Introduction
Renal cell carcinoma (RCC) accounts for 2% of adult
malignancies worldwide and for 80-85% of all malignant kidney
tumors, according to data from 2013 (1,2). Clear
cell carcinoma is the most common pathological subtype of RCC and
is highly resistant to both chemotherapy and radiation therapy
(3). Metabolic reprogramming is of
the utmost importance to oncogenesis, but the generation and
effects of this profound process remain unclear. Compared with
normal cells, glycolysis is enhanced and the mitochondrial
oxidative phosphorylation capacity is reduced in various cancer
cell types, such as in malignant glioma, leukemia and colon cancer
cell lines (4,5). It has been reported that inhibiting the
expression or activity of key glycolytic enzymes can effectively
inhibit tumor cell proliferation and even kill tumor cells. Such
enzymes include lactate dehydrogenase (LDHA), hexokinase 2 (HK2),
phosphofructokinase (PFK) and M2 type acetone kinase (PKM2)
(6,7). Given the increasing incidence of this
cancer and its lack of effective therapeutic targets, there is an
urgent requirement to identify the potential mechanisms by which
clear cell RCC (ccRCC) behavior is regulated.
Hypoxia has long been implicated in genetic
instability and tumor progression. Hypoxia inducible factors (HIFs)
are central players in cellular hypoxia adaptation, and the HIF-1
signaling pathway is critical to tumor development and progression
(8,9). HIF-1 [also known as aryl hydrocarbon
receptor nuclear translocator (ARNT)] is a heterodimeric
transcription factor composed of HIF-1α and HIF-1β (10). ARNT is a member of the basic
helix-loop-helix PER-ARNT-SIM family (11); it is a central player in two cellular
signaling pathways, namely the aryl hydrocarbon receptor and HIF
pathways (12,13). Previous studies have indicated that
the expression of ARNT is decreased in pancreatic islets from
humans with type 2 diabetes (14–16).
ARNT expression is critical to normal angiogenesis and glycolysis,
and the prevention of apoptosis (17). However, no evidence has been reported
concerning the expression and potential functions of ARNT in
ccRCC.
The present study aimed to investigate the role of
ARNT and the possible mechanisms by which it influences the
glycolytic pathway in ccRCC using cBioPortal analysis and in
vitro experiments, in order to provide evidence regarding the
role of ARNT in ccRCC.
Materials and methods
Dataset collection and samples from
patients with ccRCC
A total of 58 patients with ccRCC who had never
previously been treated with radiotherapy or chemotherapy were
enrolled for the study. According to ARNT expression, patients were
divided into three groups, and their clinical characteristics are
shown in Table I. Tumor tissues were
taken from the surgically resected tissues of patients with ccRCC
treated at The Second Hospital of Shandong University (Jinan,
China) between April 2018 and April 2019. All patients provided
written informed consent. Furthermore, this study was approved by
the Human Research Ethics Committee of The Second Hospital of
Shandong University.
 | Table I.Patient clinical characteristics. |
Table I.
Patient clinical characteristics.
| Characteristic | Group 1 (n=12) | Group 2 (n=17) | Group 3 (n=29) |
|---|
| ARNT expression | Weak | Moderate | Strong |
| Male/Female, n | 5/7 | 11/6 | 20/9 |
| Mean age (range),
years | 56.8 (39–76) | 55.0 (38–70) | 58.2 (35–82) |
Oncomine (http://www.oncomine.org), an online microarray
database, was used to analyze differences in mRNA expression for
ARNT and key enzymes involved in the glycolysis pathway [LDHA, HK2,
PKM2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3
(PFKFB3)] between tumor and normal tissues from patients with
ccRCC. Genes were considered to be significantly differentially
expressed using P<0.05 and |LogFC|>2.
Preprocessed level 3 RNA-seq data were downloaded
from The Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov). For TCGA data, the
edgeR package version 3.30.3 (https://bioconductor.org/packages/edgeR/) was used for
differentially expressed gene (DEG) screening. P<0.05 and
|LogFC|>2 were chosen as the cut-off criteria.
Alterations in the ARNT gene occurring in ccRCC were
investigated using cBioPortal (http://www.cbioportal.org) to generate a network of
the interactions between ARNT and its neighboring genes.
Immunohistochemistry
Tumor tissues from patients with ccRCC were fixed in
4% paraformaldehyde for 24–48 h at room temperature, dehydrated
using 70, 80 and 95% alcohol (45 min each), followed by 3 washes
using 100% alcohol (1 h each), and cleared using 2 washes of xylene
(1 h each), and then embedded in paraffin. Paraffin-embedded
tissues were cut into 4-µm-thick sections. Subsequently, the
sections were stained using the Mouse two-step test kit (Mouse
reinforced Polymer test System; cat. no. PV-9002; OriGene
Technologies, Inc.) according to the manufacturers protocol.
Briefly, to inactivate endogenous peroxidase, an appropriate amount
of endogenous peroxidase blocker (included in the Mouse two-step
test kit) was added at room temperature for 10 min, while to block
non-specific antibody binding, the slides were incubated with 5%
bovine serum albumin (cat. no. 0332; GBCBIO Technologies, Inc.) for
30 min at room temperature. Subsequently, the slides were incubated
with an anti-ARNT primary antibody (1:200; cat. no. ab2771; Abcam)
at 4°C overnight. The slides were washed 3 times (3 min each) in
PBS buffer (Gibco; Thermo Fisher Scientific, Inc.). Next, the
Enhance enzyme-labeled secondary antibody (goat anti-mouse IgG)
from the kit was applied to the slides and incubated in a
humidified chamber at room temperature for 1 h. Images were taken
using a light microscope (magnification, ×200). Nuclei were
counterstained using a hematoxylin counterstain reagent (Roche
Diagnostics) for 1–2 min at room temperature. Scoring was based on
the color intensity: 0, no staining; 1, light yellow; 2,
yellow-brownish; and 3, brown. Scoring of the percentage of
positive cells was performed as follows: 0, 1-10%; 1, 11-25%; 2,
26-50%; 3, 51-75%; and 4, 76-100%. Finally, the two scores were
multiplied to obtain the final score: 0, negative; 1-4, weak; 5-8,
moderate; and 9-12, strong expression.
