Introduction
Extracellular vesicles (EVs) are cell-derived lipid
bilayer-enclosed vesicles with a diameter of nanoscale, which can
mediate intercellular communication through transferring proteins
and nucleic acids derived from donor cells. Almost all types of
cells can produce and release EVs, which play complicated roles in
various diseases, including cancer (1). Increasing evidence has indicated that
EVs secreted by host or tumor cells are extensively involved in
tumor development and progression (1–3). An
increasing number of studies have been focusing on the diagnostic
value of EV detection in various diseases, particularly cancer
(4–7).
EVs can carry various cargoes, such as proteins,
lipids and nucleic acids (8).
Furthermore, previous findings have revealed that EV-derived
nuclear DNA (EV nDNA) covers the entire nuclear genome and reflects
the mutational status of parental cells (9,10). To
date, several studies have detected mutations of EV DNA as a means
for early cancer screening, and findings have shown an advantage of
EV DNA over cell-free DNA (11,12), as
EV DNA it remains relatively intact due to protection of lipid
envelop from degradation by DNase (13). Therefore, the characterization of EV
DNA in plasma may provide useful biomarkers for the diagnosis and
clinical monitoring of cancer.
The mitochondrion is a double membrane-bound
organelle found in almost all eukaryotes, which contains its own
genetic DNA. Mitochondrial DNA (mtDNA) is a 16,569-base pair (bp)
circular chromosome that encodes 13 proteins essential for
respiratory energy metabolism. Due to the lack of histone
protection, limited DNA repair activities and oxidative stress
microenvironment in mitochondria, mtDNA is subject to sequence
mutations and copy number changes, which are closely associated
with various diseases, especially cancer (14,15). A
series of studies have observed a decrease in the mtDNA copy number
(termed mtDNA depletion) in several types of cancer, including
hepatocellular carcinoma (HCC) (16), which suggests that mtDNA depletion
may contribute to tumorigenesis. Zhao et al (17) also reported an association between
low mtDNA content in peripheral blood leukocytes and high-risk of
HBV-associated HCC. Furthermore, our previous study emphasized the
critical contributing role of somatic mtDNA D-loop mutations in
HBV-associated hepatocarcinogenesis (18). In addition, recent findings have
shown that the measurement of plasma cell-free mitochondrial tumor
DNA improves the detection of glioblastoma in patient-derived
orthotopic xenograft models (19).
These findings suggested that mtDNA may reveal unique advantages in
tumor diagnosis, treatment monitoring and evaluation of
prognosis.
The full mitochondrial genome was observed in EVs
(20). Furthermore, Sansone et
al (21) have reported that EVs
can package and transfer mtDNA into metabolically damaged breast
cancer cells, thereby restoring their metabolic activity and
leading to endocrine treatment resistance. Findings of the
aforementioned studies suggested that, compared with EV nDNA, EV
mtDNA may be a novel cancer detection marker. However, to date, the
full characteristics of mtDNA in EVs remain largely unexplored,
which greatly limits the clinical application of EV mtDNA detection
in patients with cancer.
In the present study, next-generation sequencing was
used to profile the entire EV DNA from patients with HCC. Moreover,
to the best of our knowledge, the EV mtDNA characteristics in
patients with HCC, hepatitis and healthy controls were
systematically analyzed and compared for the first time, laying a
foundation for the potential clinical application of EV mtDNA as a
liquid biopsy biomarker.
Materials and methods
Sample collection
A total of 15 patients with HBV-associated HCC, five
patients with hepatitis with HBV infection and five healthy
controls were recruited from Xijing Hospital, Fourth Military
Medical University (FMMU) in Xi'an, China between April 2018 and
September 2019. Patients with pathologically diagnosed HCC with HBV
were recruited, and there were no other comorbidities, such as HCV
or HIV infection. For patients with hepatitis with HBV infection,
no cirrhosis was observed by B-ultrasound.
Peripheral venous blood (10 ml per subject) was
collected from patients with hepatitis, healthy controls and
patients with HCC who had not received any treatment (such as
radiofrequency ablation, hepatectomy or transcatheter arterial
chemoembolization) prior to blood sample collection. Paired tumor
tissues and adjacent non-HCC tissues were collected in five
patients HCC who had undergone hepatectomy. The study was approved
by the Ethics Committee of FMMU and written consent was obtained
from each subject.
The clinical data of all subjects was obtained from
medical records for analysis, including: Personal data (age at
diagnosis and sex), blood test results (alphafetoprotein, aspartate
aminotransferase, alanine aminotransferase, γ-glutamyl
transpeptidase, total bilirubin, alkaline phosphatase, albumin),
Tumor-Node-Metastasis (TNM) stage and cirrhosis status. TNM stage
referred to TNM Staging System of AJCC (8th version) (22). Patient characteristics are listed in
Table SI.
Isolation of EVs from plasma
samples
Peripheral blood was drawn from the median cubital
vein in the antecubital fossa into EDTA-containing tubes and
centrifuged at 300 × g for 15 min to collect plasma within 2 h.
Plasma samples were centrifuged again at 11,200 × g for 30 min to
remove apoptotic bodies, mitochondrial particles and large cell
debris. Next, ~4 ml of plasma was centrifuged at 110,000 × g for 8
h. All centrifugation was performed at 4°C. The resulting EV pellet
was suspended in 50 µl PBS and stored at −20°C for further use. The
identification of EV was performed by transmission electron
microscopy, nanoparticle tracking analysis and western blot
analysis (Fig. S1).
Transmission electron microscopy
Similar to the previous description (23), freshly isolated plasma EVs were
dissolved in PBS buffer, dropped into a carbon-coated copper grid,
where they dried at room temperature. Then, the EVs were subjected
to negative staining with 1% uranyl acetate at room temperature for
1 min and washed twice with deionized water. After dried at room
temperature, the EVs were imaged by JEM-1400Plus transmission
electron microscope (JEOL, Inc).
Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) was to detect
the size distribution and concentration of the plasma EVs using
ZetaView version 8.04.02 (Particle Metrix, Inc). Isolated plasma
EVs were diluted with PBS buffer to 1:100 and resuspended before
being injected into the sample cell chamber.
Western blotting
Plasma EV isolations were lysed with RIPA buffer
(Beijing Solarbio Science & Technology Co., Ltd.). The protein
concentration in the EV lysate solution was determined using the
BCA protein assay kit (Beyotime Biotechnology, Inc). Proteins (30
µg) from plasma EVs were loaded per lane onto a 10% gel, resolved
using SDS-PAGE and then transferred to PVDF membranes (Bio-Rad,
Inc). The PVDF membranes were blocked with 5% non-fat milk in PBS
for 1 h at room temperature, and then probed separately with
anti-human CD9 (1:2,000, cat. no. ab92726, Abcam), anti-human CD63
(1:1,000, cat. no. ab134045, Abcam) and anti-human CD81 (1:1,000,
cat. no. ab109201, Abcam) for 16 h at 4°C. After washing, the blots
were incubated with horseradish peroxidase-conjugated goat
anti-rabbit secondary antibodies (1:10,000, cat. no. SA00001-2,
ProteinTech Group, Inc) for 1 h at room temperature. Protein
signals were visualized using Chemiscope 3000 mini
chemiluminescence imaging system and Chemi Capture software
(version 16.12.03A. Clinx, Inc. URL: http://www.clinx.cn/case_cat/info?id=49).
