Introduction
Cancer is a major public health problem in China.
Laryngeal squamous cell carcinoma (LSCC) is the most common
malignant disease of the head and neck area, and is responsible for
99% of primary laryngeal carcinoma worldwide (1). Due to the increasing incidence rate (7
cases/100,000) (2), LSCC has
gradually caused extensive concern among researchers and medical
professionals for intensified research (3,4) and
clinical trials (5–8). Although surgery combined with
chemoradiotherapy has significantly improved the survival rate of
patients (5-year survival rate is 63%) (2), recurrence and metastasis are still
common and the prognosis remains poor, particularly in patients at
the advanced stage. Therefore, the discovery of novel specific
markers for early diagnosis and prognosis is urgently needed to
improve patient survival.
Melanoma-associated antigen (MAGE), which was first
discovered by Van der Bruggen (9)
and termed MAGE-I, is a group of well-differentiated members of
cancer/testicular antigens (CTA) (9). Thus far, ~60 members of MAGE have been
discovered and investigated (10).
The MAGE gene family encodes tumor antigens recognized by
autologous cytotoxic T lymphocytes (11,12).
Based on the difference in gene expression and genetic structure,
the MAGE family is categorized into two subfamilies,
MAGE-I (MAGE-A, MAGE-B and MAGE-C) and MAGE-II
(MAGE-D) (13). The most
widely studied gene is MAGE-A, which is strictly
tumor-specific, and includes 12 family members, termed
MAGE-A1-12 (13).
MAGE-A is expressed in several types of tumors, such as
breast and gastric cancer as well as glioma (12,14).
The expression of MAGE-A genes in the
peripheral blood of patients with LSCC remains unclear. Due to the
high similarity in the sequences of MAGE-A1, -A2, -A3, -A4
and-A6, it is difficult to design a unique primer to detect
the different genes. In order to investigate the expression of the
MAGE-A family genes in the peripheral blood of patients with
LSCC and its association with prognosis, multiple MAGE-A
genes in the peripheral blood of 104 patients with LSCC and 30
healthy volunteers were detected by multiple nested reverse
transcription (RT)-PCR and restriction endonuclease treatment. The
aim of the present study was to explore whether the expression of
MAGE-As in the peripheral blood circulating tumor cells
(CTCs) may be used as a biomarker for guiding clinical treatment
and monitoring prognosis in patients with LSCC.
Materials and methods
Patients and clinical parameters
A total of 104 patients with LSCC were recruited
from the Department of Otolaryngology, The Fourth Hospital of Hebei
Medical University between June 2011 and June 2012. In addition, 30
healthy volunteers with no history of carcinoma were enrolled in
the present study during the same period. None of the patients
underwent chemotherapy or radiotherapy prior to surgery. Written
informed consent was provided by all participants before
enrollment. The study protocol was approved by the Medical Ethics
Committee of The Fourth Hospital of Hebei Medical University
(approval no. 2011KY112).
The clinicopathological data of the patients were
retrospectively collected, including age, smoking history, tumor
size, clinical stage (8th edition of the American Joint Committee
on Cancer) (15), clinical
classification, pathological degree and lymph node metastasis.
Blood and tissue sample
collection
Fresh blood samples (5 ml) were collected from the
patients before surgery, as well as from the volunteers. All blood
samples were immediately stored at 4°C, and RNA extraction was
performed on the day of sample collection. Blood samples were
processed within 1 to 4 h after collection. Blood samples of
healthy volunteers were used as the negative control for the RT-PCR
assay.
Normal testicular tissue samples were collected from
two patients undergoing castration at the Department of Urinary
Surgery, The Fourth Hospital of Hebei Medical University. Written
informed consent was provided by the two patients. The samples were
stored at −80°C until subsequent experiments.