Cell culture and transfection
The normal human kidney HK-2 cell line and the RCC
cell lines A498, OS-RC-2 and 786-O were purchased from The Cell
Bank of Type Culture Collection of the Chinese Academy of Science.
The HK-2 cell line was used as a normal control of RCC cell lines.
A498, OS-RC-2 and 786-O cells were used to study the function of
ARNT in RCC cell lines. In a humidified 5% CO2
environment at 37°C, HK-2 and OS-RC-2 cells were cultured in DMEM
supplemented with 10% FBS (both Gibco; Thermo Fisher Scientific,
Inc.). A498 and 786-O were cultured in RPMI 1640 (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS. To generate
ARNT-overexpressing stable cell populations (OV-ARNT), OS-RC-2
cells were infected with an empty vector control and an ARNT
lentivirus vector, which were constructed by Shanghai GeneChem Co.,
Ltd. OS-RC-2 cells were seeded until they reached 60-80% confluency
the following day. After the cells were attached to the walls, they
were infected with ARNT lentivirus or control lentivirus for 16 h,
the medium was replaced with fresh DMEM supplemented with 10% FBS
and subsequent experiments were performed after 48 h. The virus was
used at a multiplicity of infection of 10 to infect OS-RC-2. The
efficiency of infection was assessed via western blot analysis. To
generate ARNT-knockdown stable cell populations (si-ARNT), three
siRNAs to ARNT and a negative control (NC) siRNA were ordered from
Guangzhou RiboBio Corporation Co., Ltd. A498 cells were seeded for
24 h until they reached 60-80% confluency, and were then
transfected with 20 µM siRNAs using Oligofectamine™ reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturers protocol. Subsequent experiments were conducted 6 h
after transfection. The sequences used for the knockdown
experiments were as follows: NC sense, 5-UUCUCCGAACGUGUCACGUTT-3
and antisense, 5-ACGUGACACGUUCGGAGAATT-3; ARNT-homo-926
(si-ARNT-homo-1) sense, 5-GGCUCAAGGAGAUCGUUUATT-3 and antisense,
5-UAAACGAUCUCCUUGAGCCTT-3; ARNT-homo-1442 (si-ARNT-homo-2) sense,
5-CGGUCUAAGAACCAAGAAUTT-3 and antisense, 5-AUUCUUGGUUCUUAGACCGTT-3;
and ARNT-homo-1942 (si-ARNT-homo-3) sense,
5-GGCAGAGAAUUUCAGGAAUTT-3 and antisense,
5-AUUCCUGAAAUUCUCUGCCTT-3.
Cell migration and invasion
assays
Cell migration and invasion assays were performed as
previously described (18). Briefly,
5×105 cells were placed in 6-well plates. The migration
rate of OS-RC-2 and A498 cells pre-transfected with ARNT or control
was assessed by wound-healing assays, with 1% FBS used in the
culture medium. Next, photomicrographs were captured of OS-RC-2 and
A498 cells under a light microscope (Leica DFC300FX; Leica
Microsystems, Inc.; magnification, ×40) at 0 and 24 h. Transwell
invasion assays were performed in OS-RC-2 and A498 cells
pre-transfected with ARNT or control (3×104 cells/well).
Cells were placed in an 8-mm Transwell cell culture chamber
(Corning Inc.) FBS-free DMEM was added in the upper chamber, while
DMEM with 10% FBS was used in the lower chamber. The cells and
Matrigel (Becton, Dickinson and Company) were incubated in a 37°C
incubator for 36 h. After 36 h, the cells were fixed with methanol
for 30 min at room temperature and stained using crystal violet
solution (1X PBS, 0.05% w/v crystal violet, 1% formaldehyde and 1%
methanol) for 30 min at room temperature. Cells in the upper
chamber were removed using a cotton swab. The migrated cells were
observed under a light microscope (Olympus Corporation;
magnification, ×100) and counted manually from five randomly
selected fields.
Cell proliferation assays
To investigate the sensitivity to sorafenib (LC
Laboratories), the Cell Counting Kit-8 (CCK-8; Beyotime Institute
of Biotechnology) assay was used to detect the cell survival rate,
according to the manufacturers protocol. Cells were treated with
different concentrations of sorafenib (5, 10 or 20 µM) for 48 h at
37°C. DMSO group served as a control. Briefly, 5×103
OS-RC-2 or A498 cells were seeded in a 96-well plate and cell
proliferation was evaluated after 48 h of sorafenib treatment.
Next, 10 µl WST-1 reagent was added per well and the cultures were
incubated for 2 h, after which the absorbance was measured at 450
nm using a microplate reader (BioTek Instruments Inc.).
Western blot analysis and
antibodies
Western blotting was performed as previously
described (19). Briefly, cultured
cells (HK-2, A498, OS-RC-2 and 786-O) were lysed using
radioimmunoprecipitation buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology) and centrifuged at 13,000 × g for 20
min at 4°C. Protein concentrations of the extracts were determined
using a BCA protein assay kit (cat. no. 23227; Pierce; Thermo
Fisher Scientific, Inc.). Soluble lysates (20 µg/lane) were
subjected to 10% SDS-PAGE and transferred to nitrocellulose
membranes (cat. no. 10402495; Whatman plc; Cytiva). Membranes were
incubated with primary antibodies overnight at 4°C and then with
HRP-conjugated secondary antibodies for 1 h at room temperature.