DNA extraction from EV, plasma and
tissue samples
To eliminate the contamination of ss- and ds-DNA,
each EV pellet sample was treated with 2U DNaseI (Thermo Fisher
Scientific, Inc.) for 30 min at 37°C prior to DNA extraction, and
then DNaseI was heat-inactivated by incubation for 10 min at 70°C.
Subsequently, DNA was extracted from EV pellets using the QIAamp
DNA micro kit (Qiagen AB) and cfDNA was extracted from an average
of 2 ml plasma using the QIAamp Circulating Nucleic Acid kit
(Qiagen AB), following the manufacturer's instructions. In
addition, DNA was extracted from tissue samples using the E.N.Z.A
DNA kit (Omega Bio-Tek, Inc.) following the manufacturer's
instructions. DNA quantity and quality were analyzed using Qubit
3.0 (Thermo Fisher Scientific, Inc.) and Agilent 2100 bioanalyzer
system (Agilent Technologies GmbH).
Library construction and whole genome
sequencing (WGS)
NEB ultra v2 kit (New England Biolabs, Inc.) was
used for the construction of a sequencing library of tissue DNA,
cfDNA and EV DNA, following the manufacturer's instructions. WGS
was carried out for cfDNA and EV DNA sequencing libraries of five
HCC, five hepatitis and five healthy control samples on the Hiseq X
Ten platform (Illumina, Inc.) with a paired-end read length of 150
bp.
Capture-based mtDNA sequencing
Sequencing libraries of matched cfDNA, EV DNA and
tissue DNA from 5 HCC patients were mixed with home-made
biotinylated capture probes at a ratio of 1:800 for hybridization,
as previously described (18), with
minor modifications. Probes with an average length of 250 bp
specifically targeted to mtDNA and 3 nuclear genes were captured.
Finally, the captured libraries which contained cfmtDNA, EV mtDNA
and tissue mtDNA were sequenced on the Hiseq X Ten platform
(Illumina, Inc.) using paired-end runs of 2×150 bp.
mtDNA copy number analysis
Based on WGS data, the absolute mtDNA copy number
was calculated as the average sequencing depth of mtDNA divided by
the average sequencing depth of total DNA ×2.
Bioinformatics analysis of sequencing
data
Raw data were trimmed using Fastp (version 0.20.0)
(24) to remove adapter sequences
and reads whose lengths were <50 bp and mean quality <Q30.
BWA (version 0.7.10-r789) was used to map clean data (25). To minimize the effect of nuclear
mtDNA segments, clean data were initially mapped to the Revised
Cambridge Reference Sequence of mtDNA and whole genome hg19. Next,
Picard tools (version 2.18.27 http://github.com/broadinstitute/picard/releases/tag/2.18.27)
were used to mark and remove the duplicate reads. Local realignment
was performed using IndelRealigner in GATK (version 3.2–2)
(26). Finally, the pileup files
were generated by Samtools (version 1.8) (25) for heteroplasmic variants screening
under the following filter conditions: i) A minimum allele
frequency of ≥1% on both strands; ii) ≥3 reads in each strand
supporting the alternative allele; iii) a total sequencing coverage
of ≥100X.
Statistical analysis
The data are presented as the mean ± standard error
of the mean. GraphPad Prism 7.0 (GraphPad Software, Inc.) and SPSS
21.0 (IBM, Inc.) were used for statistical analysis. Statistical
differences between two groups were analyzed by unpaired t-test.
Differences between multiple groups were analyzed using one-way
ANOVA with post hoc test by least-significant difference. All
P-values were two-tailed. P<0.05 was considered to indicate a
statistically significant difference.
Results
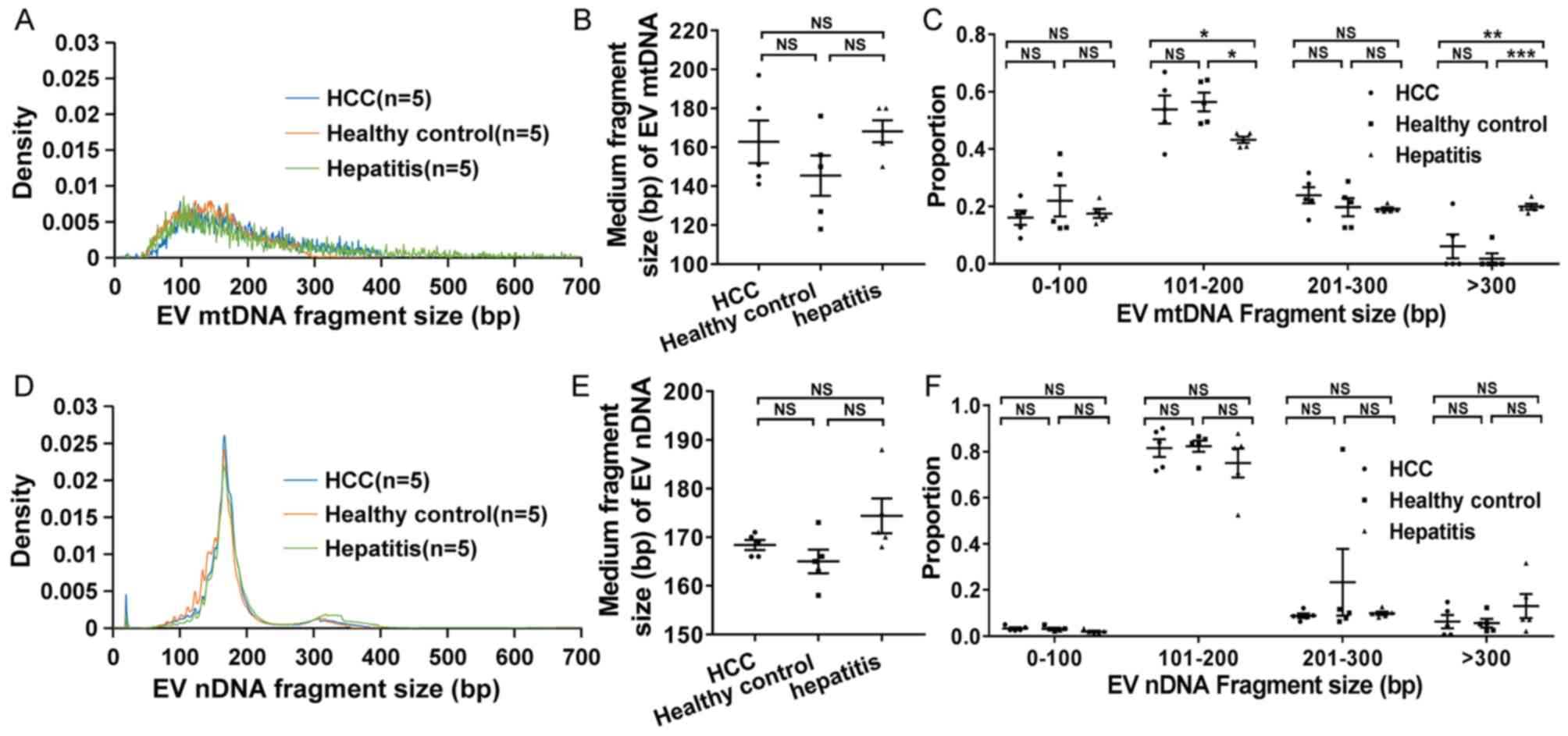
EV mtDNA has a larger fragment size
compared with cell-free mtDNA (cfmtDNA)
Based on WGS data, the distribution of DNA fragment
size was analyzed. The results showed that the cfmtDNA and EV mtDNA
had a dominant fragment size peak at ~80 and ~130 bp, respectively
(Fig. 1A). The EV mtDNA exhibited a
larger median fragment size than cfmtDNA (159 bp vs. 109 bp;
P<0.001; Fig. 1B). Furthermore,
the data indicated that EV mtDNA exhibited a significantly smaller
proportion of short fragments (0-100 bp) compared with cfmtDNA
(P<0.001; Fig. 1C). By contrast,
more long fragments (201–300 and >300 bp) were significantly
enriched in EV mtDNA compared to cfmtDNA (P<0.001 and P=0.001,
respectively; Fig. 1C). Similar to
mtDNA, EV nDNA also exhibited a significantly larger fragment size
compared with cfDNA (P<0.001; Fig.
