Introduction
The acylation of proteins is one of the
modifications, which occurs following translation and can modulate
the function of the proteins by changing the surface charge and
regulating protein conformations or protein-protein interactions,
which is similar to phosphorylation (1). Histone deacetylases (HDACs) can remove
the acetylated group from the N-acetyl-lysine of histones or
non-histones. To date, four types of histone deacetylases have been
discovered, class I include HDAC1-3 and HDAC8, class II include
HDAC4-7 and HDAC9-10, and HDAC11 belongs to class IV (1,2).
Sirtuins are defined as class III HDACs and are dependent on
NAD+, which distinguishes them from the classes I, II
and IV, which are zinc-dependent (3). In the 1970s, the silent information
regulator 2 gene (SIRT) was first discovered in budding yeast, as a
regulator of chromatin structure (4). This group of proteins are now known to
be also distributed in a wide range of mammal species, such as
drosophila, murine and cattle (4,5). In
humans, the SIRT family consists of seven members, SIRT1-7, which
are divided into four classes with respect to their sequence
information. SIRT-1, −2, and −3 are class I proteins, SIRT4 is
class II, SIRT5 is class III, and SIRT-6 and −7 are class IV
(6,7). All of the sirtuins share a highly
conserved NAD+-binding catalytic core composed of ~270
amino acids but vary in their N- and C-terminal sequences. The
different N- and C-terminal sequences are important for subcellular
localization and enzyme activities (8,9).
These seven mammalian sirtuins have different
protein structures, subcellular localizations, functional
properties, enzymatic activities, and substrate specificities
(Table I) (10). SIRT-1, −6, and −7 are primarily
located in the nucleus, SIRT2 is primarily located in the
cytoplasm, and SIRT3-5 are predominately found in the mitochondria
(11). However, some of these
proteins are known to translocate from their primary location in
different tissues or under various physiological conditions
(12). For example, in myoblast cell
line, SIRT1 moves to the cytosol under the action of some kinases
and PI3K signal cascade (8). and
SIRT2 moves into the nucleus during the G2/M cell cycle
transition (13). Each sirtuin also
has unique catalytic activities, which enable the sirtuins to
exhibit broad and important regulatory functions in numerous
biological processes, such as life span, gene transcription, cell
proliferation, differentiation, apoptosis, genome stability,
cellular metabolism, tumorigenesis, energy homeostasis, DNA damage,
and stress responses (7,14–17).
 | Table I.Characteristics and properties of the
sirtuins family. |
Table I.
Characteristics and properties of the
sirtuins family.
| Name | Class | Length, aa | Location of
catalytic domain, aa | Primary
location | Catalytic
activities |
|---|
| SIRT1 | I | 747 | 244-498 | Nucleus | Deacetylation |
|
|
|
|
|
| Deacylation |
| SIRT2 | I | 389 | 65-340 | Cytoplasm | Deacetylation |
|
|
|
|
|
| Deacylation |
|
|
|
|
|
|
Demyristoylation |
| SIRT3 | I | 399 | 126-382 | Mitochondria | Deacetylation |
|
|
|
|
|
|
Decrotonylation |
| SIRT4 | II | 314 | 45-314 | Mitochondria |
ADP-ribosylation |
|
|
|
|
|
| Deacetylation |
|
|
|
|
|
| Deacylation |
|
|
|
|
|
| Delipoylation |
| SIRT5 | III | 310 | 41-309 | Mitochondria | Deacetylation |
|
|
|
|
|
| Demalonylation |
|
|
|
|
|
|
Desuccinylation |
| SIRT6 | IV | 355 | 35-274 | Nucleus | Deacetylation |
|
|
|
|
|
|
ADP-ribosylation |
|
|
|
|
|
|
Demyristoylation |
| SIRT7 | IV | 400 | 90-331 | Nucleus | Deacetylation |
|
|
|
|
|
|
Desuccinylation |
In contrast to other sirtuins, research on
mitochondrial SIRT4 is relatively limited. However, it is
well-known that SIRT4 is widely expressed in human organs and
tissues, particularly in the heart, liver, kidney, spleen,
prostate, testis, and ovaries (18).
Unlike other sirtuins, SIRT4 possesses a conserved deacetylase
domain, but earlier reports suggested it lacked detectable histone
deacetylase activity (19). In 2006,
Haigis et al (20) first
discovered that SIRT4 possesses ADP-ribosyltransferase activity.
Subsequently, it was found to have substrate-specific deacetylase,
lipoamidase, and long-chain deacylase activities (21). These catalytic activities provide
SIRT4 with the ability to play a vital role in insulin secretion,
fatty oxidation, leucine catabolism, ATP homeostasis, lipid
catabolism, tumorigenesis, neurological disorders and
cardiovascular diseases (7,21). However, SIRT4 is still the least
well-known sirtuin, as these activities are weak. It has been
suggested that SIRT4 will become a novel therapeutic target and
molecules which modulate its activity will be developed to treat
various diseases in the future (22,23). In
the present review, SIRT4 will be discussed and its functions in
cellular metabolism and human cancer.
Structural characteristics of the sirtuin
family
As aforementioned, every member of the sirtuin
family contains a highly conserved catalytic center comprised of
~270 amino acids and divergent N- or C-terminal sequences (24,25).
Therefore, the structure of sirtuins was divided into the
non-enzymatic and the enzymatic parts. The non-enzymatic part
includes seven specific N- and C-terminal segments, which
determines the subcellular localization and catalytic function of
the sirtuin (25). The presence of
an N-terminal mitochondrial targeting sequence ensures that
mitochondrial SIRT3-5 localize within the mitochondrial matrix.
When these signal sequences are cleaved, the enzymatic functions of
these proteins are activated (26,27).
The enzymatic part of the seven sirtuins is
characterized by a high degree of fidelity. In 2001, Finnin et
al (28) first identified the
structure of sirtuins, and SIRT2 was the first reported subtype,
which almost represents the structural basis of all sirtuins in the
enzymatic part. The catalytic core consists of two main parts: A
conserved large Rossmann fold domain and a variable small domain
(28). The large inverted Rossmann
fold domain consists of 6 β-strands and 6 α-helices. The small
domain contains a zinc finger module and a helical module (29). These two modules of the small domain
are joined to the large Rossmann fold domain by four polypeptide
chains, which form a large groove between the small and large
domains. This junctional groove includes the
NAD+-binding site, and most of the residues are highly
conserved. The catalytic core of sirtuins are also included in the
junctional groove, and mutations within these sites result in the
loss of deacetylation catalytic activity (28,30–33).
Moreover, when the substrate is close to the center of SIRTs, a
conformational rearrangement occurs, and the small domain is
rotated by ~25, thereby changing the conformation of the binding
site to allow the catalytic reaction to proceed in a continuous
process (34,35).
With respect to SIRT4, the non-enzymatic part has no
C-terminal sequence and only contains 28 positively charged amino
acids in the N-terminal sequence, which serves as a mitochondrial
localization signal with low sequence conservation (36,37). In
the enzymatic part, the typical sirtuin structure is located, which
consists of a large Rossmann fold domain with a conserved His160
catalytic residue and a small domain. There are 2 specific
structures, which separate SIRT4 from the other isoforms. The first
one is a flexible loop, which contains an additional 12 residues
(residues 195-206) in the Zn2+-binding module. This loop
is located deep within the catalytic core, which contributes to
substrate binding and restrains relevant active site dynamics. The
second unusual structure is the channel in the catalytic core,
which starts from the acyl-Lys binding tunnel and terminates at the
protein surface. This channel has a small positively charged area
on the outer entrance and is predominantly hydrophobic. This
channel serves as the binding site for longer substrate acyls and
regulatory metabolites (37,38). The positive potential of the channel
accounts for its weak NAD+-dependent deacetylase, which
is within the 14-600 µM range, and also explains why the positively
charged NADH was easier to combine with SIRT4 and the mitochondrial
concentration of free NADH, NAD+/NADH ratio could act as
a physiological SIRT4 regulator (39,40).