ECL Prime Western Blotting Detection Reagent (cat. no. RPN2232; GE
Healthcare) was used to visualize protein bands via Image Quant
LAS4000 Imaging Systems (GE Healthcare). Relative protein contents
were quantified using the Quantity One software (v462 version.
Bio-Rad Laboratories, Inc.). Proteins were assessed using the
following primary antibodies (diluted in 5% BSA): ARNT (1:4,000;
cat. no. ab2771), LDHA (1:1,000; cat. no., ab101562), HK2 (1:500;
cat. no. ab104836), PKM2 (1:1,000; cat. no. ab85555), PFKFB3
(1:1,000; cat. no. ab181861), tubulin (1:500, cat. no. ab6046) and
β-actin (1:200; cat. no. ab115777) (all Abcam). HRP-linked
secondary antibodies included anti-rabbit IgG (cat. no. 7074) and
anti-mouse IgG (cat. no. 7076) (both diluted 1:1,000 in 5% BSA;
Cell Signaling Technology, Inc.).
Statistical analysis
All data were analyzed using SPSS version 18.0
(SPSS, Inc.). Data are presented as the mean ± standard error.
Statistical differences were calculated by two-tailed unpaired
Students t-test or one-way ANOVA and Tukeys post hoc test.
P<0.05 was used to indicate a statistically significant
difference.
Results
Expression profiles of ARNT in
patients with ccRCC and in different RCC cell lines
ARNT is a transcription factor that plays a critical
role in the response to environmental stresses, such as dioxin
exposure and hypoxia (11). However,
few studies have explored its functions, particularly its
physiological role in ccRCC. Therefore, the present study examined
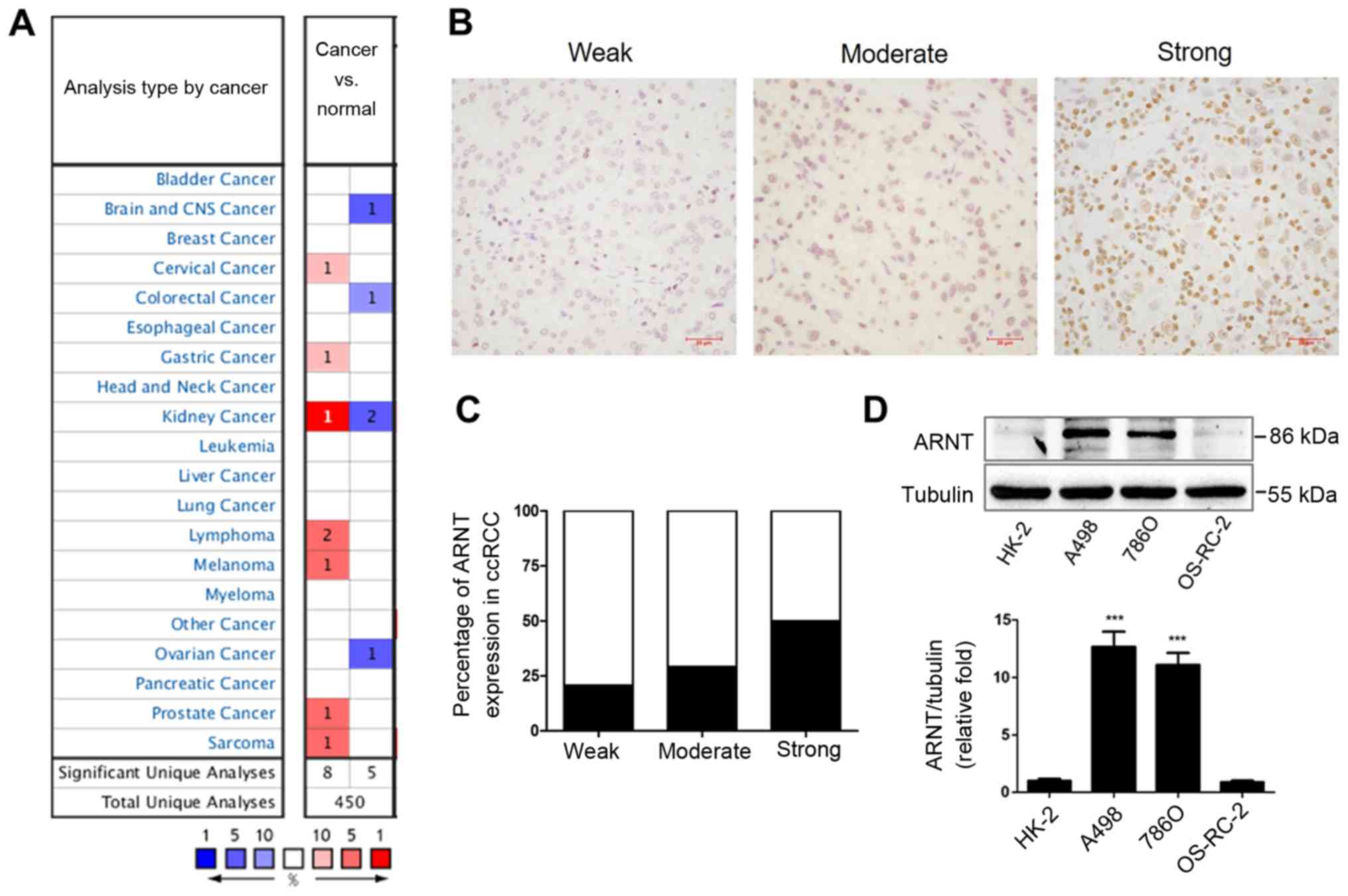
the expression profiles of ARNT using the Oncomine database. ARNT
expression was found to be significantly upregulated in 8 analyses
and downregulated in 5 analyses in different cancer types (Fig. 1A). Next, to validate the expression
profiles of ARNT, immunohistochemical staining was performed to
examine ARNT expression in tumor tissues from 58 patients with
ccRCC (Fig. 1B). Notably, nearly 50%
of the clinical samples showed that ARNT was strongly upregulated
(Fig. 1C). Furthermore, varying
levels of ARNT expression were detected in different RCC cell lines
in vitro. A498 and 786-O RCC cell lines showed significantly
elevated expression of ARNT compared with the human renal tubular
epithelial cell line (HK-2), while there was no significant
difference in expression in the OS-RC-2 cell line (Fig. 1D).