1D-E). In addition, an obvious peak in fragment size at ~330 bp
was observed in EV nDNA, but not cfDNA, which might be due to
double-nucleosome DNA fragmentation (Fig. 1D). As expected, more long fragments
(201–300 and >300 bp) were significantly enriched in EV nDNA
than in cfDNA (P=0.049 and P=0.001; Fig.
1F).
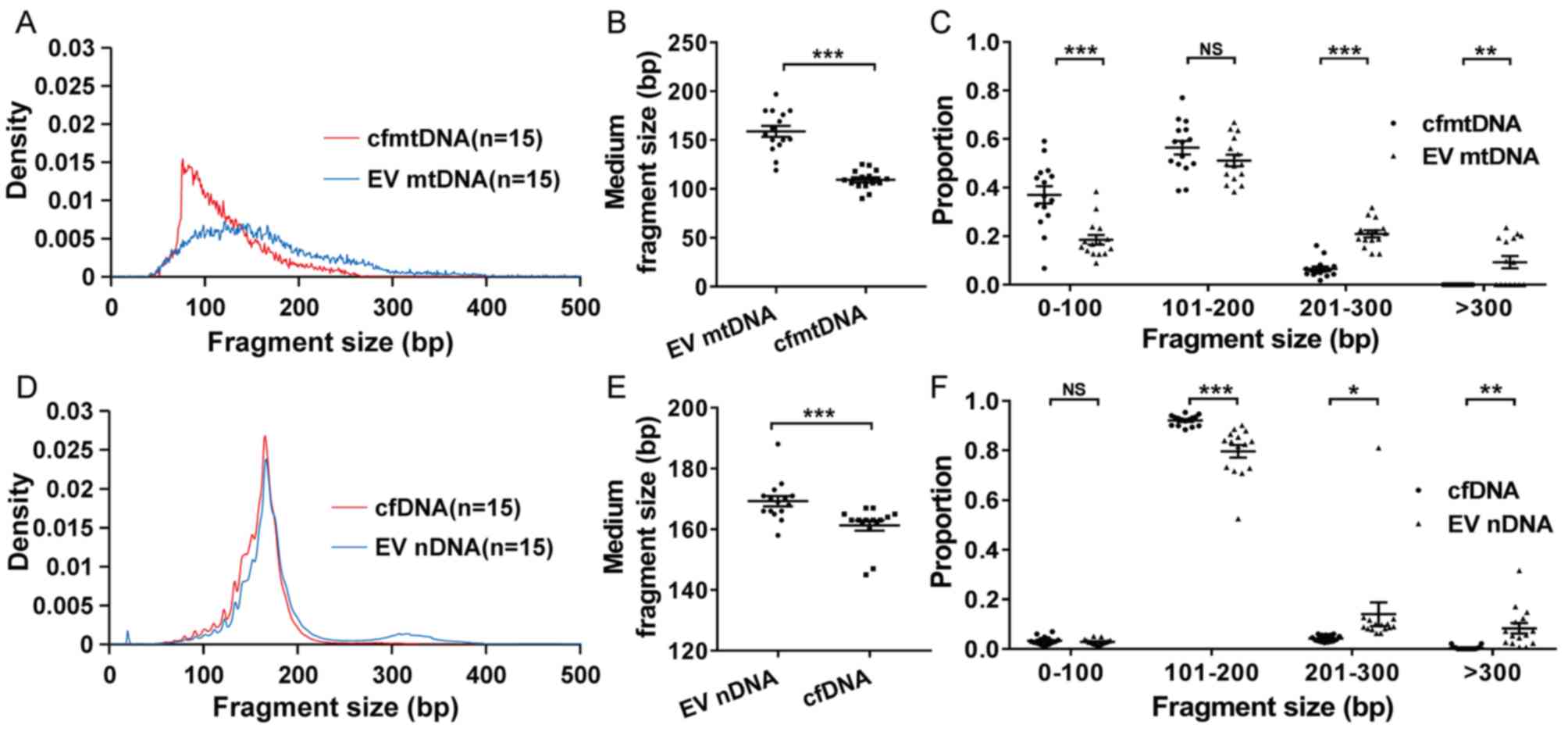 | Figure 1.Distribution of DNA fragment size in
plasma and EV samples. (A) Density, (B) medium fragment size and
(C) proportion of cfmtDNA and EV mtDNA fragments with different
sizes. Density was defined as the fragment number of a certain
length divided by the total fragment number. Proportion was defined
as the fragment number of a certain length range divided by the
total fragment number. (D) Density, (E) medium fragment size and
(F) proportion of cfDNA and EV nDNA fragments with different sizes.
EV nDNA, extracellular vesicle nuclear DNA. *P<0.05, **P<0.01
and ***P<0.001. NS, no significance; EV, extracellular vesicles;
mtDNA, mitochondrial DNA; cfmtDNA, cell-free mtDNA; EV mtDNA,
extracellular vesicle mtDNA; bp, base pair; nDNA, nuclear DNA. |
EV mtDNA fragment size in patients
with hepatitis is larger compared with that in patients with HCC
and healthy controls
The distribution of EV DNA fragment size was further
compared in plasma samples from healthy controls, patients with
hepatitis and patients with HCC. The data indicated that EV mtDNA
from patients with hepatitis exhibited a significantly larger
fragment size compared with that from patients HCC and healthy
controls (Fig. 2A and C). The medium
fragment size of EV mtDNA was 163, 145 and 168 bp in HCC patients,
healthy controls and hepatitis patients, respectively (Fig. 2B). Furthermore, the interval
distribution of the fragment size was analyzed (Fig. 2C), and it was found that the
proportion of EV mtDNA fragments with a length of 101–200 bp in
patients with hepatitis was significantly lower compared with that
in HCC patients (P=0.049) and healthy controls (P=0.019).
