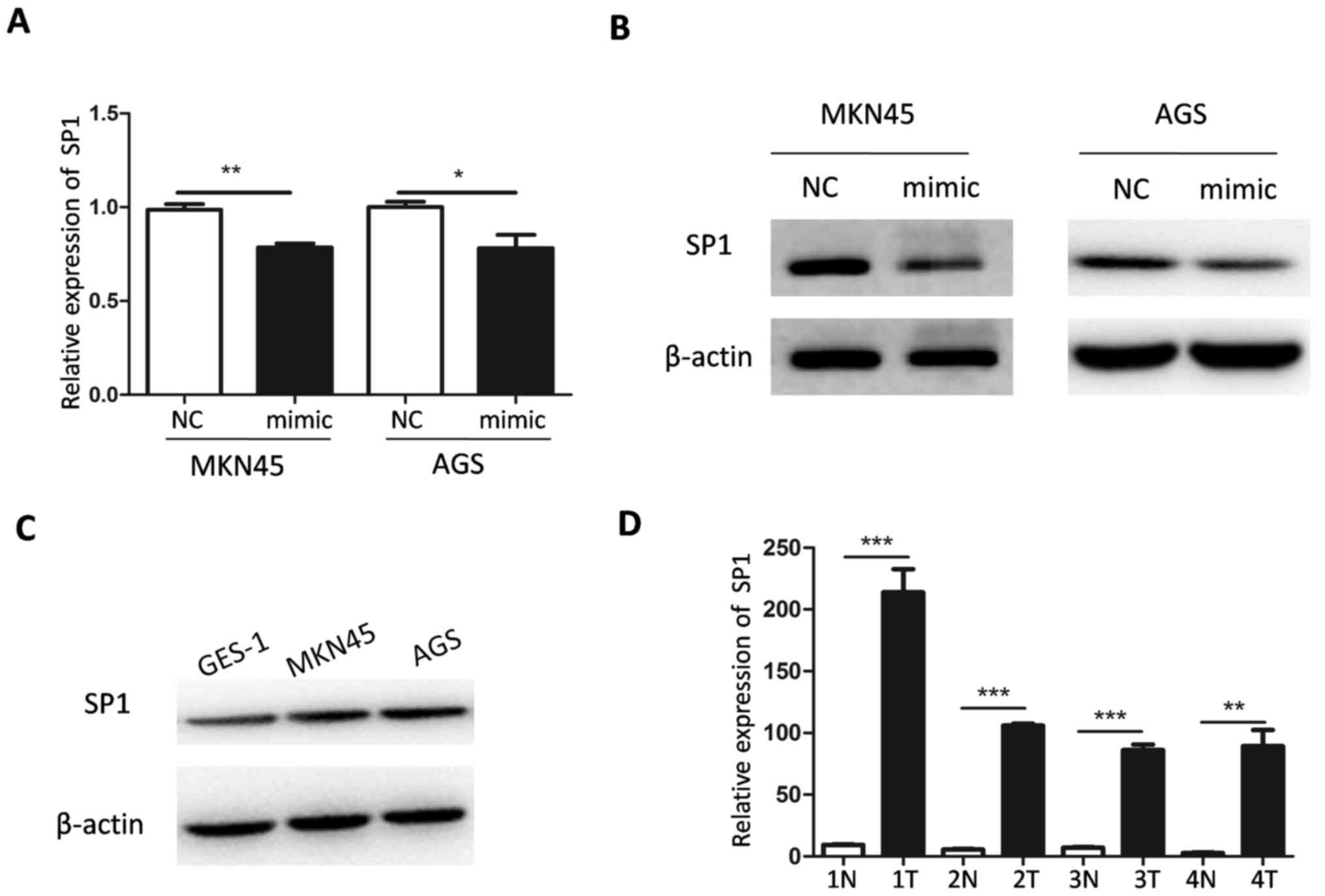
miR-502-5p targets SP1
To investigate the molecular mechanism underlying
miR-502-5p-mediated inhibition of GC progression, target genes of
miR-502-5p were predicted using miRNA prediction software and
databases. Target Scan 7.2, miR and a 2010 and miR Base 22.1
predicted that SP1 is a target gene of miR-502-5p. The mRNA level
of SP1 decreased significantly in AGS and MKN45 cells transfected
with miR-502-5p mimic (P<0.05 and P<0.01, respectively,
Fig. 3A). In addition, western
blotting demonstrated that the protein level of SP1 was lower in
AGS and MKN45 cells transfected with miR-502-5p mimics compared
with the controls. SP1 levels were also evaluated in GC cells.
Compared with GES cells, the expression of miR-502-5p in AGS and
MKN45 cells was lower (Fig. 1B), but
the expression of SP1 was higher (Fig.
3C). The mRNA expression level of SP1 in four sets of tumor
tissues was significantly higher compared with that in normal
paracancerous tissues (Fig. 3D).