RNA extraction and cDNA synthesis
Red blood cell lysis buffer was used to collect the
peripheral blood cells. Total RNA was extracted from the peripheral
blood cells and tissues using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Ultraviolet spectrophotometry and
agarose gel electrophoresis were used to determine the quality of
the RNA. The isolated RNA was stored at −80°C. RNA (2 µg) was used
to synthesize the first-strand cDNA using the RevertAid First
Strand cDNA Synthesis kit (Fermentas; Thermo Fisher Scientific,
Inc.) at 42°C for 1 h.
RT-PCR
cDNA was used for PCR amplification by using Go Taq
Green Master Mix (Promega Corporation) with primers for the
MAGE-A9 and MAGE-A11 genes. RNA integrity was
confirmed by performing PCR amplification with the primer for the
GAPDH gene. Agarose gel electrophoresis (2%) was used to
identify the RT-PCR products, and the bands were observed under
ultraviolet light. Each sample were measured three times.
GAPDH was used as the internal reference gene. The primer
sequences, product lengths, PCR cycle conditions, initial
denaturation and final extension steps are presented in Table I.
 | Table I.Primer sequences for reverse
transcription-PCR. |
Table I.
Primer sequences for reverse
transcription-PCR.
| Gene | Primer sequences
(5′→3′) | PCR cycle
conditions | No. of cycles | PCR product length,
bp |
|---|
| MAGE-A9 | F:
GTCTCTCGAGCAGAGGAGTCCGC | 95°C 30 sec; 58°C
30sec, 72°C 45 sec | 35 | 340 |
|
| R:
CTCAGCCACCTTCAATTTCAGT |
|
|
|
| MAGE-A11 | F:
ATGGAGACTCAGTTCCGAGA | 95°C 30 sec, 52°C
30 sec, 72°C 45 sec | 35 | 878 |
|
| R:
AAGAACTTTCATCTTGCTGG |
|
|
|
| GAPDH | F:
ACCTGACCTGCCGTCTAGAA | 95°C 15 sec, 58°C
15 sec, 72°C 20 sec | 28 | 247 |
|
| R:
TCCACCACCCTGTTGCTGTA |
|
|
|
Multiplex semi-nested PCR
GoTaq® Green Master mix (Promega
Corporation) was used to amplify the cDNA. In the first cycle of
PCR, the reaction mixture comprised 5 µl cDNA product of the
reverse transcription, 0.5 µl MAGE-F1 (10 µM), 1 µl MAGE-R1 (10
µM), 0.2 µl MAGE-F2 (10 µM), 0.2 µl MAGE-R2 (10 µM), 25 µl 2X PCR
Master mix and deionized water added up to 50 µl. In the second
cycle of PCR, the reaction mixture comprised 5 µl external PCR
product, 0.5 µl MAGE-F3 (10 µM), 1 µl MAGE-R3 (10 µM), 0.2 µl
MAGE-F4 (10 µM), 0.2 µl MAGE-R4 (10 µM), 50 µl 2X PCR Master mix
and deionized water added up to 100 µl. The primers are presented
in Table II. GAPDH was used
as the internal reference gene. The thermocycling conditions were
as follows: (1)95°C for 5 min;
(2)32 cycles of 95°C for 45 sec,
65°C for 45 sec and 72°C for 90 sec and 31 cycles of (2); (3)72°C
for 6 min. PCR product (6 µl) was used for electrophoresis on a
1.5% agarose gel. The amplification results were observed in the
gel imaging system (Syngene Inc.).
 | Table II.Primer sequences for multiplex
semi-nested PCR. |
Table II.