Conversely, the proportion of EV mtDNA fragments with a length of
>300 bp in patients with hepatitis was significantly higher
compared with that in patients HCC (P=0.003) and healthy controls
(P<0.001). However, no significant difference in fragment size
distribution in EV nDNA was observed among patients HCC, hepatitis
and healthy controls (Fig.
2D-F).
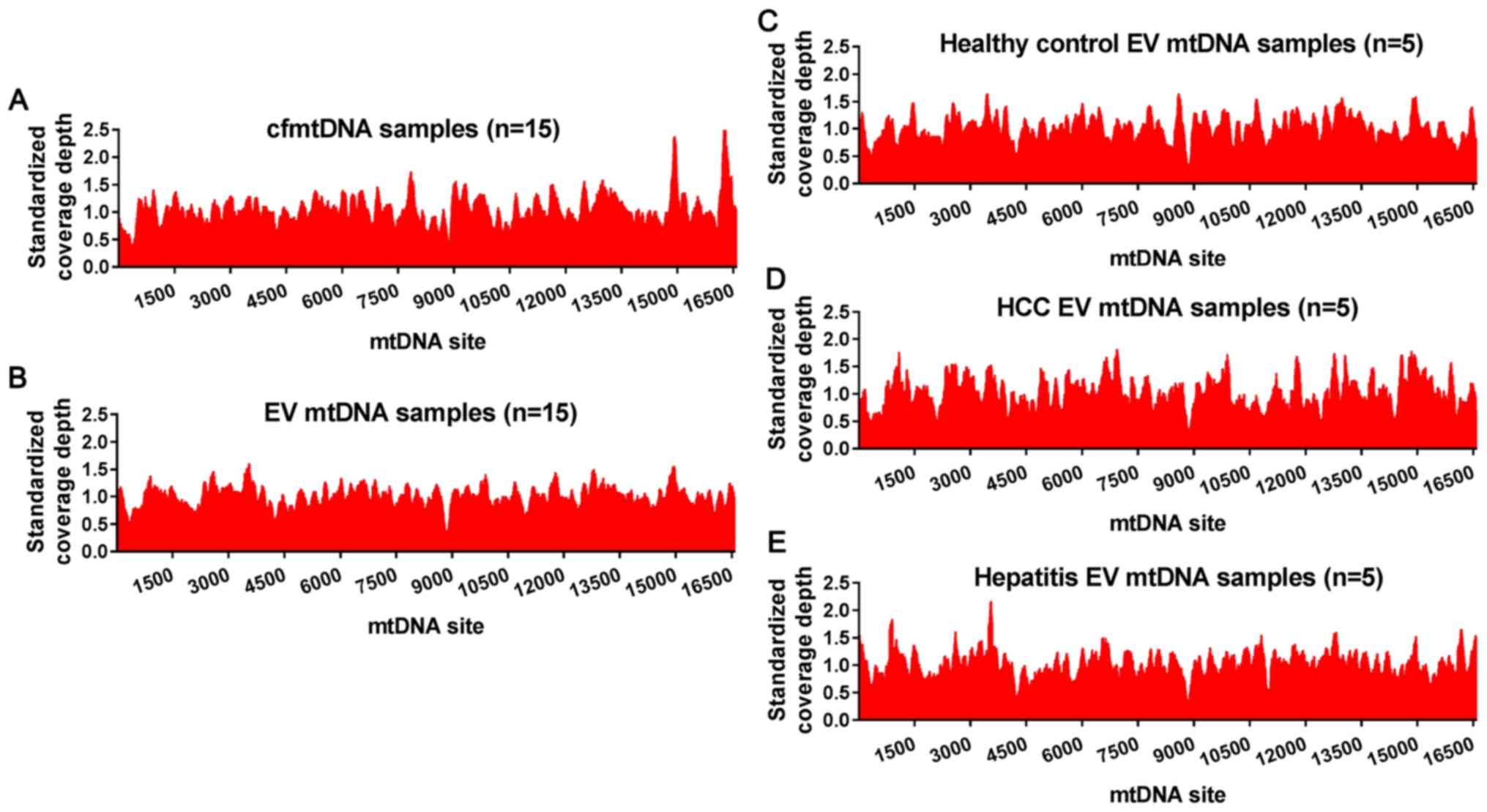
EV mtDNA covers the whole
mitochondrial genome
To confirm whether the whole mitochondrial genome
can be encapsulated in EVs from patients HCC and healthy controls,
the sequencing reads of EV mtDNA were mapped to the human reference
mitochondrial genome, and the coverage depth of each EV mtDNA site
was then divided by the average sequencing depth of the whole mtDNA
to eliminate the deviation caused by the different sequencing
depths among samples. Similar to cfmtDNA (Fig. 3A), EV mtDNA covered the whole
mitochondrial genome (Fig. 3B) with
no obvious preference in DNA regions, suggesting a non-specific
encapsulation; however, the mechanism underlying mtDNA
encapsulation remains largely unknown. Furthermore, no significant
difference in mtDNA encapsulation was observed among patients HCC,
healthy controls and patients with hepatitis (Fig. 3C-E).