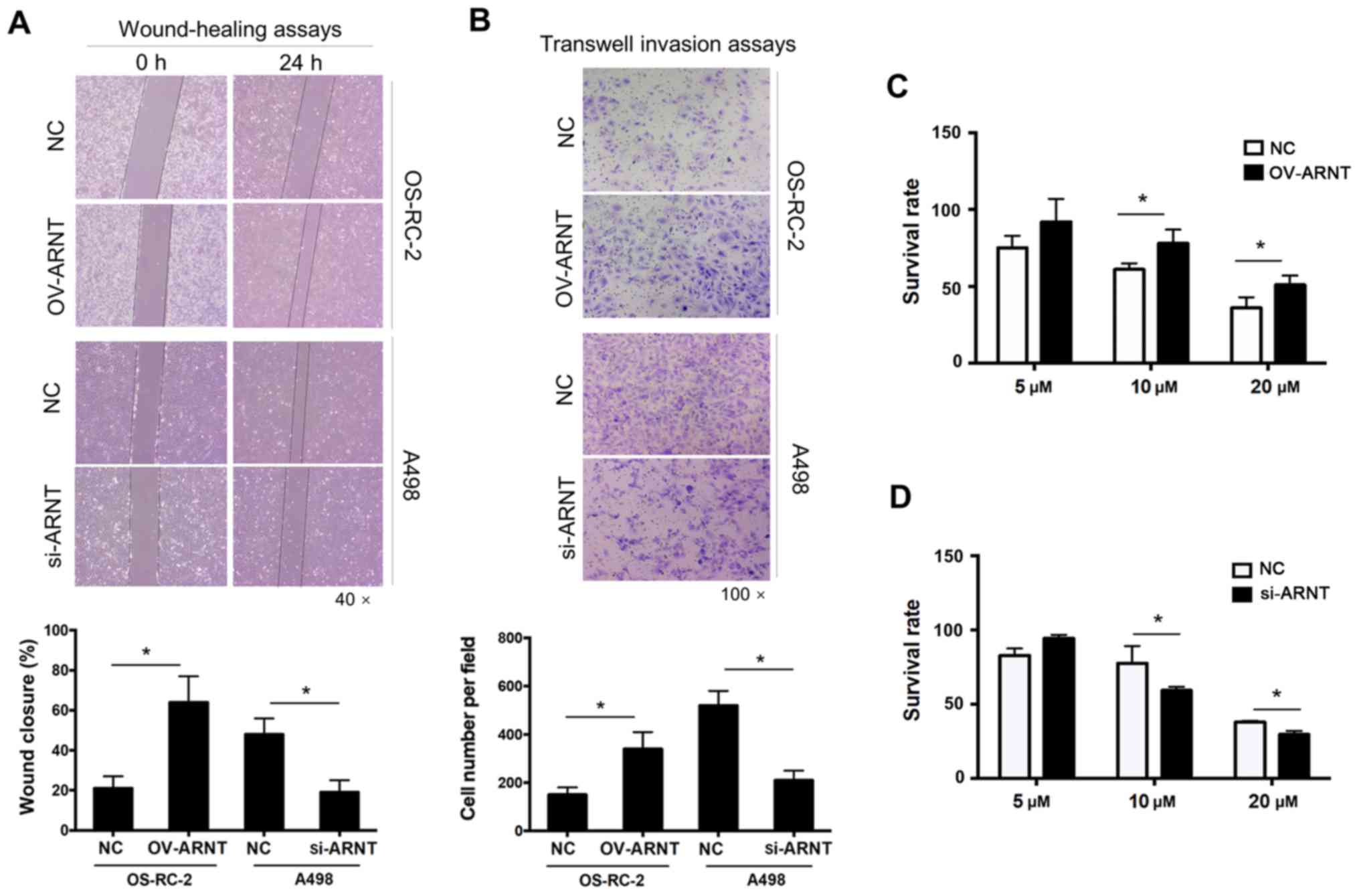
ARNT promotes the proliferation,
migration and invasion of human ccRCC cells
To investigate the roles of ARNT in the biological
behavior of ccRCC cells, overexpression and knockdown experiments
were performed. In Fig. 1D, the A498
and 786-O RCC cell lines showed significantly upregulated ARNT
expression compared with the human renal tubular epithelial cell
line (HK-2), while there was no significant difference in ARNT
expression in the OS-RC-2 cell line. Therefore, the OS-RC-2 cell
line was selected for ARNT overexpression, and A498 cells were
chosen for ARNT-knockdown, which involved transfection of ARNT into
OS-RC-2 cells and silencing of endogenous ARNT expression in A498
cells using a lentiviral vector. Compared with silencing ARNT in
A498 cells, overexpression of ARNT in OS-RC-2 cells resulted in
stronger migratory abilities and accelerated cell proliferation.
The wound-healing assay showed greater wound closure in the OS-RC-2
OV-ARNT group compared with that in the control group at 24 h. By
contrast, less wound closure was noted in the A498-siRNA-ARNT group
compared with that in the control group (Fig. 2A). Significantly higher migration
rates were observed in OS-RC-2 OV-ARNT cells, as revealed by
Transwell migration assays (Fig.