Primer sequences for multiplex
semi-nested PCR.
| Gene | Primer sequences
(5′→3′) | Fragment length,
bp |
|---|
| MAGE-As first
cycle | F1:
ACTGGCCCTGGCTGCAAC | 993 |
|
| R1:
GCCCTGACCAGAGTCATCAT |
|
|
| F2:
ACTGGCCTTGGCTGCAAC | 965 |
|
| R2:
CGAGAGTCATCATG |
|
| MAGE-As second
cycle | F3:
ACTGGCCCTGGCTGCAAC | 914 |
|
| R3:
AGGCCCTGGGCCTGGTG |
|
|
| F4:
ACTGGCCTTGGCTGCAAC | 893 |
|
| R4:
AGGCCCTGGGCTTGGTG |
|
| GAPDH | F:
ACCTGACCTGCCGTCTAGAA | 247 |
|
| R:
TCCACCACCCTGTTGCTGTA |
|
Restriction endonuclease
treatment
The products of multiplex semi-nested PCR were
purified using the QIAquick PCR Product Purification kit (Qiagen
China Co., Ltd.) according to the manufacturer's instructions. The
purified products were digested by restriction endonucleases
BclI, SphI, EcoRI, Eco47III and
AflIII, and gene fragments of MAGE-A1, -A2, -A3, -A4
and -A6 were obtained, respectively (Table III). The restriction fragment (6
µl) was analyzed by electrophoresis using a 1.5% agarose gel. The
bands were observed in the gel imaging system.
 | Table III.Restriction endonuclease mixtures,
multi-MAGE-A products and restriction fragments for each tested
MAGE-A gene. |
Table III.
Restriction endonuclease mixtures,
multi-MAGE-A products and restriction fragments for each tested
MAGE-A gene.
| Restriction
endonuclease | MAGE gene | PCR product length,
bp | Fragment length,
bp |
|---|
| BclI | A1 | 893 | 106,787 |
| SphI | A2 | 914 | 21,22,151,720 |
| EcoRI | A3 | 914 | 167,747 |
| Eco47III | A4 | 917 | 375,542 |
| AflIII | A6 | 914 | 22,172,282,438 |
Statistical analysis
SPSS v20.0 software (IBM Corp.) was used to analyze
the data. The display strip is defined as high expression group and