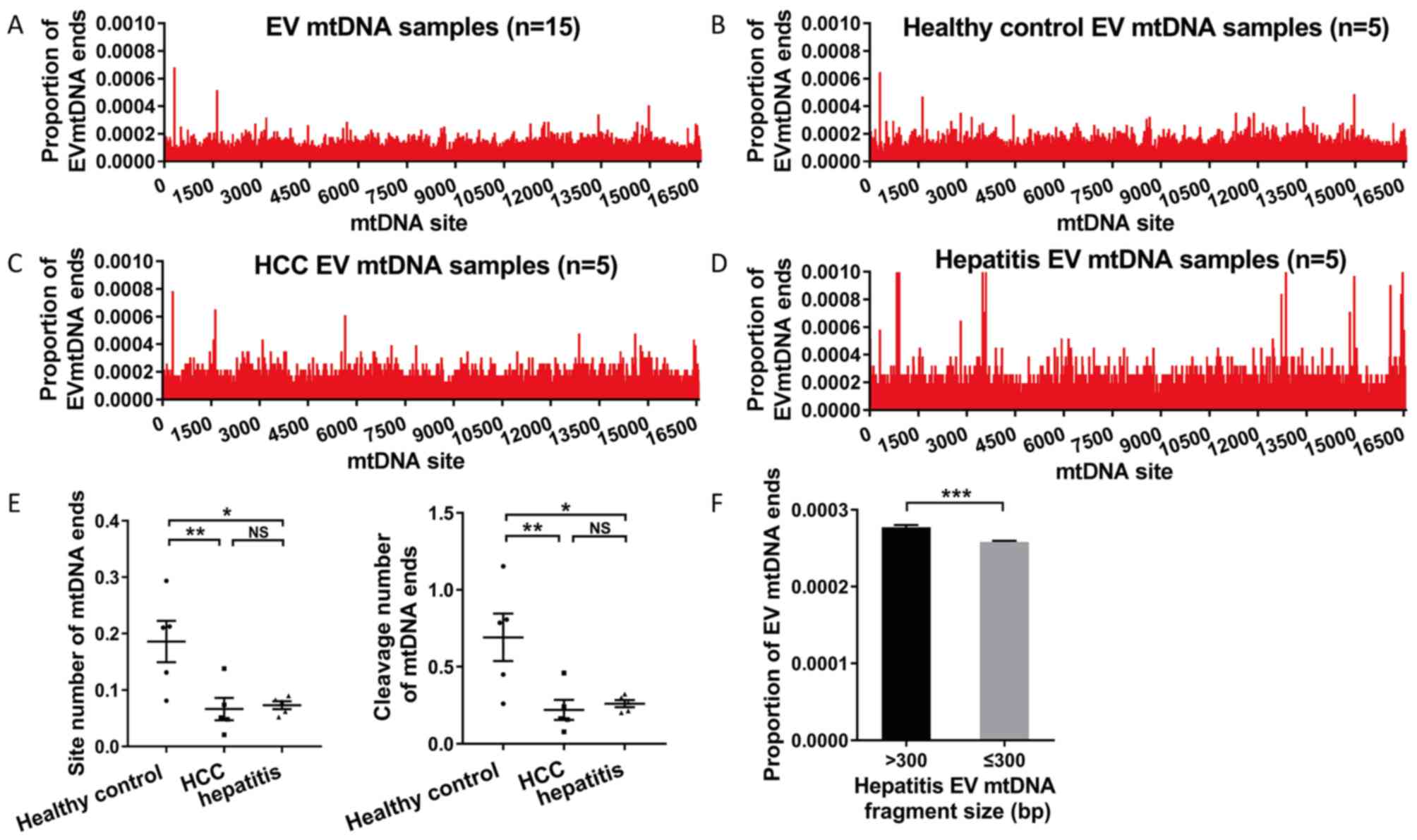
Distribution of mtDNA ends in EV
DNA
The distribution of mtDNA fragment cleavage position
was explored and mtDNA ends were defined as the cleavage position
of mtDNA fragments at the mitochondrial genome. When compared with
the majority of mtDNA sites, a small part of mtDNA sites showed a
higher proportion of ends (range, 0.0004–0.0010) in EV mtDNA
(Fig. 4A-D). However, these mtDNA
sites did not exist at the same time in different samples. The
number of mtDNA end sites that were cleaved more than twice and the
cleavage number of these sites in EV mtDNA from different sample
groups were further compared. It was found that healthy controls
had a significantly higher site and cleavage number of mtDNA ends
compared with patients with HCC (P=0.005 and P=0.003) and hepatitis
(P=0.021 and P=0.020; Fig. 4E). EV
mtDNA fragments of >300 bp exhibited a significantly higher
proportion of EV mtDNA ends compared with those with ≤300 bp in
patients with hepatitis (P<0.001; Fig. 4F).
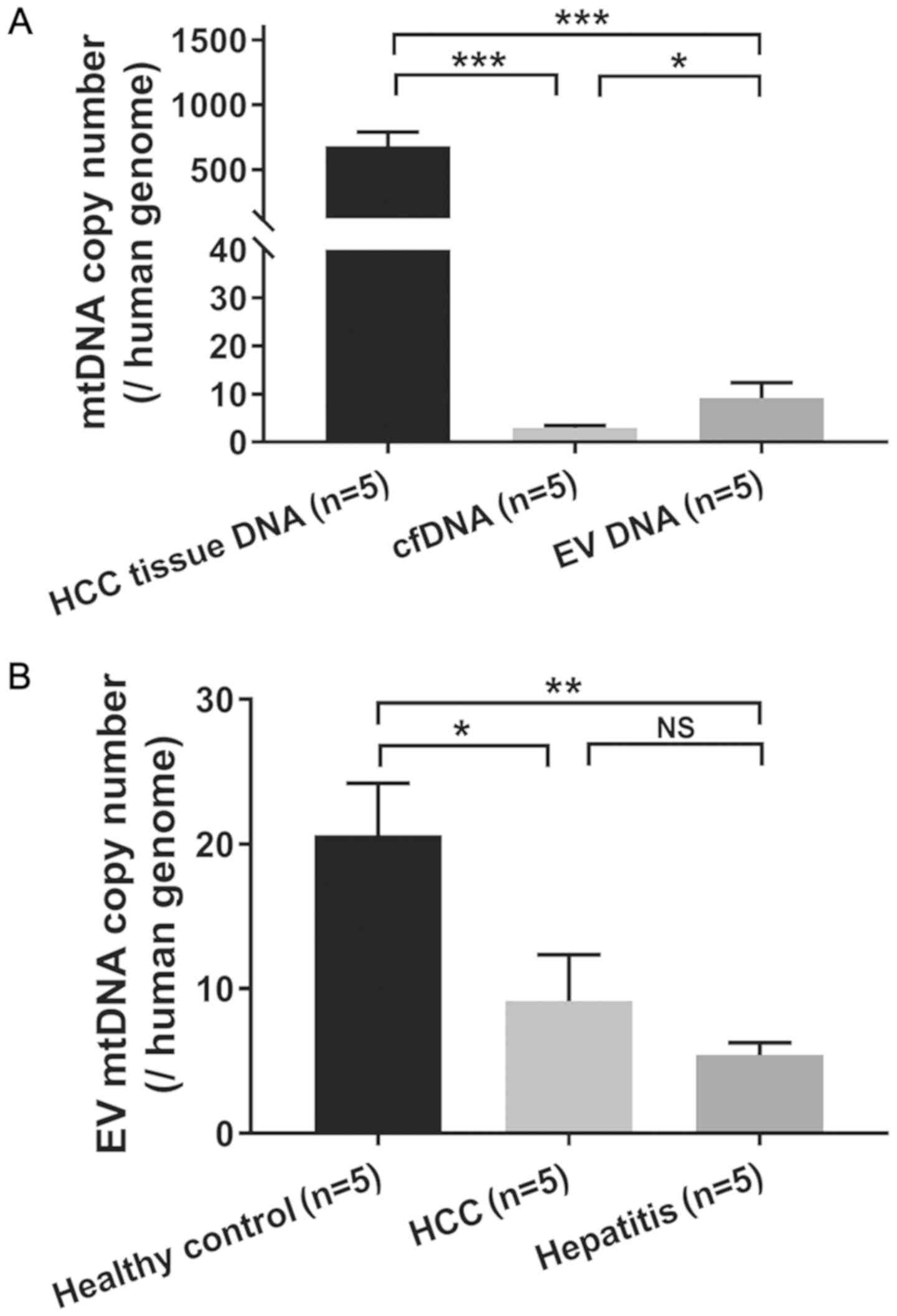
mtDNA copy number analysis of plasma
EV DNA
The mtDNA copy number was compared among different
types of DNA samples, and it was found that HCC tissues had a
significantly higher mtDNA copy number (mean, 681.17) compared with
cfDNA (mean, 2.89; P<0.001) and EV DNA (mean, 9.14; P<0.001;
Fig. 5A). It appeared that EV DNA
had a significantly higher mtDNA copy number than cfDNA (P=0.017).