2B). Sorafenib is the first oral multi-kinase inhibitor that
targets Raf and affects tumor cell proliferation and tumor
angiogenesis (20). It has been
reported that sorafenib treatment prolongs survival rates in
patients with ccRCC in whom previous therapy has failed (21). To investigate whether the increased
sensitivity to Sorafenib in ccRCC is associated with ARNT
expression, the cell survival rate was examined 48 h after
treatment with sorafenib using the CCK-8 assay. After ARNT
overexpression, the sensitivity of OS-RC-2 OV-ARNT cells to
sorafenib was decreased and the cell survival rate was increased
compared with that of OS-RC-2 NC cells at 10 and 20 µM (Fig. 2C). By contrast, the survival rate of
the A498-siRNA-ARNT cells was adversely affected compared with that
of the NC (Fig. 2D). These results
suggest ARNT may be a target of sorafenib, the effects of which
could improve the treatment of drug-resistant ccRCC.
ARNT is involved in the glycolysis
pathway
Next, the possible mechanism influencing ARNT
function was investigated. A network of the interactions of ARNT
with several frequently altered neighboring genes was drawn using
cBioPortal (Fig. 3A). The network
reflected a clear interaction between ARNT and several glycolytic
genes, including LDHA, HK2, PKM2 and PFKFB3. Moreover, in the
Oncomine database, key glycolysis pathway enzymes (LDHA, HK2 and
PKM2) were significantly upregulated in kidney cancer (Fig. 3B). Additionally, based on TCGA
dataset, a set of glycolysis pathway enzymes, including LDHA, HK2,
PKM2 and PFKFB3, were identified from the DEGs, all of which were
significantly upregulated (Fig. 3C).
These results suggest that ARNT may contribute to the cancer
program through effects on the glycolysis pathway.
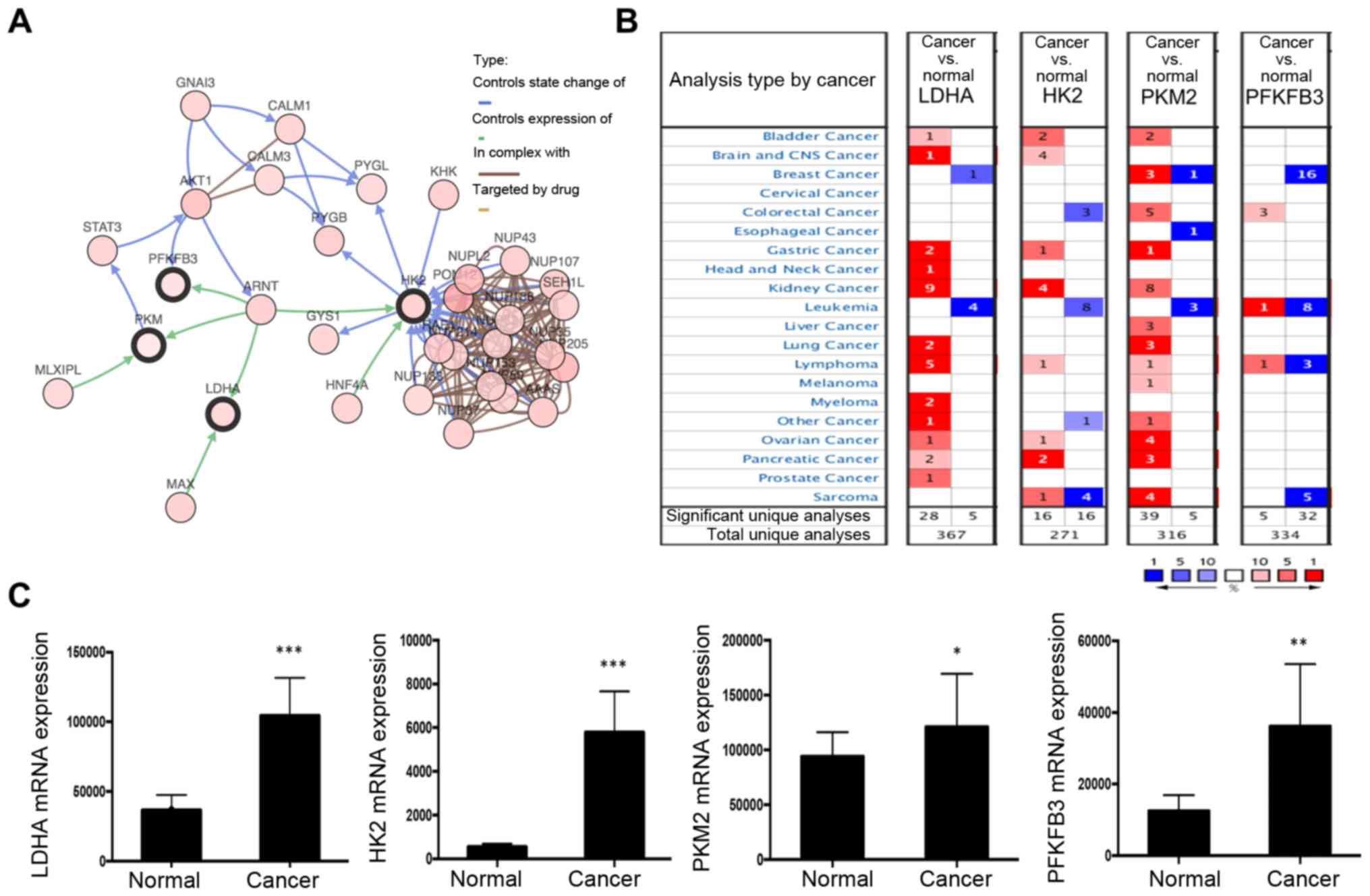 | Figure 3.Network of ARNT and altered
neighboring genes. (A) Interaction of ARNT with other genes was
analyzed in the cBioPortal database. (B) Summary of LDHA, HK2, PKM2
and PFKFB3 expression in different cancer types. (C) The clinical
significance of glycolysis pathway protein (LDHA, HK2, PKM2 and
PFKFB3) expression in ccRCC, based on The Cancer Genome Atlas data,
was investigated. *P<0.05, **P<0.01 and ***P<0.001 vs.