vice versa. χ2 or Fisher's exact test were used to
evaluate the potential association between the expression of all
MAGE-A genes or single MAGE-A genes and patient
clinicopathological characteristics. The Kaplan-Meier method was
used to estimate the overall survival time of patients with LSCC.
The Cox regression model was used for univariate and multivariate
analysis of overall survival and prognostic factors. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of MAGE-A genes in the
peripheral blood of patients with LSCC and healthy donors
Multiplex semi-nested RT-PCR and RT-PCR were used to
detect the expression of MAGE-A genes, including MAGE-A1,
-A2, -A3, -A4, -A6, -A9 and -A11, in two normal
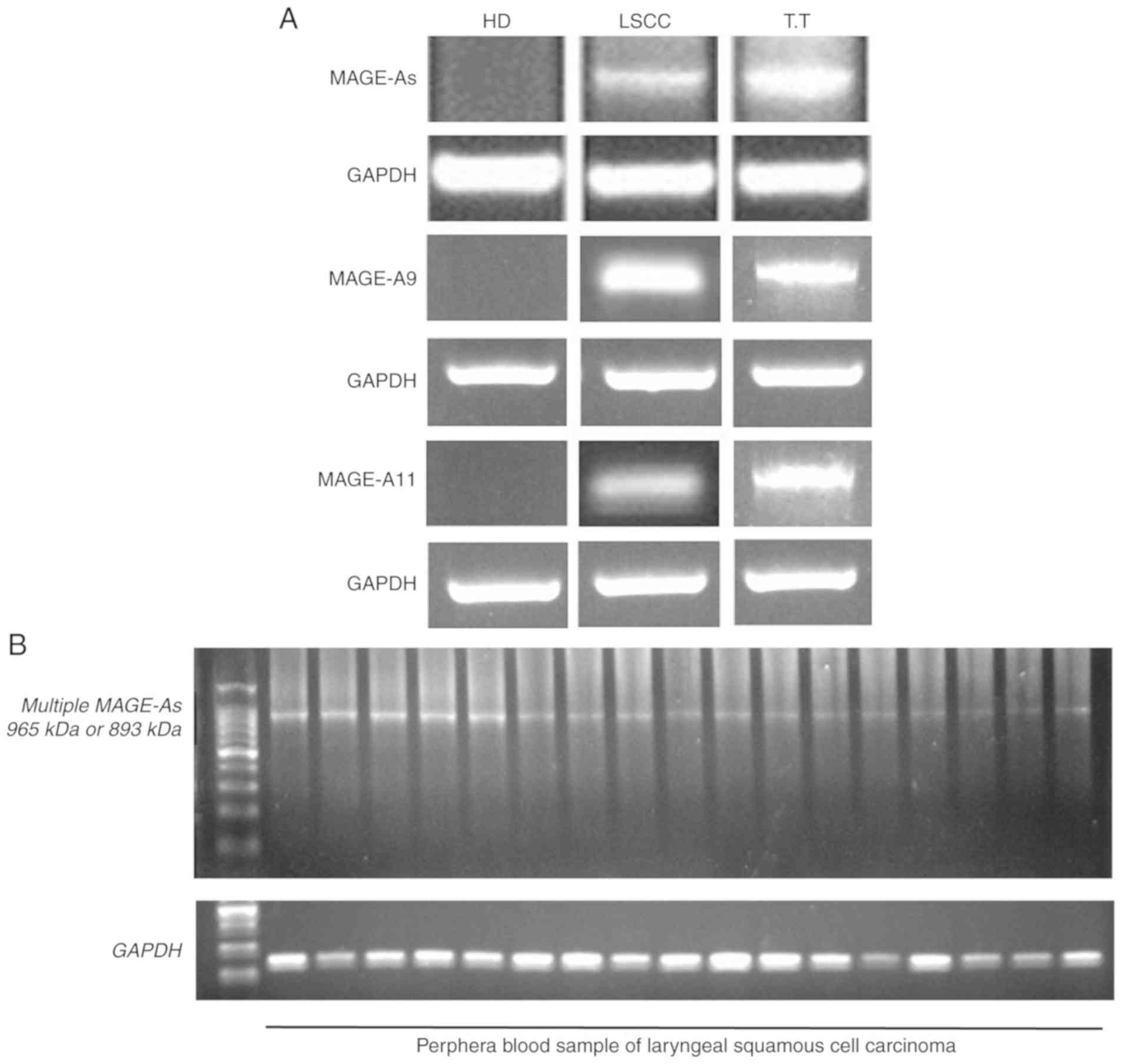
testicular tissue samples (Fig. 1A).
As the MAGE-A gene belongs to the CTA gene family, it
is only expressed in normal testis and other germ cells, but not in
other normal tissues (9). Therefore,
testicular tissue was used in the present study as a positive
control. Subsequently, the expression of MAGE-A in the blood
samples of 104 patients with LSCC and 30 healthy donors was
examined. The representative samples of the control and
MAGE-A products in the peripheral blood of healthy donors
(negative control) and patients with LSCC, as well as normal
testicular tissue (positive control) are presented in Fig. 1A. The MAGE-A product was
observed in the normal testicular tissues and in the blood samples
of a number of patients with LSCC, but not in the blood samples
from the healthy donors. The representative LSCC blood samples with
positive MAGE-A gene expression after internal PCR (second
PCR cycle) are presented in Fig.
1B.
As presented in Fig.