Enzymatic activities, substrates, and
cellular functions of SIRT4
The robust enzymatic activities of SIRT4 are not
fully understood; however, SIRT4 is associated with numerous
cellular metabolic processes, such as insulin secretion, glutamine
metabolism, and fatty acid metabolism (21). The principal enzymatic activities of
SIRT4, involved in these biological processes, are
ADP-ribosyltransferase, NAD+-dependent deacetylase,
lipoamidase, and long-chain deacylase (Table II). These functions will be
described in more detail and are also summarized in Fig. 1.
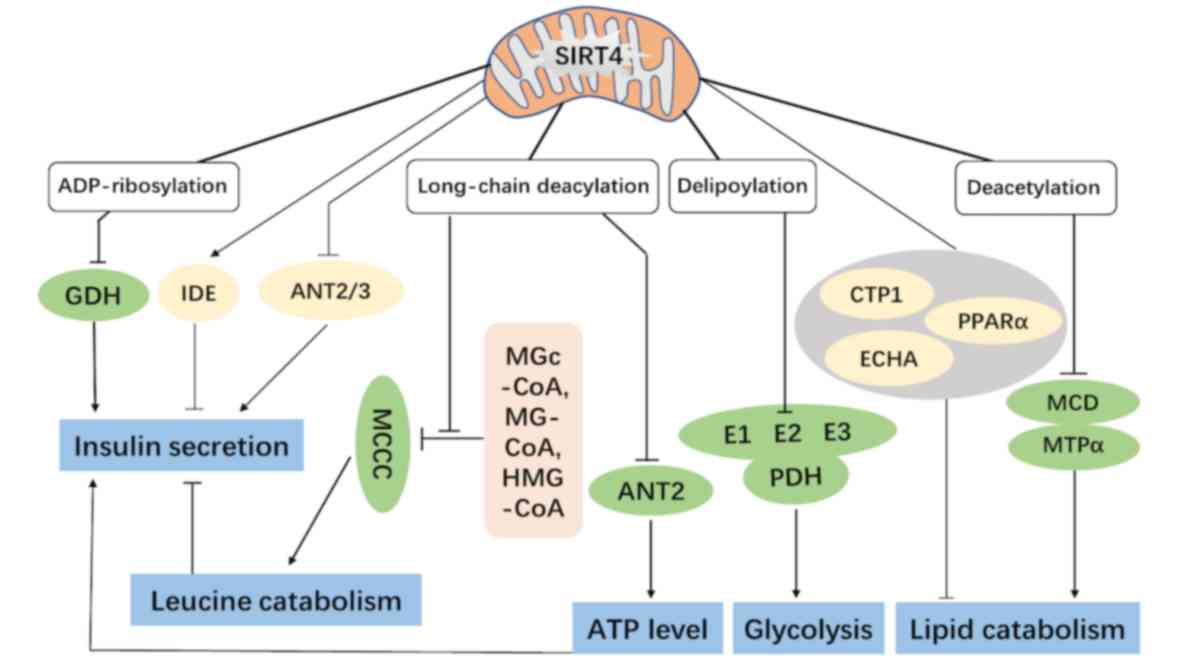 | Figure 1.The functions of mitochondrial SIRT4
in cellular metabolism. The ADP-ribosylation on GDH and
interactions with IDE and ANT2/3 can regulate insulin secretion in
pancreatic β cells. The long-chain deacylation on MCCC can regulate
leucine catabolism and affect insulin secretion indirectly. The
delipoylation on the E2 component of the PDH complex further
impacts cellular energy metabolism. The NAD+-dependent
deacetylation on MCD and MTPα can inhibit fatty acid β-oxidation.
In addition, SIRT4 also modulates some proteins associated with
lipid metabolism (CTP1, PPARα and ECHA) and finally represses lipid
catabolism collectively. GDH, glutamate dehydrogenase; IDE, insulin
degrading enzyme; ANT2/3, ADP/ATP translocase 2/3; MCCC,
methylcrotonyl-CoA carboxylase; PDH, pyruvate dehydrogenase; MCD,
malonyl CoA decarboxylase; MTPα, mitochondrial trifunctional
protein α; MGc, 3-methylglutaconyl; MG, 3-methylglutaryl; HMG,
3-hydroxy-3-methylglutaryl. |
 | Table II.Enzymatic activities, substrates and
metabolic function of SIRT4. |
Table II.
Enzymatic activities, substrates and
metabolic function of SIRT4.
| Enzymatic
activities | Substrates | Metabolic
functions | (Refs.) |
|---|
|
ADP-ribosyltransferase | GDH | Insulin
secretion | (20) |
|
|
| Tumor
suppressor |
|
|
NAD+-dependent deacetylase | MCD | Fatty acids
oxidation | (52,57) |
|
| MTPα |
|
|
| Lipoamidase | PDH | Glycolysis, | (59) |
|
|
| Cellular
metabolism |
|
| Long-chain
deacylase | MCCC | Leucine
metabolism, | (62,66) |
|
| ANT2 | ATP
homeostasis |
|
| Debiotinylase | No native substrate
has been identified |
| (59) |
ADP-ribosyltransferase and insulin
secretion
Glutamine is the most abundant free cytoplasmic
amino acid and has been found to be the staple nitrogen donor for
the synthesis of nucleotides and amino acids (41). It can be converted into glutamate and
α-ketoglutarate (α-KG) by two consecutive reactions and
subsequently enters the TCA cycle. Glutamine is first converted
into glutamate by glutaminase, then it is further converted into
the TCA cycle intermediate α-KG either by glutamate dehydrogenase
(GDH) or transamination-coupled reactions (42,43). GDH
was the first mitochondrial protein to be identified to be
inactivated through mono-ADP-ribosylation, which was found to be
catalyzed by SIRT4. SIRT4 directly and enzymatically transfers the
ADP-ribosyl group from NAD+ to the C172 histone residue
of GDH, thereby inhibiting its function and ultimately inhibiting
the metabolism of glutamine in the mitochondria and reducing ATP
production (20,42,44).
The inhibitory effect of SIRT4 on GDH represses
insulin secretion in response to glucose and amino acids in
pancreatic β cells. As insulin secretion is an ATP-dependent
process, the inhibition of GDH will lead to a decrease in ATP
production in the mitochondria (36,44).
Glucose and amino acid metabolism lead to an increased ATP/ADP
ratio. Increased levels of ATP can close ATP-sensitive potassium
channels which results in the depolarization of the plasma
membrane, thus opening voltage-gated L-type calcium channels,
facilitating the fusion of insulin-containing secretory vesicles to
the plasma membrane to cause insulin exocytosis (45,46).
SIRT4 inhibits the metabolism of glutamate, resulting in decreased
production of ATP in mitochondria and inhibition of insulin
secretion in pancreatic β cells (20). In addition, SIRT4 can interact with
insulin-degrading enzyme (IDE) and ADP/ATP translocase 2/3 (ANT2/3)
to synergistically inhibit insulin secretion (36).