Furthermore, healthy controls exhibited a significantly higher
mtDNA copy number (mean, 20.63) than both hepatitis (mean, 5.41;
P=0.002) and HCC patients (mean, 9.14; P=0.014). However, no
significant difference in the mtDNA copy number was observed
between patients with HCC and hepatitis (P=0.367; Fig. 5B).
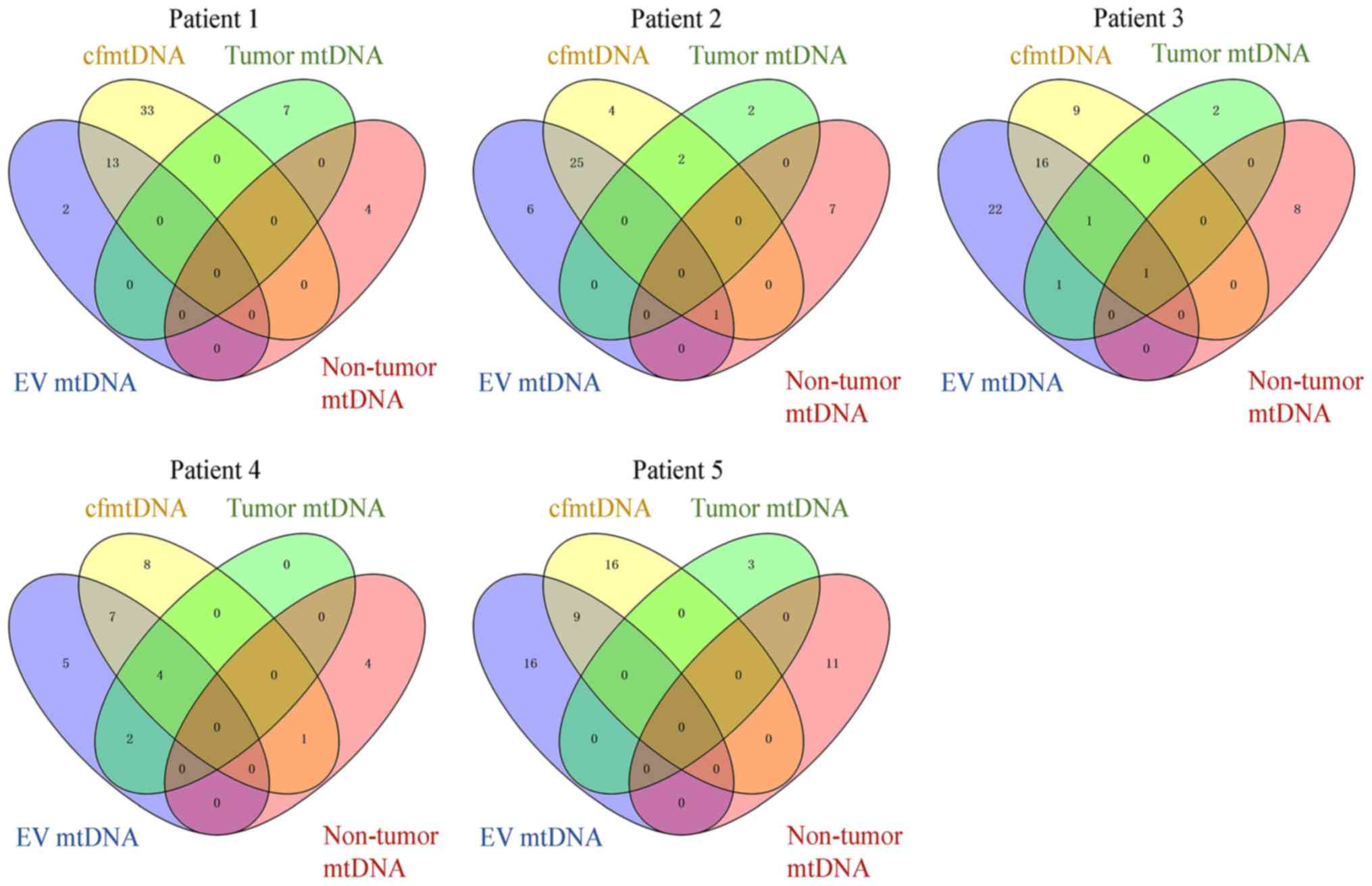
Consistent analysis of mtDNA
heteroplasmic variants in the tissue, plasma and EVs of the same
patient
The consistency of mtDNA heteroplasmic variants was
investigated in different samples from five patients with HCC
(Fig. 6). A total of 252 mtDNA
heteroplasmic variants were detected in all the samples. The
majority of mtDNA heteroplasmic variants were identified in plasma
cfmtDNA (131 variants) and EV mtDNA (150 variants). Among them, 77
heteroplasmic variants existed in both EV mtDNA and cfmtDNA. A
total of 25 mtDNA heteroplasmic variants were found in tumor
tissues, seven of which were detected in the plasma and 8 in EVs.
Only three out of 37 heteroplasmic variants that derived from
non-HCC tissues could be detected in plasma and EVs.
Discussion
In the present study, it was found that EV mtDNA
covered the whole mitochondrial genome. The characteristics of EV
mtDNA were significantly different from those of cfmtDNA. Disease
status in the liver (HBV-associated hepatitis and HCC) markedly
affected the EV mtDNA characteristics. In addition, mtDNA mutation
detection of EV DNA was complementary to cfDNA and tissue DNA
samples. To the best of our knowledge, the present study is the
first study to systematically analyze the characteristics of EV
mtDNA in patients with HCC.
The present data showed that EV mtDNA had a larger
fragment size compared with cfmtDNA, and a consistent result was
also observed in EV nDNA. Similar to the result of the present
study, two previous studies have reported a larger fragment size in
EV nDNA compared with in cfDNA, and even fragments with a size
range of 2.5–10 kbp have been found to exist in EV nDNA (9,10). These
findings are likely to be associated with the unique vesicle
structure of EVs, which protects internal DNA from external
interference (27). In addition, the
different fragment size distribution between mtDNA and nuclear DNA
in EVs and plasma suggests the existence of different degradation
mechanisms. cfDNA fragments commonly show a prominent mode average
at ~160 bp, suggesting release from apoptotic caspase-dependent
cleavage (28). However, the
underlying mechanism of mtDNA degradation needs to be further
explored.
Evidence has shown that cfDNA analysis is a
promising tool with potential for broad application in clinical
settings, especially cancer detection. Recent findings have shown
that the cell of origin and the mechanism of cfDNA release into the
blood can mark cfDNA with specific fragmentation signatures,
demonstrating that differences in fragment lengths of cfDNA may be
exploited to enhance sensitivity in detecting the presence of
circulating tumor DNA (29,30). However, whether fragment size
analysis of mtDNA in cfDNA and EV DNA contributes to cancer
detection is largely unknown. Previous studies have found that the
release of mtDNA from mitochondria depends on the mitochondrial
permeability transition pore (MPTP), BAK and BAX proteins (31–33).