This suggests that SP1 is targeted by miR-502-5p in GC.
Discussion
In GC, miRNAs are considered as novel potential
diagnostic biomarkers, prognostic factors and therapeutic targets
(16). Abnormal expression of miRNAs
in GC has previously been reported (13,14).
However, the specific role and subsequent underlying molecular
mechanisms of miR-502-5p in GC remain unclear. To the best of our
knowledge, the present study is the first to report the expression,
biological function and molecular mechanism of miR-502-5p in GC.
The level of miR-502-5p was downregulated in GC tissues and cell
lines, and, in functional analyses, miR-502-5p significantly
decreased the proliferative, migratory and invasive properties of
GC cells.
Aberrant expression of miR-502-5p is associated with
tumorigenesis and is a tumor suppressor in hepatocellular
carcinoma, breast and colon cancer (17–19). In
hepatocellular carcinoma cells, miR-502-5p significantly inhibits
proliferation in vitro and tumor growth in vivo by
targetingphophatidylinositor-4, 5-bisphosphate 3-kinase catalytic
subunit ɣ (17). It has been
reported that miR-502-5p expression in MCF-7 and MDA-MB-231 cells
is low, and miR-502-5p can promote apoptosis and inhibit the
proliferation of breast cancer cells in vitro by binding to
the tumor necrosis factor receptor-associated factor 2 (TRAF2) gene
in breast cancer (18). Another
study indicated that miR-502 can inhibit autophagy, proliferation
and cell cycle progression in colon cancer cells in vitro.
Furthermore, miR-502 can inhibit colon cancer progression in mouse
tumor xenografts models in vivo (19). These previous studies suggest that
miR-502 may be implicated in tumor progression.
SP1 is a ubiquitous transcription regulator in human
cells that regulates proliferation, apoptosis and embryonic
development (20). SP1 also promotes
the invasion and metastasis of tumor cells by regulating cell
adhesion protein matrix metalloproteinase, urokinase-type
plasminogen activator and micro-vessel density in tumors (21). SP1 is abnormally expressed in gastric
cancer cells and participates in the proliferation and apoptosis of
these cells (22); however, the
relationship between SP1 and tumor metastasis is complex. For
example, in certain tumors, such as glioma (23) and colon cancer (24), the effect of SP1 on tumor metastasis
can be reduced by inhibiting the expression of SP1 in tumor cells.
However, in GC (25) and lung
adenocarcinoma (26), inhibiting the
expression of SP1 can promote the metastasis and invasion ability
of tumor cells.
The present study investigated the molecular
mechanisms underlying miR-502-5p function in GC. First,
bioinformatics was used to predict the potential target genes of
miR-502-5p, which identified SP1 as a candidate. Then, mRNA
expression levels of SP1 were measured in four GC tissue sets. It
was determined that SP1 mRNA levels were significantly higher
compared with those in normal adjacent tissues. In addition,
RT-qPCR and western blotting demonstrated that overexpression of
miR-502-5p decreased the expression levels of SP1 mRNA and protein
in GC cells, respectively. These results suggest that SP1 is a
downstream target gene of miR-502-5p.
In conclusion, the present study demonstrated that
miR-502-5p is a novel tumor suppressor, as overexpression of
miR-502-5p inhibited the proliferation, migration and invasion of
GC cells. Thus, downregulation of miR-502-5p may be necessary for
GC carcinogenesis via SP1 regulation. The present findings may
improve our understanding of the molecular pathogenesis of GC and
highlight the potential of miR-502-5p as a target for antitumor
therapy. However, the present study also has some limitations. For
example, the effect of miR-502-5p on the biological behavior of GC
cells at the cellular level was only investigated in vitro.
In the future, further studies should be conducted to demonstrate
the effect ofmiR-502-5p on the biological behavior of GC in
vivo.
Acknowledgements
Not applicable.
Funding
The present study was funded by The National Natural
Science Foundation of China (grant no. 81672892).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XP and MW designed the study. WL and CG performed
the data analysis. CG sorted out the experimental data. XP, LZ and
XZ performed the data analyses and wrote the manuscript. LZ taught
the experimental protocols. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The Changhai Hospital Ethics Committee (Shanghai,
China) approved the present study (approval no. CHEC2016-157). All
patients who agreed to participate in the study provided written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bria E, De Manzoni G, Beghelli S,
Tomezzoli A, Barbi S, Di Gregorio C, Scardoni M, Amato E, Frizziero
M, Sperduti I, et al: A clinical-biological risk stratification
model for resected gastric cancer: Prognostic impact of Her2, Fhit,
and APC expression status. Ann Oncol. 24:693–701. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng PL, Zhou XY, Yi GD, Chen PF, Wang F
and Dong WG: Identification of a novel gene pairs signature in the
prognosis of gastric cancer. Cancer Med. 7:344–350. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Akhondi-Meybodi M, Ghane M,
Akhondi-Meybodi S and Dashti G: Five-year survival rate for gastric
cancer in Yazd Province, Central Iran, from 2001 to 2008. Middle
East J Dig Dis. 9:39–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bass AJ, Thorsson V, Shmulevich I,
Reynolds SM, Miller M, Bernard B, Hinoue T, Laird PW, Curtis C,
Shen H, et al: Comprehensive molecular characterization of gastric
adenocarcinoma. Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lei T, Zhu Y, Jiang C, Wang Y, Fu J, Fan Z
and Qin H: MicroRNA-320 was downregulated in non-small cell lung
cancer and inhibited cell proliferation, migration and invasion by
targeting fatty acid synthase. Mol Med Rep. 14:1255–1262. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hagan JP and Croce CM: MicroRNAs in
carcinogenesis. Cytogenet Genome Res. 118:252–259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia E, Kanematsu S, Suenaga Y, Elzawahry
A, Kondo H, Otsuka N, Moriya Y, Iizasa T, Kato M, Yoshino I and
Yokoi S: MicroRNA induction by copy number gain is associated with
poor outcome in squamous cell carcinoma of the lung. Sci Rep.