NAD+-dependent deacetylase
and lipid metabolism
Reversible acetylation of lysine residues is an
important post-translational modification, which changes the charge
of lysine residues and potentially alters enzyme activity,
structure, specificity, and subcellular localization of the protein
(47). Deacetylation is the most
important and common activity of sirtuins. They catalyze
deacetylation by breaking the bonds between NAD+ and
niacinamide (NAM) ribosomes, transferring the acetylated groups
from proteins to ADP-ribose, then releasing the deacetylated
products and NAM. 2-O-acetyl-ADP-ribose is generated as a result of
the transfer of the acetyl group onto the ADP-ribose residue. NAM
is an inhibitor of deacetylation, which can also reverse the
reaction to reproduce NAD+ (25,48).
As a member of the sirtuin family, SIRT4 also
possesses typical NAD+-dependent deacetylation activity,
but this activity is weak, has substrate specificity, and is
associated with cellular lipid metabolism (49). In 2010, Nasrin et al (50) found that downregulating the
expression of SIRT4, using adenoviral shRNA, in hepatocytes and
myocytes significantly enhanced the expression of the genes, which
are associated with fatty acid oxidation (FAO), such as MCAD, PDK4,
CTP1, PPARα, PGC1α, ERRα, and CoxV. Meanwhile, the FAO of
hepatocytes and myocytes was also significantly increased. During
cellular lipid metabolism, malonyl CoA provides the carbon skeleton
for lipogenesis and also inhibits fat oxidation. Therefore, the
regulation of malonyl CoA levels can control the balance between
lipid anabolism and catabolism. Both acetyl CoA carboxylase (ACC)
and malonyl CoA decarboxylase (MCD) regulate the cellular malonyl
CoA level. ACC converts acetyl CoA to malonyl CoA, and MCD converts
it back to acetyl CoA (51–53). ACC activity is regulated by
phosphorylation from the AMP-activated protein kinase (AMPK)
signaling pathway (51). MCD
activity is regulated via deacetylation by SIRT4. For example, the
deacetylation of the K471 residue of MCD by SIRT4 inhibited the
activity of MCD, which further repressed the oxidative
decomposition of intracellular fatty acids and promoted the
synthesis of lipids in white adipose tissue (52). In vivo, SIRT4-knockout mice
had a greater exercise tolerance and protection against
diet-induced obesity (52).
With the progress of research, it has been found
that the regulation of lipid metabolism by SIRT4 involves several
mechanisms. Carnitine O-palmitoyltransferase 1, liver isoform
(CPT1) is the rate-limiting enzyme to transfer fatty acids into
both the inner and outer mitochondrial membranes. The inhibition of
SIRT4 by MCD increased the concentration of malonyl CoA, which
downregulated CPT1 activity and inhibited fatty acid oxidation
(FAO) (53). Peroxisome
proliferator-activated receptor-α (PPARα) is one of the mediators
of the hepatic response to fasting and a ligand-activated
transcription factor, which promotes the transcription of genes
involved in fatty acid catabolism, such as Lipg, Acot3, Pdk4,
Acox1, Cpt1a and Acadm (54,55). Accompanied by the high expression of
SIRT1 both at mRNA and protein levels, SIRT4 decreased the rate of
FAO by inhibiting the expression of PPARα and downstream genes
associated with fatty acid catabolism (55). In addition, the deacetylase activity
of SIRT4 also acts on the mitochondrial trifunctional protein α
(MTPα) in hepatocytes, which is a critical enzyme for fatty acid
β-oxidation (56). Deacetylation by
SIRT4 increased the activity of MTP-α, which inhibited the
oxidation of fatty acids and eventually led to hepatic steatosis.
In contrast, low expression of SIRT4 promoted the acetylation of
MTP-α, increased cellular FAO, and prevented the development of
non-alcoholic fatty liver disease. In addition, SIRT4 might inhibit
the activity of enoyl-CoA hydratase α-subunit (ECHA), which is an
enzyme involved in branched-chain amino acid (BCAA) catabolism, to
suppress FAO (57).
Lipoamidase and glycolysis
The pyruvate dehydrogenase (PDH) complex is a
mitochondrial complex, and is the rate-limiting enzyme that
catalyzes the decarboxylation of pyruvate to produce acetyl CoA. It
also links glycolysis to the TCA cycle and consists of three
catalytic subunits: E1, a pyruvate decarboxylase; E2, a
dihydrolipoyllysine acetyltransferase (DLAT), and E3, a
dihydrolipoyl dehydrogenase (58).
Mathias et al (59)
demonstrated that SIRT4 had a higher NAD+-dependent
lipoamidase activity compared with the catalytic efficiency for
deacetylation, and exhibited a greater enzymatic activity for
lipoyl- and biotinyl-lysine modifications. The E2 component of the
PDH is a biological substrate of SIRT4 lipoamidase activity. SIRT4
enzymatically hydrolyzes lipoamide cofactors from DLAT, which
results in a reduction in PDH lipoyl levels and inhibition of its
function (59). PDH controls
pyruvate decarboxylation, fueling multiple downstream pathways,
such as aerobic oxidation of glucose to the tricarboxylic acid
cycle and oxidative phosphorylation (58). These findings suggest that SIRT4 is a
critical modulator of mitochondrial function and cellular
metabolism by regulating glycolysis via the inhibition of PDH.
Long-chain deacylase and leucine
metabolism, ATP homeostasis
Leucine is a BCAA and an effective stimulator of
insulin secretion, as it can allosterically activate GDH, thereby
promoting glutamine metabolism and providing energy (60). 3-Methylglutaconyl (MGc)-CoA,
3-methylglutaryl (MG)-CoA, and 3-hydroxy-3-methylglutaryl (HMG)-CoA
are intermediates of leucine catabolism catalyzed by
methylcrotonyl-CoA carboxylase (MCCC) (60,61).
MGc-CoA, MG-CoA, and HMG-CoA function as reactive acyl species to
covalently modify and inhibit MCCC using a negative feedback loop.
The highly conserved α-helical region of SIRT4 removes this
modification and reduces insulin secretion by promoting leucine
catabolism (62). This effect on
insulin secretion in SIRT4-knock-out mice led to elevated glucose-
and leucine-stimulated basal insulin levels, which resulted in the
development of accelerated age-induced glucose intolerance and
insulin resistance. Notably, this is varied with genetic
backgrounds, SIRT4KO mice on a C57BL/6NJ genetic background have
elevated leucine-stimulated insulin levels; however, SIRT4KO mice
on a C57BL/6J background do not (62,63). The
repression of SIRT4 activity promoted insulin secretion partly via
GDH activation. On the other hand, the loss of SIRT4 function
resulted in the accumulation of acyl modified MCCC, which repressed
leucine catabolism and allowed for leucine to function as an
allosteric activator of GDH. In general, SIRT4 regulates insulin
secretion in pancreatic β cells through ADP-ribosylation of GDH and
deacylation of MCCC in a coordinated way (20,64).
The deacyl function of SIRT4 plays a crucial role in
ATP homeostasis. ANT2 is a transmembrane protein located in the
inner mitochondrial membrane, and the acylation of ANT2 is known to
uncouple mitochondria and reduce the efficiency of oxidative
phosphorylation (65). SIRT4
regulates oxygen consumption via modulating the coupling efficiency
in an ANT2-dependent manner. In the absence of SIRT4,
ANT2-dependent uncoupling led to a decrease in cellular ATP levels
and activation of a feedback loop, which involved a reverse
signaling response from the mitochondria to the nucleus through
AMPK, CPT1, PGC1α, and ACC (66).
Functions of SIRT4 in human cancer
Metabolic reprogramming is an important aspect of
cancer cell metabolism, in which glucose and glutamine metabolic
reprogramming are two primary forms. In 1927, Warburg et al
(67) found that the rate of
glycolysis was much faster compared with that in normal cells, even
in the presence of sufficient oxygen. This phenomenon of aerobic
glycolysis, to achieve rapid cell proliferation and energy supply,
was termed the Warburg Effect. Glutamine is the most abundant
non-essential amino acid in cell plasma (68). Studies have found that the majority
of cancer cells can utilize glutamine at a high rate, and some
types of cancer cells, such as breast cancer cells, HeLa cervical
carcinoma cells and hepatocellular cells cannot survive without an
exogenous glutamine supply (69).