MPTP is located in the mitochondrial inner membrane. Inhibition of
MPTP opening by cyclosporine A decreases leakage of mtDNA into
cytosol (31). In hepatitis caused
by HBV infection, HBV induces MPTP opening to regulate the release
of mitochondrial contents, including mtDNA (32). When BAK and BAX are activated, they
oligomerize in the mitochondrial outer membrane, which increases
the permeability of mitochondrial membrane and leads to the release
of mtDNA and cytochrome c (33). In
the present study, it was found that the long mtDNA fragment more
frequently existed in EV DNA from patients with hepatitis compared
with HCC and healthy controls. This suggested that hepatitis may
affect the permeability of the mitochondrial membrane, resulting in
the release of longer mtDNA fragments into the cytosol and reducing
the degradation of mtDNA in EVs. In addition, the analysis of mtDNA
ends in EVs also indicated that patients with hepatitis and HCC
exhibited a lower site and cleavage number of mtDNA ends compared
with healthy controls, suggesting a more uneven degradation.
Furthermore, it was found that long EV mtDNA fragments (>300 bp)
exhibited a significantly higher proportion of EV mtDNA ends in
patients with hepatitis, suggesting that a non-random end model may
exist in inflammatory cells. These findings indicated that the
fragment size and end analysis of EV mtDNA may be used as a
biomarker for liver inflammation. Based on the present findings, it
was speculated that EV mtDNA may also have fragment length and
mtDNA terminal end characteristics in other tumors. However,
because of hepatitis stage during the development of HBV-associated
HCC, EV mtDNA characteristics in other tumors may be different from
those in HCC (34,35). In addition, whether mtDNA in EVs have
a critical role in inflammatory diseases of the liver needs to be
further explored.
Compared with nuclear DNA, mtDNA has higher copy
numbers in each cell, a shorter genome length and an active
transcription and translation. Changes in the mtDNA copy number
have been extensively explored in a variety of tumors, including
HCC. Previous findings have shown a decrease in mtDNA copy number
in HCC tissues, which may contribute to HCC tumorigenesis and
progression (36). Similar results
were observed in the present study, where the mtDNA copy number was
significantly decreased in EV DNA from patients with HCC and
hepatitis compared with that from healthy controls. In addition,
both EV DNA and cfDNA exhibited a significantly lower mtDNA copy
number, when compared with HCC tissue DNA, suggesting a faster
degradation of mtDNA over nuclear DNA in both EVs and plasma.
Consistent with the protective role of EVs, EV DNA had a slightly
higher copy number compared with cfDNA. A recent study also showed
a higher concentration of mtDNA in EVs compared with plasma
(37). The present study also
confirmed that EV DNA covered the whole mitochondrial genome,
suggesting that EV mtDNA can carry all the information from tumor
cells and may be used as a potential cancer biomarker. Overall,
these findings suggest that detection of the EV mtDNA copy number
should be considered in the clinical diagnosis of HCC. An increased
sample size is required to validate the present results in the
future.
Recently, an increasing number of studies have
focused on the detection of EVs and its potential clinical
application value. Bernard et al (38) reported that KRAS mutations in exosome
DNA are more consistent with KRAS mutations in tissue DNA compared
with cfDNA in primary pancreatic cancer. Two more studies have also
indicated that EV-derived DNA is superior to cfDNA for specific
mutation detection in pancreatic (12) and non-small-cell lung cancer
(11). At present, there is no study
investigating the profile of mtDNA mutations in EVs. The present
study systematically described the mtDNA mutations in different
type of samples from patients with HCC (Fig. 6). The data showed that more mtDNA
heteroplasmic mutations were detected in EVs and plasma compared
with in paired tissues. However, compared with the HCC and adjacent
non-HCC tissue, the majority of mtDNA heteroplasmic variants were
found specifically in the plasma and EVs, which may come from white
blood cells or other tissues. However, this remains to be
confirmed. A recent study reported that tumor-derived mtDNA
mutations are rarely detected in cfDNA (39). The present results also indicated
that only a small number of tumor-derived mtDNA heteroplasmic
variants can be detected in cfDNA and EV DNA. These findings
suggested that EV mtDNA mutation detection may provide an
alternative choice for cancer diagnosis and clinical monitoring.
These data also showed that EV DNA was a good complement to plasma
cfDNA in the heteroplasmic variant detection of mtDNA.
In conclusion, EV mtDNA exhibited different
characteristics among patients with HCC and hepatitis and healthy
controls, indicating its potential clinical application value as a
diagnostic biomarker that complements cfmtDNA. However, the present
study also had limitations. For example, the sample size was small,
therefore the present results need to be validated using a larger
sample size to further demonstrate the application of EV mtDNA
characteristics in the diagnosis of HCC. Meanwhile, due to the
large amount of data and high cost of the whole genome sequencing,
capture-based mtDNA sequencing could be used to detect EV mtDNA
characteristics, and the value of this technique needs to be
verified. Further functional studies are also needed to understand
the underlying mechanisms suggested by the present findings.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant nos. 81602463 and 8183000102)
and the Science and Technology Co-ordinate Innovation Project of
Shaanxi Province, China (grant nos. 2016KTZDSF-01-02, 2017SF-188
and 2018ZDXM-SF-061).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX and KT conceived and designed the study. YJL, YW
and YL performed the experiments. JA, LC and MJ collected clinical
samples and experimental data. YJL, XG and SG analyzed the data and
interpreted the data. YJL, LC, MJ and JA wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Fourth Military Medical University (Xi'an, China; approval no.
KY20163277-1). Written consent was provided by each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tkach M and Thery C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li X, Li C, Zhang L, Wu M, Cao K, Jiang F,
Chen D, Li N and Li W: The significance of exosomes in the
development and treatment of hepatocellular carcinoma. Mol Cancer.
19:12020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu R, Rai A, Chen M, Suwakulsiri W,
Greening DW and Simpson RJ: Extracellular vesicles in
cancer-implications for future improvements in cancer care. Nat Rev
Clin Oncol. 15:617–638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cappello F, Logozzi M, Campanella C,
Bavisotto CC, Marcilla A, Properzi F and Fais S: Exosome levels in
human body fluids: A tumor marker by themselves? Eur J Pharm Sci.