8:153632018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kundu A, Quirit JG, Khouri MG and
Firestone GL: Inhibition of oncogenic BRAF activity by
indole-3-carbinol disrupts microphthalmia-associated transcription
factor expression and arrests melanoma cell proliferation. Mol
Carcinog. 56:49–61. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma X, Feng J, Lu M, Tang W, Han J, Luo X,
Zhao Q and Yang L: microRNA-501-5p promotes cell proliferation and
migration in gastric cancer by downregulating LPAR1. J Cell
Biochem. 121:1911–1922. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu H, Zhang J, Wen Q, Dai Y, Zhang W, Li F
and Li J: MicroRNA-6852 suppresses cell proliferation and invasion
via targeting forkhead box J1 in gastric cancer. Exp Ther Med.
16:3249–3255. 2018.PubMed/NCBI
|
|
14
|
Liu J, Wei Y, Li S, Li Y, Liu H, Liu J and
Zhu X: MicroRNA-744 promotes cell apoptosis via targeting B cell
lymphoma-2 in gastric cancer cell line SGC-7901. Exp Ther Med.
16:3611–3616. 2018.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cai H, Lin H, Cao W, Sun J, Huang Y and
Fang Y: Downregulation of miR-519a predicts poor prognosis and
contributes to tumor progression in gastric cancer. Oncol Res
Treat. 43:19–26. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen S, Li F, Chai H, Tao X, Wang H and Ji
A: miR-502 inhibits cell proliferation and tumor growth in
hepatocellular carcinoma through suppressing phosphoinositide
3-kinase catalytic subunit gamma. Biochem Biophys Res Commun.
464:500–505. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun LL, Wang J, Zhao ZJ, Liu N, Wang AL,
Ren HY, Yang F, Diao KX, Fu WN, Wan EH and Mi XY: Suppressive role
of miR-502-5p in breast cancer via downregulation of TRAF2. Oncol
Rep. 31:2085–2092. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhai H, Song B, Xu X, Zhu W and Ju J:
Inhibition of autophagy and tumor growth in colon cancer by
miR-502. Oncogene. 32:1570–1579. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Beishline K and Azizkhan-Clifford J: Sp1
and the ‘hallmarks of cancer’. FEBS J. 282:224–258. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chakraborty G, Rangaswami H, Jain S and
Kundu GC: Hypoxia regulates cross-talk between Syk and Lck leading
to breast cancer progression and angiogenesis. J Biol Chem.
281:11322–11331. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang X, Jiang Y, Xie Y, Leng X, He M and
Song F: Long noncoding RNA TRPM2-AS induced by SP1 inhibits cell
apoptosis via MAPK and STAT3 in gastric cancer. J Gastroenterol
Hepatol. May 18–2020.(Epub ahead of print). View Article : Google Scholar
|
|
23
|
Guan H, Cai J, Zhang N, Wu J, Yuan J, Li J
and Li M: Sp1 is upregulated in human glioma, promotes
MMP-2-mediated cell invasion and predicts poor clinical outcome.
Int J Cancer. 130:593–601. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kou XX, Hao T, Meng Z, Zhou YH and Gan YH:
Acetylated Sp1 inhibits PTEN expression through binding to PTEN
core promoter and recruitment of HDAC1 and promotes cancer cell
migration and invasion. Carcinogenesis. 34:58–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee HS, Park CK, Oh E, Erkin ÖC, Jung HS,
Cho MH, Kwon MJ, Chae SW, Kim SH, Wang LH, et al: Low SP1
expression differentially affects intestinal-type compared with
diffuse-type gastric adenocarcinoma. PLoS One. 8:e555222013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC,
Chang WC and Hung JJ: Sp1 expression regulates lung tumor
progression. Oncogene. 31:3973–3988. 2012. View Article : Google Scholar : PubMed/NCBI
|