Glutamine metabolic reprogramming in tumor cells primarily occurs
by increased glutamine uptake and glutamine catabolism (68,70). In
proliferating cells, the Krebs cycle metabolite, citrate is
exported to the cytoplasm for the generation of acetyl CoA.
Glutamine serves as an important factor for the replenishment of
TCA intermediates which are required due to the continual loss of
citrate. In addition, it can also promote the production of
intracellular NADH and glutathione to stabilize the intracellular
redox balance and regulate the activity of some signal transduction
systems, including mTORC1 signaling, the ERK pathway and signaling
associated with regulation of mitochondrial ROS production
(71–73).
Every member of the sirtuin family has been
associated with tumorigenesis (74).
SIRT4 serves as a tumor suppressor as it inhibits glutamine
metabolism in the mitochondria of cancer cells. Early studies found
that SIRT4 mRNA levels were reduced in several malignant tumor
tissues, such as bladder cancer, T-cell leukemia, lung cancer,
ovarian cancer, and thyroid cancer (75–77). In
2013, Jeong et al (78)
discovered that SIRT4 acted as a tumor suppressor by regulating
glutamine metabolism in HepG2 cells and PC3 human prostate cancer
cells, and SIRT4 knock-out mice spontaneously developed lung
cancer. As a result, the tumor suppressor function of SIRT4 had
been confirmed in a variety of malignant tumors (Table III). In these studies, the
expression level of SIRT4 in normal tissues was significantly
higher compared with that in corresponding tumors in the majority
of the cancers, and SIRT4 suppressed the biological function of
tumor cells in vitro. In addition, several studies have
shown that the low expression levels of SIRT4 was significantly
associated with the advanced stage of cancer and the poor prognosis
of patients (79–95).
 | Table III.Summary of research on tumor
suppressor function of SIRT4. |
Table III.
Summary of research on tumor
suppressor function of SIRT4.
| Neoplasms | Expression of SIRT4
in the cancer tissuesa | Role of SIRT4 in
the cancer cell in vitro | Association of
expression with advanced stage and metastasisb | Association between
expression level and overall survival timea, b | (Refs.) |
|---|
| HNSCC | Low | NA | NA | Negative
correlation | (79) |
| HCC | Low | Inhibition | Yes | Negative
correlation | (80) |
| NSCLC | Low | Inhibition | Yes | Negative
correlation | (81) |
| EAC | Low | NA | Yes | NA | (82) |
| NB | Low | Inhibition | Yes | Negative
correlation | (83) |
| GC | Low | Inhibition | Yes | Negative
correlation | (84–87) |
| CRC | Low | Inhibition | Yes | Negative
correlation | (88–90) |
| IBC | Low | NA | NA | Negative
correlation | (91) |
| ESCC | Low | Inhibition | NA | Negative
correlation | (92) |
| BCL | NA | Inhibition | NA | NA | (93) |
| TC | Low | Inhibition | NA | NA | (94) |
| PC | NA | Inhibition | NA | NA | (95) |
In terms of the molecular mechanism, SIRT4 notably
represses the metabolism of glutamine through the ADP-ribosylation
of GDH, which decreases the energy and material supply required for
nucleic acid and protein synthesis to the rapidly proliferating
tumor cells (78). This has been
confirmed in breast cancer, colorectal cancer, esophageal squamous
cell carcinoma, myc-induced B cell lymphoma and thyroid cancer
(78,88,92–94).
Furthermore, stress-induced DNA damage causes genomic instability
in carcinogenic genes, such as TP53, ATM and CDKN2A, which induces
SIRT4 expression to inhibit glutamine metabolism, and results in
cell cycle arrest (96). This
provides sufficient time for DNA damage repair (DDR) and protects
the stability of the genome in HeLa cells (97). SIRT4 enhances E-cadherin expression
and inhibits the expression of N-cadherin and vimentin, thus
inhibiting the process of epithelial-mesenchymal transition, and
decreasing the migratory and invasive abilities of gastric and
colorectal cancer cells (84,88). In
addition, aerobic glycolysis leads to the accumulation of
intracellular acidic metabolites, such as pyruvate and lactic acid,
and ammonia produced by glutamine metabolism can alleviate this pH
imbalance to maintain the homeostasis of the intracellular
environment. However, SIRT4 disturbs this pH balance and produces
an acidic environment in breast cancer (98). This regulatory effect of SIRT4 was
modulated by C-terminal binding protein and varied with glucose
metabolic levels (98,99). Furthermore, overexpression of SIRT4
blocks cell cycle progression and decreases cancer cell replication
by inactivating ERK, p-ERK, cyclin D, and cyclin E in thyroid
cancer and gastric cancer cells (87,94).
In addition, mammalian target of rapamycin complex 1
(mTORC1) was associated with the nutritional status and metabolism
of cells, and it repressed the protein level of SIRT4 by
destabilizing cAMP-dependent transcription factor ATF-4 (CREB2). On
the other hand, low mRNA levels of SIRT4 increased the expression
of mTORC downstream genes, such as MYC, CCND1, HIF1A and SREBP1.
The mutual inhibition between SIRT4 and mTORC1 was important for
the proliferation and survival of colon carcinoma cell line DLD1
and prostate cancer cell line DU145 (100,101).
In hepatocellular carcinomas, SIRT4 deletion by shRNA of HL7702
cell line increased mTOR signaling by inhibiting AMPK through the
regulation of glutamine catabolism and the AMP/LKB1 pathway
(80). In non-small cell lung cancer
cell lines, SIRT4 reduced mitochondrial fission by interacting with
the Fis-1/Drp1 axis and regulated cell invasive abilities by
repressing MEK/ERK activity (81). A
study also demonstrated that SIRT4 is a substrate of ubiquitin-like
with plant homeodomain and ring finger domains 1 (UHRF1) and
negatively regulated aerobic glycolysis, tumor proliferation, and
metastasis of pancreatic cancer cells (95). A recent study also suggested that
SIRT4 overexpression could heighten the sensitivity of ER-positive
breast cancer to tamoxifen via inhibiting the interleukin-6/STAT3
pathway (102).
Notably, SIRT4 plays a dual role in tumorigenesis.
SIRT4 plays an important role in DDR and protects the stability of
the genome, preventing tumorigenic transformation. In the same way,
SIRT4 also protects cancer cells against stresses, such as DNA
damage and caloric restriction. For example, the knock-out of SIRT4
in HepG2 cells resulted in decreased cell survival and tumor growth
following the DNA damage caused by gamma-radiation, and inversely,
the overexpression of SIRT4 in HepG2 cells increased drug and
radiation resistance (103).
Moreover, SIRT4 promotes cancer cell survival by degrading
phosphatase and tensin homolog (PTEN) using the lysosome pathway,
which is mediated by IDE (104).
PTEN is a lipid phosphatase, which inhibits cancer cell survival
and proliferation, and the degradation of PTEN accelerates
autophagy during nutritional starvation stresses, such as glucose
deprivation. During autophagy, autosomes fuse with lysosomes to
form autophagic lysosomes (105,106).
It is a salvage pathway to produce proteins, lipids, and
carbohydrates for cells to survive in unfavorable conditions
(107). Therefore, we hypothesize
that SIRT4 is a tumor suppressor prior to the development of
cancer; however, once cancer has developed, SIRT4 can still exert a
tumor suppressor effect by regulating energy metabolism. Therefore,
it has a certain protective effect against cancer cells.