96:93–98. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Melo SA, Luecke LB, Kahlert C, Fernandez
AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari
N, et al: Glypican-1 identifies cancer exosomes and detects early
pancreatic cancer. Nature. 523:177–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duijvesz D, Luider T, Bangma CH and
Jenster G: Exosomes as biomarker treasure chests for prostate
cancer. Eur Urol. 59:823–831. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Niel G, D'Angelo G and Raposo G:
Shedding light on the cell biology of extracellular vesicles. Nat
Rev Mol Cell Biol. 19:213–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thakur BK, Zhang H, Becker A, Matei I,
Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, et
al: Double-stranded DNA in exosomes: A novel biomarker in cancer
detection. Cell Res. 24:766–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kahlert C, Melo SA, Protopopov A, Tang J,
Seth S, Koch M, Zhang J, Weitz J, Chin L, Futreal A and Kalluri R:
Identification of double-stranded genomic DNA spanning all
chromosomes with mutated KRAS and p53 DNA in the serum exosomes of
patients with pancreatic cancer. J Biol Chem. 289:3869–3875. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wan Y, Liu B, Lei H, Zhang B, Wang Y,
Huang H, Chen S, Feng Y, Zhu L, Gu Y, et al: Nanoscale
extracellular vesicle-derived DNA is superior to circulating
cell-free DNA for mutation detection in early-stage non-small-cell
lung cancer. Ann Oncol. 29:2379–2383. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Allenson K, Castillo J, San Lucas FA,
Scelo G, Kim DU, Bernard V, Davis G, Kumar T, Katz M, Overman MJ,
et al: High prevalence of mutant KRAS in circulating
exosome-derived DNA from early-stage pancreatic cancer patients.
Ann Oncol. 28:741–747. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin Y, Chen K, Wang Z, Wang Y, Liu J, Lin
L, Shao Y, Gao L, Yin H, Cui C, et al: DNA in serum extracellular
vesicles is stable under different storage conditions. BMC Cancer.
16:7532016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu M: Somatic mitochondrial DNA mutations
in human cancers. Adv Clin Chem. 57:99–138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu M: Generation, function and diagnostic
value of mitochondrial DNA copy number alterations in human
cancers. Life Sci. 89:65–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reznik E, Miller ML, Şenbabaoğlu Y, Riaz
N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi
AA and Sander C: Mitochondrial DNA copy number variation across
human cancers. ELife. 5:e107692016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao SY, Yang YF, Liu J, Liu HQ, Ge NJ,
Yang HS, Zhang HX and Xing JL: Association of mitochondrial DNA
content in peripheral blood leukocyte with hepatitis B
virus-related hepatocellular carcinoma in a Chinese Han population.
Cancer Sci. 102:1553–1558. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yin C, Li DY, Guo X, Cao HY, Chen YB, Zhou
F, Ge NJ, Liu Y, Guo SS, Zhao Z, et al: NGS-based profiling reveals
a critical contributing role of somatic D-loop mtDNA mutations in
HBV-related hepatocarcinogenesis. Ann Oncol. 30:953–962. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mair R, Mouliere F, Smith CG, Chandrananda
D, Gale D, Marass F, Tsui DWY, Massie CE, Wright AJ, Watts C, et
al: Measurement of plasma cell-free mitochondrial tumor DNA
improves detection of glioblastoma in patient-derived orthotopic
xenograft models. Cancer Res. 79:220–230. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guescini M, Genedani S, Stocchi V and
Agnati LF: Astrocytes and glioblastoma cells release exosomes
carrying mtDNA. J Neural Transm (Vienna). 117:1–4. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sansone P, Savini C, Kurelac I, Chang Q,
Amato LB, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly
L, et al: Packaging and transfer of mitochondrial DNA via exosomes
regulate escape from dormancy in hormonal therapy-resistant breast
cancer. Proc Natl Acad Sci U S A. 114:E9066–E9075. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Satala CB, Jung I, Kobori L, Kovacs Z,
Fodor D, Szodorai R and Gurzu S: Benefits of the 8th American joint
committee on cancer system for hepatocellular carcinoma staging. J
Gastrointest Cancer. 2020.(Online ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu X, Shen H, Yin X, Yang M, Wei H, Chen
Q, Feng F, Liu Y, Xu W and Li Y: Macrophages derived exosomes
deliver miR-223 to epithelial ovarian cancer cells to elicit a
chemoresistant phenotype. J Exp Clin Cancer Res. 38:812019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen S, Zhou Y, Chen Y and Gu J: Fastp: An
ultra-fast all-in-one FASTQ preprocessor. Bioinformatics.
34:i884–i890. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R; 1000 Genome
Project Data Processing Subgroup, : The sequence alignment/map
format and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Maas SLN, Breakefield XO and Weaver AM:
Extracellular vesicles: Unique intercellular delivery vehicles.
Trends Cell Biol. 27:172–188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: Quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.PubMed/NCBI
|
|
29
|
Mouliere F, Chandrananda D, Piskorz AM,
Moore EK, Morris J, Ahlborn LB, Mair R, Goranova T, Marass F,
Heider K, et al: Enhanced detection of circulating tumor DNA by
fragment size analysis. Sci Transl Med. 10:eaat49212018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Underhill HR, Kitzman JO, Hellwig S,
Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP
and Shendure J: Fragment length of circulating tumor DNA. PLoS
Genet. 12:e10061622016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Patrushev M, Kasymov V, Patrusheva V,
Ushakova T, Gogvadze V and Gaziev A: Mitochondrial permeability
transition triggers the release of mtDNA fragments. Cell Mol Life
Sci. 61:3100–3103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qu C, Zhang S, Li Y, Wang Y, Peppelenbosch
MP and Pan Q: Mitochondria in the biology, pathogenesis, and
treatment of hepatitis virus infections. Rev Med Virol.
29:e20752019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McArthur K, Whitehead LW, Heddleston JM,
Li L, Padman BS, Oorschot V, Geoghegan ND, Chappaz S, Davidson S,
Chin HS, et al: BAK/BAX macropores facilitate mitochondrial
herniation and mtDNA efflux during apoptosis. Science.
359:eaao60472018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Wu X, Hu Q, Wu J, Wang G, Hong Z
and Ren J; Lab for Trauma and Surgical Infections, : Mitochondrial
DNA in liver inflammation and oxidative stress. Life Sci.
236:1164642019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hsu CC, Lee HC and Wei YH: Mitochondrial
DNA alterations and mitochondrial dysfunction in the progression of
hepatocellular carcinoma. World J Gastroenterol. 19:8880–8886.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee HC, Li SH, Lin JC, Wu CC, Yeh DC and
Wei YH: Somatic mutations in the D-loop and decrease in the copy
number of mitochondrial DNA in human hepatocellular carcinoma.
Mutat Res. 547:71–78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Baysa A, Fedorov A, Kondratov K, Ruusalepp
A, Minasian S, Galagudza M, Popov M, Kurapeev D, Yakovlev A, Valen
G, et al: Release of mitochondrial and nuclear DNA during on-pump
heart surgery: Kinetics and relation to extracellular vesicles. J
Cardiovasc Transl Res. 12:184–192. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bernard V, Kim DU, San Lucas FA, Castillo
J, Allenson K, Mulu FC, Stephens BM, Huang J, Semaan A, Guerrero
PA, et al: Circulating Nucleic Acids Are Associated With Outcomes
of Patients With Pancreatic Cancer. Gastroenterology. 156:108–118.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Weerts MJA, Timmermans EC, van de Stolpe
A, Vossen RHAM, Anvar SY, Foekens JA, Sleijfer S and Martens JWM:
Tumor-specific mitochondrial DNA variants are rarely detected in
cell-free DNA. Neoplasia. 20:687–696. 2018. View Article : Google Scholar : PubMed/NCBI
|