normal. ARNT, aryl hydrocarbon receptor nuclear translocator;
PFKFB3, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3;
LDHA, lactate dehydrogenase; HK2, hexokinase; PFK,
phosphofructokinase; PKM2, M2 type acetone kinase. |
Effects of ARNT on the glycolysis
pathway in RCC cell lines
The effect of ARNT on glycolysis pathway-related
proteins was assessed in RCC cell lines. Western blot analysis
revealed markedly increased ARNT protein levels in OS-RC-2 OV-ARNT
cells and significantly decreased ARNT protein levels in
A498-si-ARNT-2/3 cells, compared with the respective NCs (Fig. 4A and B). Overexpression of ARNT in
RCC cells led to upregulated HK2 and PFKFB3 expression compared
with the NC (Fig. 4A). By contrast,
downregulation of ARNT caused significantly decreased protein
levels for certain glycolysis pathway members, namely PFKFB3 and
HK2, compared with the NC (Fig.
4B-F). These results suggest that ARNT is involved in the
regulation of glycolysis.
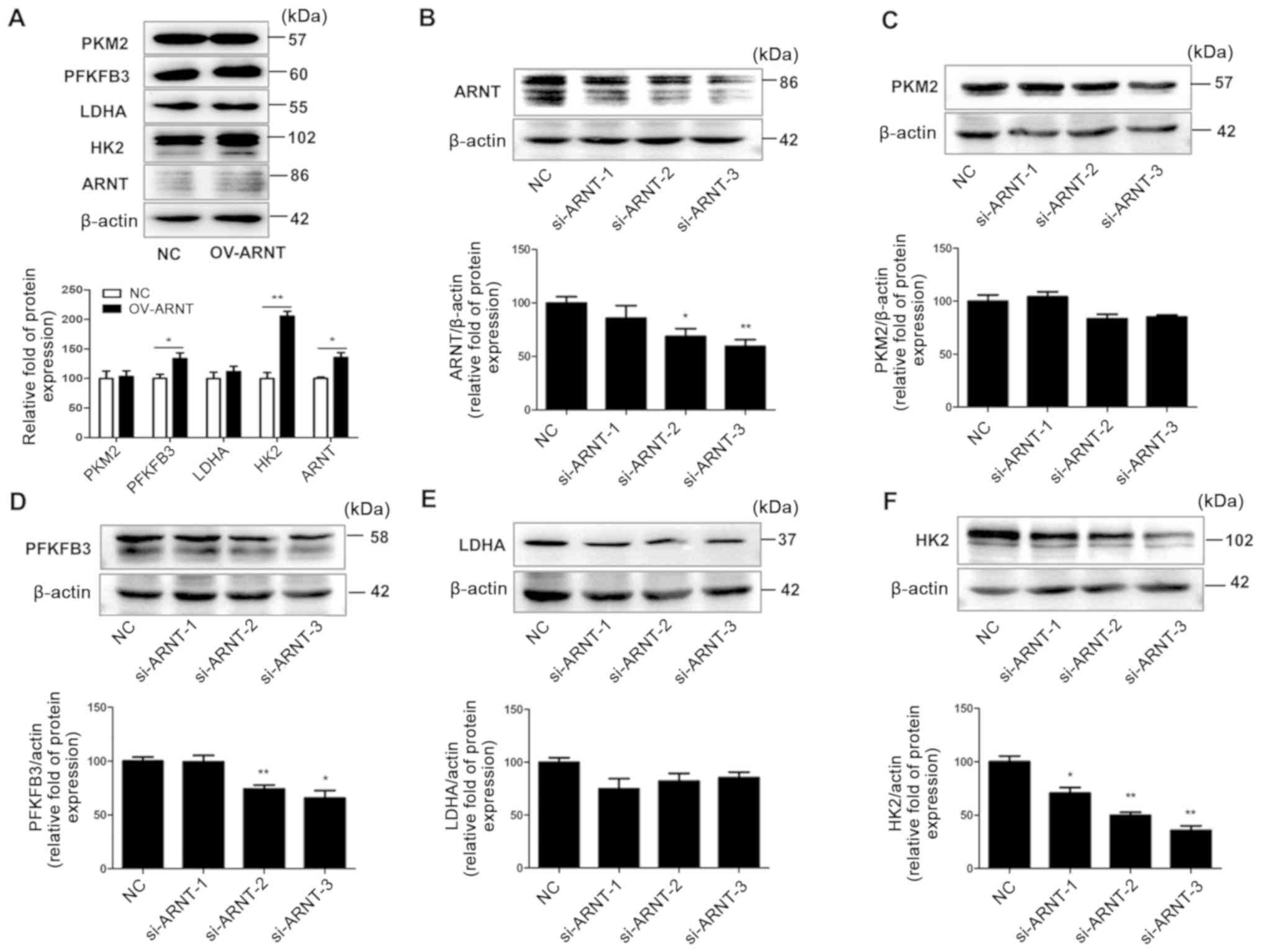 | Figure 4.ARNT is associated with the glycolysis
signaling pathway in RCC cells. OS-RC-2 cells were transfected with
ARNT to overexpress ARNT, while A498 cells were transfected with a
lentiviral vector carrying siRNA to silence endogenous ARNT. (A)
Protein levels of key glycolysis pathway proteins in OS-RC-2
OV-ARNT cells. Protein levels of (B) ARNT, (C) PKM2, (D) PFKFB3,
(E) LDHA and (F) HK2 in A498 si-ARNT cells. β-actin was used as an
internal control. *P<0.05 and **P<0.01 vs. NC. ARNT, aryl
hydrocarbon receptor nuclear translocator; PFKFB3,
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3; LDHA,
lactate dehydrogenase; HK2, hexokinase; PFK, phosphofructokinase;
PKM2, M2 type acetone kinase. |
Discussion
It has been reported that the HIF signaling pathway
consists of three α subunits (HIF-1α, HIF-2α and HIF-3α) and two β
subunits (ARNT and ARNT2) (22). The
major difference between the α and β subunits is the type of
regulation; the regulation of HIF-1α is influenced by oxygen
tension, whereas ARNT is constitutively and ubiquitously expressed.