A number of studies have found that SIRT4 is
associated with several human diseases. In angiotensin II-induced
cardiac hypertrophy in mice, SIRT4 inhibited manganese superoxide
dismutase activity and promoted the accumulation of reactive oxygen
species in cardiomyocytes, which eventually led to cardiac
hypertrophy through the activation of the MAPK/ERK pathway
(108,109). Furthermore, the overexpression of
SIRT4 protein levels can protect against myocardial
ischemia-reperfusion injury by decreasing myocardial infarct size,
serum creatine phosphokinase levels, and myocardial apoptosis
(110). In the central nervous
system, glutamate transport is an energy-dependent process. SIRT4
positively regulated ATP production in neural cells by inhibiting
GDH. Specifically, it enhanced the function of the
sodium/potassium-ATPase, which led to increased glutamate uptake
and glutamate transport to maintain the normal functions of
neurons. In vivo, SIRT4-knock-out mice have enhanced seizure
phenotypes compared with that in wild-type mice following treatment
with a potent excitotoxin kainic acid (111,112).
With respect to urogenital diseases, a study demonstrated that
Leydig cells treated with lipopolysaccharide led to impaired
steroidogenesis and enhanced cellular apoptosis via suppression of
SIRT4 by the activation of JNK (113). In addition, SIRT4 controls energy
metabolism and meiotic apparatus during oocyte maturation, and
mouse oocytes with overexpressed SIRT4 protein levels are unable to
undergo meiosis completely (114).
These data suggest that the roles of SIRT4 in human diseases are
both important and complex. Future studies are required to identify
connections and detailed mechanisms between them.
Conclusion
Mitochondria are vital for maintaining an energy
balance in cellular and organismal physiology, and aberrant
mitochondrial function has been associated with cellular
dysfunctions and metabolic diseases. Mitochondrial dysfunction
diseases are a group of genetically heterogeneous diseases that can
involve any organs, onset at any age, and be inherited from an
autosome, the X chromosome, or maternally (115). The sirtuin family is evolutionary
conserved and exhibits a wide range of biological functions in life
span, gene transcription, cell proliferation, differentiation,
apoptosis, genome stability, cellular metabolism, tumorigenesis,
energy homeostasis, DNA damage and stress responses (14–17).
Among them, SIRT4 is predominately located in mitochondria and
regulates its functions through some known and unknown mechanisms.
Recent studies have unraveled numerous biological processes and
human diseases, which are regulated by SIRT4 (116,117).
However, there are still two crucial issues that require further
investigation to provide a more comprehensive understanding of
SIRT4.
The first problem is that SIRT4 is still the least
well-known mammalian sirtuin; no convincing enzymatic activity has
been verified for SIRT4, and there are limited studies on SIRT4
modulators. Its key roles in metabolism and several catalytic
activities have been reported; however, more structural information
is required to uncover the robust activity and investigate the
regulatory mechanisms of SIRT4. To address this gap in the
knowledge, the development of effective chemical modulators of
SIRT4 is important. Then, these modulators could be applied to
scientific research to elucidate the detailed functions of SIRT4,
and eventually, for use in clinical diagnosis, treatment or
prognosis prediction (7,21,118).
The second problem relates to all seven members of
the sirtuin family. Biologically, each of them has a complicated
regulatory mechanism via acting on substrates or being modulated by
upstream proteins. In fact, their functions are not independent of
each other, and numerous studies have confirmed that there is a
tight interaction between the sirtuins family. For example, the
mRNA level of SIRT4 in granulocytes and monocytes of patients with
type 2 diabetes was lower, while the mRNA level of SIRT1 was
higher, compared with that in healthy individuals; however, the
interaction between these two SIRTs remains unknown (119). In addition, SIRT1 plays a
complicated and important role in cardiovascular metabolic diseases
(23). Furthermore, SIRT4
ribosylated GDH and decreased its activity; however, SIRT3
deacetylated GDH and increased its activity (120). Both SIRT-4 and −6 have
anti-inflammatory functions (121),
whereas the activities of SIRT-4 and −5 partly overlap, and
collectively regulate the metabolism of BCAA (122). Therefore, a valid network exists
among the seven sirtuins, in which they are involved in similar
pathways. However, their specific mechanisms remain unknown, and in
the future, an extensive amount of investigation is required to
clarify this complex network.
Acknowledgements
Not applicable.
Funding
This study was partly supported by the National
Natural Science Foundation of China (grant no. 81672523) and the
Shenyang Plan Project of Science and Technology (grant no.
17-230-098).
Availability of data and materials
Not applicable.
Authors' contributions
CW drafted the original manuscript. CW and YL
created the tables, designed the figures and performed the
literature review. YZ was involved in drafting the initial
manuscript, read and approved the final manuscript. CK reviewed and
revised the manuscript for important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang Y, He J, Liao M, Hu M, Li W, Ouyang
H, Wang X, Ye T, Zhang Y and Ouyang L: An overview of Sirtuins as
potential therapeutic target: Structure, function and modulators.
Eur J Med Chem. 161:48–77. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jing H and Lin H: Sirtuins in epigenetic
regulation. Chem Rev. 115:2350–2375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirschey MD: Old enzymes, new tricks:
Sirtuins are NAD(+)-dependent de-acylases. Cell Metab. 14:718–719.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Klar AJ, Fogel S and Macleod K: MAR1-a
Regulator of the HMa and HMalpha Loci in SACCHAROMYCES CEREVISIAE.
Genetics. 93:37–50. 1979.PubMed/NCBI
|
|
5
|
Michan S and Sinclair D: Sirtuins in
mammals: Insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Frye RA: Phylogenetic classification of
prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res
Commun. 273:793–798. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Zhou Y, Wang F, Chen X, Wang C, Wang
J, Liu T, Li Y and He B: SIRT4 is the last puzzle of mitochondrial
sirtuins. Bioorg Med Chem. 26:3861–3865. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tanno M, Sakamoto J, Miura T, Shimamoto K
and Horio Y: Nucleocytoplasmic shuttling of the NAD+-dependent
histone deacetylase SIRT1. J Biol Chem. 282:6823–6832. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liszt G, Ford E, Kurtev M and Guarente L:
Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J
Biol Chem. 280:21313–21320. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumar S and Lombard DB: Mitochondrial
sirtuins and their relationships with metabolic disease and cancer.
Antioxid Redox Signal. 22:1060–1077. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Michishita E, Park JY, Burneskis JM,
Barrett JC and Horikawa I: Evolutionarily conserved and
nonconserved cellular localizations and functions of human SIRT
proteins. Mol Biol Cell. 16:4623–4635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mei Z, Zhang X, Yi J, Huang J, He J and
Tao Y: Sirtuins in metabolism, DNA repair and cancer. J Exp Clin
Cancer Res. 35:1822016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vaquero A, Scher MB, Lee DH, Sutton A,
Cheng HL, Alt FW, Serrano L, Sternglanz R and Reinberg D: SirT2 is
a histone deacetylase with preference for histone H4 Lys 16 during
mitosis. Genes Dev. 20:1256–1261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeong SM and Haigis MC: Sirtuins in
cancer: A balancing act between genome stability and metabolism.
Mol Cells. 38:750–758. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
German NJ and Haigis MC: Sirtuins and the
metabolic hurdles in cancer. Curr Biol. 25:R569–R583. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grabowska W, Sikora E and Bielak-Zmijewska
A: Sirtuins, a promising target in slowing down the ageing process.
Biogerontology. 18:447–476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kupis W, Palyga J, Tomal E and
Niewiadomska E: The role of sirtuins in cellular homeostasis. J
Physiol Biochem. 72:371–380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frye RA: Characterization of five human
cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins
(sirtuins) metabolize NAD and may have protein
ADP-ribosyltransferase activity. Biochem Biophys Res Commun.
260:273–279. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
North BJ, Marshall BL, Borra MT, Denu JM
and Verdin E: The human Sir2 ortholog, SIRT2, is an NAD+-dependent
tubulin deacetylase. Mol Cell. 11:437–444. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Haigis MC, Mostoslavsky R, Haigis KM,
Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos
GD, Karow M, Blander G, et al: SIRT4 inhibits glutamate
dehydrogenase and opposes the effects of calorie restriction in
pancreatic beta cells. Cell. 126:941–954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar S and Lombard DB: For certain, SIRT4
activities! Trends Biochem Sci. 42:499–501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi JX, Wang QJ, Li H and Huang Q: SIRT4
overexpression protects against diabetic nephropathy by inhibiting
podocyte apoptosis. Exp Ther Med. 13:342–348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kane AE and Sinclair DA: Sirtuins and
NAD(+) in the development and treatment of metabolic and
cardiovascular diseases. Circ Res. 123:868–885. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Howitz KT, Bitterman KJ, Cohen HY, Lamming
DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL,
et al: Small molecule activators of sirtuins extend Saccharomyces
cerevisiae lifespan. Nature. 425:191–196. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McGuinness D, McGuinness DH, McCaul JA and
Shiels PG: Sirtuins, bioageing, and cancer. J Aging Res.
2011:2357542011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Onyango P, Celic I, McCaffery JM, Boeke JD
and Feinberg AP: SIRT3, a human SIR2 homologue, is an NAD-dependent
deacetylase localized to mitochondria. Proc Natl Acad Sci USA.
99:13653–13658. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schwer B, North BJ, Frye RA, Ott M and
Verdin E: The human silent information regulator (Sir)2 homologue
hSIRT3 is a mitochondrial nicotinamide adenine
dinucleotide-dependent deacetylase. J Cell Biol. 158:647–657. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Finnin MS, Donigian JR and Pavletich NP:
Structure of the histone deacetylase SIRT2. Nat Struct Biol.
8:621–625. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bellamacina CR: The nicotinamide
dinucleotide binding motif: A comparison of nucleotide binding
proteins. FASEB J. 10:1257–1269. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sanders BD, Jackson B and Marmorstein R:
Structural basis for sirtuin function: What we know and what we
don't. Biochim Biophys Acta. 1804:1604–1616. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moniot S, Weyand M and Steegborn C:
Structures, substrates, and regulators of Mammalian
sirtuins-opportunities and challenges for drug development. Front
Pharmacol. 3:162012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yuan H and Marmorstein R: Structural basis
for sirtuin activity and inhibition. J Biol Chem. 287:42428–42435.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moniot S, Schutkowski M and Steegborn C:
Crystal structure analysis of human Sirt2 and its ADP-ribose
complex. J Struct Biol. 182:136–143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou Y, Zhang H, He B, Du J, Lin H,
Cerione RA and Hao Q: The bicyclic intermediate structure provides
insights into the desuccinylation mechanism of human sirtuin 5
(SIRT5). J Biol Chem. 287:28307–28314. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao
C, Dai H, Choy W, Bemis JE, Jirousek, et al: Crystal structures of
human SIRT3 displaying substrate-induced conformational changes. J
Biol Chem. 284:24394–24405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ahuja N, Schwer B, Carobbio S, Waltregny
D, North BJ, Castronovo V, Maechler P and Verdin E: Regulation of
insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase.
J Biol Chem. 282:33583–33592. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pannek M, Simic Z, Fuszard M, Meleshin M,
Rotili D, Mai A, Schutkowski M and Steegborn C: Crystal structures
of the mitochondrial deacylase Sirtuin 4 reveal isoform-specific
acyl recognition and regulation features. Nat Commun. 8:15132017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kato Y, Kihara H, Fukui K and Kojima M: A
ternary complex model of Sirtuin4-NAD+-Glutamate
dehydrogenase. Comput Biol Chem. 74:94–104. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Madsen AS, Andersen C, Daoud M, Anderson
KA, Laursen JS, Chakladar S, Huynh FK, Colaço AR, Backos DS,
Fristrup P, et al: Investigating the sensitivity of NAD+-dependent
sirtuin deacylation activities to NADH. J Biol Chem. 291:7128–7141.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Feldman JL, Dittenhafer-Reed KE, Kudo N,
Thelen JN, Ito A, Yoshida M and Denu JM: Kinetic and structural
basis for Acyl-Group Selectivity and NAD(+) Dependence in
Sirtuin-Catalyzed Deacylation. Biochemistry. 54:3037–3050. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gorrini C, Harris IS and Mak TW:
Modulation of oxidative stress as an anticancer strategy. Nat Rev
Drug Discov. 12:931–947. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fernandez-Marcos PJ and Serrano M: Sirt4:
The glutamine gatekeeper. Cancer Cell. 23:427–428. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Altman BJ, Stine ZE and Dang CV: From
Krebs to clinic: Glutamine metabolism to cancer therapy. Nat Rev
Cancer. 16:7492016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Argmann C and Auwerx J: Insulin secretion:
SIRT4 gets in on the act. Cell. 126:837–839. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lang J: Molecular mechanisms and
regulation of insulin exocytosis as a paradigm of endocrine
secretion. Eur J Biochem. 259:3–17. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ashcroft FM, Proks P, Smith PA, Ammala C,
Bokvist K and Rorsman P: Stimulus-secretion coupling in pancreatic
beta cells. J Cell Biochem. 55 (Suppl):S54–S65. 1994. View Article : Google Scholar
|
|
47
|
Glozak MA, Sengupta N, Zhang X and Seto E:
Acetylation and deacetylation of non-histone proteins. Gene.
363:15–23. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sauve AA, Wolberger C, Schramm VL and
Boeke JD: The biochemistry of sirtuins. Annu Rev Biochem.
75:435–465. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Feldman JL, Baeza J and Denu JM:
Activation of the protein deacetylase SIRT6 by long-chain fatty
acids and widespread deacylation by mammalian sirtuins. J Biol
Chem. 288:31350–31356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nasrin N, Wu X, Fortier E, Feng Y, Bare'
OC, Chen S, Ren X, Wu Z, Streeper RS and Bordone L: SIRT4 regulates
fatty acid oxidation and mitochondrial gene expression in liver and
muscle cells. J Biol Chem. 285:31995–32002. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hardie DG: Sensing of energy and nutrients
by AMP-activated protein kinase. Am J Clin Nutr. 93:891S–896. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Laurent G, German NJ, Saha AK, de Boer VC,
Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran
B, et al: SIRT4 coordinates the balance between lipid synthesis and
catabolism by repressing malonyl CoA decarboxylase. Mol Cell.
50:686–698. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Saggerson D: Malonyl-CoA, a key signaling
molecule in mammalian cells. Annu Rev Nutr. 28:253–272. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kersten S, Seydoux J, Peters JM, Gonzalez
FJ, Desvergne B and Wahli W: Peroxisome proliferator-activated
receptor alpha mediates the adaptive response to fasting. J Clin
Invest. 103:1489–1498. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Laurent G, de Boer VC, Finley LW, Sweeney
M, Lu H, Schug TT, Cen Y, Jeong SM, Li X, Sauve AA and Haigis MC:
SIRT4 represses peroxisome proliferator-activated receptor α
activity to suppress hepatic fat oxidation. Mol Cell Biol.