In contrast to ARNT, ARNT2 is only expressed in the central nervous
system, in the kidneys and in breast cancer (11).
In the present study, it was found that ARNT
expression was significantly upregulated in patients with ccRCC.
The present research on ARNT function is limited, and the
regulation of ARNT is still poorly understood and somewhat
controversial. Most studies report that ARNT is not regulated by
oxygen tension (11). However, Wang
et al (23) reported that
ARNT mRNA and protein were specifically detected in cells exposed
to hypoxia. These findings suggest that ARNT is a hypoxia-inducible
protein similar to HIF-1α. Furthermore, Chilov et al
(10) demonstrated that the hypoxic
inducibility of ARNT occurs in specific cell lines. In the study,
ARNT was induced in L929 and Hepa1 cell lines under hypoxic
conditions; however, ARNT levels did not change in human HeLa,
Hep3B and LN229 cells when exposed to hypoxic conditions. Vavilala
et al (24) and Mandl et
al (25) provided additional
support for cell line specificity in the hypoxia-dependent
regulation of ARNT, suggesting that ARNT is upregulated under
hypoxia, for example in human melanoma cells. Previous studies
indicate that ARNT is important for normal angiogenesis and
glycolysis, and that it may impart anti-apoptotic effects (17). However, its expression and function
in ccRCC has not been reported. To the best of our knowledge, the
present study is the first to demonstrate that ARNT is highly
expressed in patients with ccRCC. ARNT expression was also
increased in A498 and 786-O RCC cell lines, and ARNT expression was
significantly upregulated in kidney cancer according to the
Oncomine database, suggesting that the regulation of ARNT may be
involved in the development of ccRCC.
Therefore, the function of ARNT in the ccRCC process
was studied. Overexpression of ARNT led to a stronger migratory
ability and accelerated cell proliferation. According to the
Guidelines for the Diagnosis and Treatment of Renal Cancer
(26), sunitinib is used as the
first-line therapy for RCC; however, National Comprehensive Cancer
Network guidelines do not recommend sorafenib as first-line
treatment for patients with ccRCC. Since sorafenib has a good
tolerance and has shown high efficiency in the Asian population, it
is still recommended as a first-line treatment in some renal cancer
patients in China. Therefore, it is meaningful to research the
mechanism of sorafenib in Chinese ccRCC patients. In the present
study, the results also showed that ARNT inhibition reduces
accelerated cell proliferation in response to sorafenib treatment,
suggesting that targeting ARNT could represent a novel approach to
kidney cancer therapy.
Previous studies have indicated that ARNT regulates
the transcription of a number of genes involved in glucose
metabolism and vascular functions, including phosphoglycerate
kinase 1, vascular endothelial growth factor, plasminogen activator
inhibitor 1 and erythropoietin (27–30).
ARNT may also regulate aldolase in breast carcinomas and in
hepatoma cells (27,28). In the present study, ARNT was shown
to regulate key enzymes involved in glycolysis. In addition, HK2
and PKM2 were upregulated in RCC cells overexpressing ARNT,
suggesting that ARNT contributes to the cancer program through
effects on the glycolytic pathway.
Furthermore, it has been reported that mutations of
the von Hippel-Lindau (VHL) gene are the main driver events in
ccRCC, and loss of VHL products will alter the expression of
HIF-1α/2α and their downstream targets (31). As continuously activated HIF forms a
pseudo-anoxic state, ccRCC cells are consequently in a
pseudo-hypoxic state: HIF-1α is stabilized and hypoxia inducible
genes are upregulated (32).
Therefore, the HIF-1α stabilization also leads to higher ARNT
levels as a compensatory mechanism. Future research will be focused
on this area.
Overall, the present study indicated that ARNT is
involved in regulating glycolysis and cell proliferation in ccRCC.
Therefore, ARNT may play an important role in kidney cancer and
could represent a new potential therapeutic target for ccRCC.
Acknowledgements
The authors would like to thank Mr Deqiang Huang
(Department of Gastroenterology, Research Institute of Digestive
Diseases, The First Affiliated Hospital of Nanchang University,
Nanchang, Jiangxi, China) for providing suggestions on the design
of the present study.
Funding
The present study was supported by grants from the
National Nature Science Foundation of China (grant no. 31460304)
and the Nature Science Foundation of Jiangxi province of China
(grant nos. 20181BAB205050, 20192BAB205072 and 20171BCB23086).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors contributions
DH and XZ designed the experiments. YZ and FH
performed the experiments. YZ and CZ analyzed the data. YZ wrote
the manuscript. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki. The Ethics Committee of The Second
Hospital of Shandong University approved the study. The
participants approved the use of clinical samples by providing
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Epidemiology, pathology, and pathogenesis
of renal cell carcinoma. UpToDate. 2020.
|
|
2
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lopez-Beltran A, Carrasco JC, Cheng L,
Scarpelli M, Kirkali Z and Montironi R: 2009 update on the
classification of renal epithelial tumors in adults. Int J Urol.
16:432–443. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng J: Energy metabolism of cancer:
Glycolysis versus oxidative phosphorylation (Review). Oncol Lett.