33:4552–4561. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ibdah JA, Paul H, Zhao Y, Binford S,
Salleng K, Cline M, Matern D, Bennett MJ, Rinaldo P and Strauss AW:
Lack of mitochondrial trifunctional protein in mice causes neonatal
hypoglycemia and sudden death. J Clin Invest. 107:1403–1409. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Guo L, Zhou SR, Wei XB, Liu Y, Chang XX,
Liu Y, Ge X, Dou X, Huang HY, Qian SW, et al: Acetylation of
mitochondrial trifunctional protein α-subunit enhances its
stability to promote fatty acid oxidation and is decreased in
nonalcoholic fatty liver disease. Mol Cell Biol. 36:2553–2567.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhou ZH, McCarthy DB, O'Connor CM, Reed LJ
and Stoops JK: The remarkable structural and functional
organization of the eukaryotic pyruvate dehydrogenase complexes.
Proc Natl Acad Sci USA. 98:14802–14807. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mathias RA, Greco TM, Oberstein A,
Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T and Cristea
IM: Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase
complex activity. Cell. 159:1615–1625. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
MacDonald MJ, Fahien LA, Brown LJ, Hasan
NM, Buss JD and Kendrick MA: Perspective: Emerging evidence for
signaling roles of mitochondrial anaplerotic products in insulin
secretion. Am J Physiol Endocrinol Metab. 288:E1–E15. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sener A and Malaisse WJ: L-leucine and a
nonmetabolized analogue activate pancreatic islet glutamate
dehydrogenase. Nature. 288:187–189. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Anderson KA, Huynh FK, Fisher-Wellman K,
Stuart JD, Peterson BS, Douros JD, Wagner GR, Thompson JW, Madsen
AS, Green MF, et al: SIRT4 is a lysine deacylase that controls
leucine metabolism and insulin secretion. Cell Metab. 25:838–855
e15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huynh FK, Hu X, Lin Z, Johnson JD and
Hirschey MD: Loss of sirtuin 4 leads to elevated glucose- and
leucine-stimulated insulin levels and accelerated age-induced
insulin resistance in multiple murine genetic backgrounds. J
Inherit Metab Dis. 41:59–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zaganjor E, Vyas S and Haigis MC: SIRT4 is
a regulator of insulin secretion. Cell Chem Biol. 24:656–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Klingenberg M: The ADP and ATP transport
in mitochondria and its carrier. Biochim Biophys Acta.
1778:1978–2021. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ho L, Titus AS, Banerjee KK, George S, Lin
W, Deota S, Saha AK, Nakamura K, Gut P, Verdin E and
Kolthur-Seetharam U: SIRT4 regulates ATP homeostasis and mediates a
retrograde signaling via AMPK. Aging (Albany NY). 5:835–849. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Vander Heiden MG, Lunt SY, Dayton TL,
Fiske BP, Israelsen WJ, Mattaini KR, Vokes NI, Stephanopoulos G,
Cantley LC, Metallo CM and Locasale JW: Metabolic pathway
alterations that support cell proliferation. Cold Spring Harb Symp
Quant Biol. 76:325–334. 2011. View Article : Google Scholar
|
|
69
|
Fuchs BC and Bode BP: Stressing out over
survival: Glutamine as an apoptotic modulator. J Surg Res.
131:26–40. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wise DR, DeBerardinis RJ, Mancuso A, Sayed
N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon
SB and Thompson CB: Myc regulates a transcriptional program that
stimulates mitochondrial glutaminolysis and leads to glutamine
addiction. Proc Natl Acad Sci USA. 105:18782–18787. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
DeBerardinis RJ, Mancuso A, Daikhin E,
Nissim I, Yudkoff M, Wehrli S and Thompson CB: Beyond aerobic
glycolysis: Transformed cells can engage in glutamine metabolism
that exceeds the requirement for protein and nucleotide synthesis.
Proc Natl Acad Sci USA. 104:19345–19350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Daye D and Wellen KE: Metabolic
reprogramming in cancer: Unraveling the role of glutamine in
tumorigenesis. Semin Cell Dev Biol. 23:362–369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lukey MJ, Wilson KF and Cerione RA:
Therapeutic strategies impacting cancer cell glutamine metabolism.
Future Med Chem. 5:1685–1700. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yuan H, Su L and Chen WY: The emerging and
diverse roles of sirtuins in cancer: A clinical perspective. Onco
Targets Ther. 6:1399–1416. 2013.PubMed/NCBI
|
|
75
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Blaveri E, Simko JP, Korkola JE, Brewer
JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P
and Waldman FM: Bladder cancer outcome and subtype classification
by gene expression. Clin Cancer Res. 11:4044–4055. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Choi YL, Tsukasaki K, O'Neill MC, Yamada
Y, Onimaru Y, Matsumoto K, Ohashi J, Yamashita Y, Tsutsumi S,
Kaneda R, et al: A genomic analysis of adult T-cell leukemia.
Oncogene. 26:1245–1255. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jeong SM, Xiao C, Finley LW, Lahusen T,
Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al:
SIRT4 has tumor-suppressive activity and regulates the cellular
metabolic response to DNA damage by inhibiting mitochondrial
glutamine metabolism. Cancer Cell. 23:450–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Mahjabeen I and Kayani MA: Loss of
mitochondrial tumor suppressor genes expression is associated with
unfavorable clinical outcome in head and neck squamous cell
carcinoma: Data from retrospective study. PLoS One.
11:e01469482016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wang YS, Du L, Liang X, Meng P, Bi L, Wang
YL, Wang C and Tang B: Sirtuin 4 depletion promotes hepatocellular
carcinoma tumorigenesis through regulating
adenosine-monophosphate-activated protein kinase alpha/mammalian
target of rapamycin axis in mice. Hepatology. 69:1614–1631. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Fu L, Dong Q, He J, Wang X, Xing J, Wang
E, Qiu X and Li Q: SIRT4 inhibits malignancy progression of NSCLCs,
through mitochondrial dynamics mediated by the ERK-Drp1 pathway.
Oncogene. 36:2724–2736. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chen X, Lai X, Wu C, Tian Q, Lei T, Pan J
and Huang G: Decreased SIRT4 protein levels in endometrioid
adenocarcinoma tissues are associated with advanced AJCC stage.
Cancer Biomark. 19:419–424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang Y, Guo Y, Gao J and Yuan X:
Tumor-suppressive function of SIRT4 in neuroblastoma through
mitochondrial damage. Cancer Manag Res. 10:5591–5603. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Sun H, Huang D, Liu G, Jian F, Zhu J and
Zhang L: SIRT4 acts as a tumor suppressor in gastric cancer by
inhibiting cell proliferation, migration, and invasion. Onco
Targets Ther. 11:3959–3968. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Huang G, Cui F, Yu F, Lu H, Zhang M, Tang
H and Peng Z: Sirtuin-4 (SIRT4) is downregulated and associated
with some clinicopathological features in gastric adenocarcinoma.
Biomed Pharmacother. 72:135–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Shen X, Li P, Xu Y, Chen X, Sun H, Zhao Y,
Liu M and Zhang W: Association of sirtuins with clinicopathological
parameters and overall survival in gastric cancer. Oncotarget.
8:74359–74370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Hu Y, Lin J, Lin Y, Chen X, Zhu G and
Huang G: Overexpression of SIRT4 inhibits the proliferation of
gastric cancer cells through cell cycle arrest. Oncol Lett.
17:2171–2176. 2019.PubMed/NCBI
|
|
88
|
Miyo M, Yamamoto H, Konno M, Colvin H,
Nishida N, Koseki J, Kawamoto K, Ogawa H, Hamabe A, Uemura M, et
al: Tumour-suppressive function of SIRT4 in human colorectal
cancer. Br J Cancer. 113:492–499. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Huang G, Cheng J, Yu F, Liu X, Yuan C, Liu
C, Chen X and Peng Z: Clinical and therapeutic significance of
sirtuin-4 expression in colorectal cancer. Oncol Rep. 35:2801–2810.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhu Y, Wang G, Li X, Wang T, Weng M and
Zhang Y: Knockout of SIRT4 decreases chemosensitivity to 5-FU in
colorectal cancer cells. Oncol Lett. 16:1675–1681. 2018.PubMed/NCBI
|
|
91
|
Shi Q, Liu T, Zhang X, Geng J, He X, Nu M
and Pang D: Decreased sirtuin 4 expression is associated with poor
prognosis in patients with invasive breast cancer. Oncol Lett.
12:2606–2612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Nakahara Y, Yamasaki M, Sawada G, Miyazaki
Y, Makino T, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S,
Mimori K, et al: Downregulation of SIRT4 expression is associated
with poor prognosis in esophageal squamous cell carcinoma.
Oncology. 90:347–355. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Jeong SM, Lee A, Lee J and Haigis MC:
SIRT4 protein suppresses tumor formation in genetic models of
Myc-induced B cell lymphoma. J Biol Chem. 289:4135–4144. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen Z, Lin J, Feng S, Chen X, Huang H,
Wang C, Yu Y, He Y, Han S, Zheng L and Huang G: SIRT4 inhibits the
proliferation, migration, and invasion abilities of thyroid cancer
cells by inhibiting glutamine metabolism. Onco Targets Ther.
12:2397–2408. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Hu Q, Qin Y, Ji S, Xu W, Liu W, Sun Q,
Zhang Z, Liu M, Ni Q, Yu X and Xu X: UHRF1 promotes aerobic
glycolysis and proliferation via suppression of SIRT4 in pancreatic
cancer. Cancer Lett. 452:226–236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability-an evolving hallmark of cancer. Nat Rev Mol
Cell Biol. 11:220–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Colombo SL, Palacios-Callender M, Frakich
N, Carcamo S, Kovacs I, Tudzarova S and Moncada S: Molecular basis
for the differential use of glucose and glutamine in cell
proliferation as revealed by synchronized HeLa cells. Proc Natl
Acad Sci USA. 108:21069–21074. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wang L, Zhou H, Wang Y, Cui G and Di LJ:
CtBP maintains cancer cell growth and metabolic homeostasis via
regulating SIRT4. Cell Death Dis. 6:e16202015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wang L, Li JJ, Guo LY, Li P, Zhao Z, Zhou
H and Di LJ: Molecular link between glucose and glutamine
consumption in cancer cells mediated by CtBP and SIRT4.
Oncogenesis. 7:262018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Csibi A, Fendt SM, Li C, Poulogiannis G,
Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T,
et al: The mTORC1 pathway stimulates glutamine metabolism and cell
proliferation by repressing SIRT4. Cell. 153:840–854. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
van de Ven RAH, Santos D and Haigis MC:
Mitochondrial Sirtuins and molecular mechanisms of aging. Trends
Mol Med. 23:320–331. 2017. View Article : Google Scholar
|
|
102
|
Xing J, Li J, Fu L, Gai J, Guan J and Li
Q: SIRT4 enhances the sensitivity of ER-positive breast cancer to
tamoxifen by inhibiting the IL-6/STAT3 signal pathway. Cancer Med.
8:7086–7097. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Jeong SM, Hwang S and Seong RH: SIRT4
regulates cancer cell survival and growth after stress. Biochem
Biophys Res Commun. 470:251–256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Liu M, Wang Z, Ren M, Yang X, Liu B, Qi H,
Yu M, Song S, Chen S, Liu L, et al: SIRT4 regulates PTEN stability
through IDE in response to cellular stresses. FASEB J.
33:5535–5547. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lee JO, Yang H, Georgescu MM, Di
Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P and Pavletich
NP: Crystal structure of the PTEN tumor suppressor: Implications
for its phosphoinositide phosphatase activity and membrane
association. Cell. 99:323–334. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Georgescu MM, Kirsch KH, Kaloudis P, Yang
H, Pavletich NP and Hanafusa H: Stabilization and productive
positioning roles of the C2 domain of PTEN tumor suppressor. Cancer
Res. 60:7033–7038. 2000.PubMed/NCBI
|
|
107
|
Mizushima N and Klionsky DJ: Protein
turnover via autophagy: Implications for metabolism. Annu Rev Nutr.
27:19–40. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Luo YX, Tang X, An XZ, Xie XM, Chen XF,
Zhao X, Hao DL, Chen HZ and Liu DP: SIRT4 accelerates Ang
II-induced pathological cardiac hypertrophy by inhibiting manganese
superoxide dismutase activity. Eur Heart J. 38:1389–1398.
2017.PubMed/NCBI
|
|
109
|
Xiao Y, Zhang X, Fan S, Cui G and Shen Z:
MicroRNA-497 inhibits cardiac hypertrophy by targeting Sirt4. PLoS
One. 11:e01680782016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zeng G, Liu H and Wang H: Amelioration of
myocardial ischemia-reperfusion injury by SIRT4 involves
mitochondrial protection and reduced apoptosis. Biochem Biophys Res
Commun. 502:15–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Shih J, Liu L, Mason A, Higashimori H and
Donmez G: Loss of SIRT4 decreases GLT-1-dependent glutamate uptake
and increases sensitivity to kainic acid. J Neurochem. 131:573–581.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Komlos D, Mann KD, Zhuo Y, Ricupero CL,
Hart RP, Liu AY and Firestein BL: Glutamate dehydrogenase 1 and
SIRT4 regulate glial development. Glia. 61:394–408. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Ramatchandirin B, Sadasivam M, Kannan A
and Prahalathan C: Sirtuin 4 regulates lipopolysaccharide mediated
leydig cell dysfunction. J Cell Biochem. 117:904–916. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zeng J, Jiang M, Wu X, Diao F, Qiu D, Hou
X, Wang H, Li L, Li C, Ge J, et al: SIRT4 is essential for
metabolic control and meiotic structure during mouse oocyte
maturation. Aging Cell. 17:e127892018. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Nunnari J and Suomalainen A: Mitochondria:
In sickness and in health. Cell. 148:1145–1159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Han Y, Zhou S, Coetzee S and Chen A: SIRT4
and its roles in energy and redox metabolism in health, disease and
during exercise. Front Physiol. 10:10062019. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Min Z, Gao J and Yu Y: The roles of
mitochondrial SIRT4 in cellular metabolism. Front Endocrinol
(Lausanne). 9:7832019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Li S and Zheng W: Mammalian Sirtuins SIRT4
and SIRT7. Prog Mol Biol Transl Sci. 154:147–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Song R, Xu W, Chen Y, Li Z, Zeng Y and Fu
Y: The expression of Sirtuins 1 and 4 in peripheral blood
leukocytes from patients with type 2 diabetes. Eur J Histochem.
55:e102011. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Kim EA, Yang SJ, Choi SY, Lee WJ and Cho
SW: Inhibition of glutamate dehydrogenase and insulin secretion by
KHG26377 does not involve ADP-ribosylation by SIRT4 or
deacetylation by SIRT3. BMB Rep. 45:458–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Lappas M: Anti-inflammatory properties of
sirtuin 6 in human umbilical vein endothelial cells. Mediators
Inflamm. 2012:5975142012. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Sebastian C, Satterstrom FK, Haigis MC and
Mostoslavsky R: From sirtuin biology to human diseases: An update.
J Biol Chem. 287:42444–42452. 2012. View Article : Google Scholar : PubMed/NCBI
|