4:1151–1157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Favaro E, Lord S, Harris AL and Buffa FM:
Gene expression and hypoxia in breast cancer. Genome Med. 3:552011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li W, Qiu Y, Hao J, Zhao C, Deng X and Shu
G: Dauricine upregulates the chemosensitivity of hepatocellular
carcinoma cells: Role of repressing glycolysis via
miR-199a:HK2/PKM2 modulation. Food Chem Toxicol. 121:156–165. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang Y, Lin D and Taniguchi CM: Hypoxia
inducible factor (HIF) in the tumor microenvironment: Friend or
foe? Sci China Life Sci. 60:1114–1124. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McKeown SR: Defining normoxia, physoxia
and hypoxia in tumours-implications for treatment response. Br J
Radiol. 87:201306762014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chilov D, Camenisch G, Kvietikova I,
Ziegler U, Gassmann M and Wenger RH: Induction and nuclear
translocation of hypoxia-inducible factor-1 (HIF-1):
Heterodimerization with ARNT is not necessary for nuclear
accumulation of HIF-1alpha. J Cell Sci. 112:1203–1212.
1999.PubMed/NCBI
|
|
11
|
Mandl M and Depping R: Hypoxia-inducible
aryl hydrocarbon receptor nuclear translocator (ARNT) (HIF-1β): Is
it a rare exception? Mol Med. 20:215–220. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abel J and Haarmann-Stemmann T: An
introduction to the molecular basics of aryl hydrocarbon receptor
biology. Biol Chem. 391:1235–1248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zagórska A and Dulak J: HIF-1: The knowns
and unknowns of hypoxia sensing. Acta Biochim Pol. 51:563–585.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gunton JE, Kulkarni RN, Yim S, Okada T,
Hawthorne WJ, Tseng YH, Roberson RS, Ricordi C, OConnell PJ,
Gonzalez FJ and Kahn CR: Loss of ARNT/HIF1beta mediates altered
gene expression and pancreatic-islet dysfunction in human type 2
diabetes. Cell. 122:337–349. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
da Silva Xavier G, Rutter J and Rutter GA:
Involvement of Per-Arnt-Sim (PAS) kinase in the stimulation of
preproinsulin and pancreatic duodenum homeobox 1 gene expression by
glucose. Proc Natl Acad Sci USA. 101:8319–8324. 2004. View Article : Google Scholar
|
|
16
|
Levisetti MG and Polonsky KS: Diabetic
pancreatic beta cells ARNT all they should be. Cell Metab. 2:78–80.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fernandez-Salguero PM, Ward JM, Sundberg
JP and Gonzalez FJ: Lesions of aryl-hydrocarbon receptor-deficient
mice. Vet Pathol. 34:605–614. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang Q, Sun Y, Ma X, Gao Y, Li X, Niu Y,
Zhang X and Chang C: Androgen receptor increases hematogenous
metastasis yet decreases lymphatic metastasis of renal cell
carcinoma. Nat Commun. 8:9182017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ju L, Zhang X, Deng Y, Han J, Yang J, Chen
S, Fang Q, Yang Y and Jia W: Enhanced expression of survivin has
distinct roles in adipocyte homeostasis. Cell Death Dis.
8:e25332017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Escudier B, Eisen T, Stadler WM, Szczylik
C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA,
et al: Sorafenib in advanced clear-cell renal-cell carcinoma. N
Engl J Med. 356:125–134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wilhelm SM, Carter C, Tang L, Wilkie D,
McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, et al:
BAY 43-9006 exhibits broad spectrum oral antitumor activity and
targets the RAF/MEK/ERK pathway and receptor tyrosine kinases
involved in tumor progression and angiogenesis. Cancer Res.
64:7099–7109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rankin EB and Giaccia AJ: The role of
hypoxia-inducible factors in tumorigenesis. Cell Death Differ.
15:678–685. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang GL, Jiang BH, Rue EA and Semenza GL:
Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS
heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci
USA. 92:5510–5514. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vavilala DT, Ponnaluri VK, Vadlapatla RK,
Pal D, Mitra AK and Mukherji M: Honokiol inhibits HIF pathway and
hypoxia-induced expression of histone lysine demethylases. Biochem
Biophys Res Commun. 422:369–374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mandl M, Kapeller B, Lieber R and Macfelda
K: Hypoxia-inducible factor-1β (HIF-1β) is upregulated in a
HIF-1α-dependent manner in 518A2 human melanoma cells under hypoxic
conditions. Biochem Biophys Res Commun. 434:166–172. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guidelines for the diagnosis and treatment
of renal cancer. National Health Commission of the Peoples Republic
of China. 2018.
|
|
27
|
Zelzer E, Levy Y, Kahana C, Shilo BZ,
Rubinstein M and Cohen B: Insulin induces transcription of target
genes through the hypoxia-inducible factor HIF-1alpha/ARNT. EMBO J.
17:5085–5094. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Salceda S, Beck I and Caro J: Absolute
requirement of aryl hydrocarbon receptor nuclear translocator
protein for gene activation by hypoxia. Arch Biochem Biophys.
334:389–394. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Forsythe JA, Jiang BH, Iyer NV, Agani F,
Leung SW, Koos RD and Semenza GL: Activation of vascular
endothelial growth factor gene transcription by hypoxia-inducible
factor 1. Mol Cell Biol. 16:4604–4613. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Okino ST, Chichester CH and Whitlock JP
Jr: Hypoxia-inducible mammalian gene expression analyzed in vivo at
a TATA-driven promoter and at an initiator-driven promoter. J Biol
Chem. 273:23837–23843. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bratslavsky G, Sudarshan S, Neckers L and
Linehan WM: Pseudohypoxic pathways in renal cell carcinoma. Clin
Cancer Res. 13:4667–4671. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Myszczyszyn A, Czarnecka AM, Matak D,
Szymanski L, Lian F, Kornakiewicz A, Bartnik E, Kukwa W, Kieda C
and Szczylik C: The role of hypoxia and cancer stem cells in renal
cell carcinoma pathogenesis. Stem Cell Rev Rep. 11:919–943. 2015.
View Article : Google Scholar : PubMed/NCBI
|