2, 31 of 104 patients with LSCC (29.8%) exhibited MAGE-A
gene expression in the peripheral blood. The expression of
MAGE-A9 and -A11 mRNA was detected by RT-PCR, whereas
the expression pattern of other individual MAGE-A genes was
identified by restriction endonuclease treatment. MAGE-A1
expression was positive in 19 of 104 (18.3%), MAGE-A2
expression was positive in 21 of 104 (20.2%), MAGE-A3
expression was positive in 21 of 104 (20.2%), MAGE-A4
expression was positive in 16 of 104 (15.4%), MAGE-A6
expression was positive in 12 of 104 (11.5%), MAGE-A9 expression
was positive in 27 of 104 (26.0%) and MAGE-A11 expression
was positive in 29 of 104 (27.9%) patients with LSCC. The frequency
of individual MAGE-A gene expression was in the following
order: A11 > A9 > A2 = A3 > A1 > A4 > A6. A total of
18 patients were positive for only one MAGE-A gene, 12
patients were positive for two genes, seven patients were positive
for three genes, eight patients were positive for four genes, eight
patients were positive for five genes and two patients were
positive for six genes. The genomic information of
MAGE-A1-12 for all 104 patients with LSCC is presented in
Table SI.
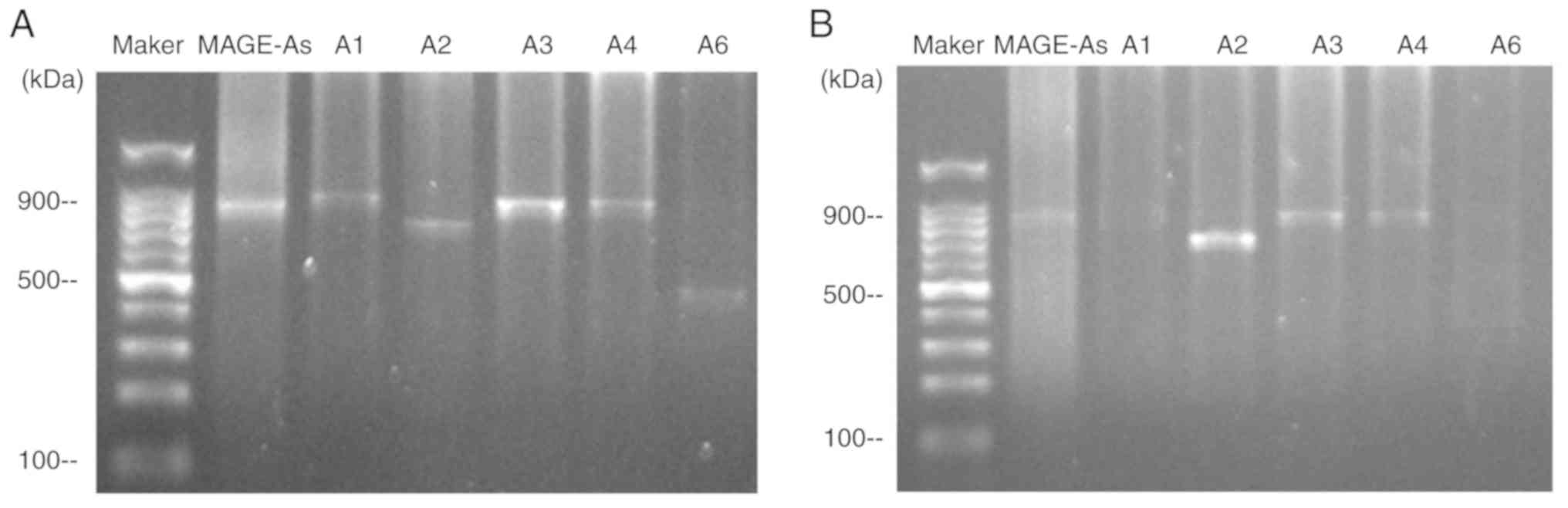
Fig. 3 demonstrates
the expression of the MAGE-A gene products from two patients
following restriction endonuclease treatment. A multiple
MAGE-As product was observed in the peripheral blood of
patients no. 17 and 45. Subsequently, the MAGE-A product
(second PCR cycle) was digested with BclI, SphI,
EcoRI, Eco47III and AflIII, and the digested
products were separated by agarose gel electrophoresis. The
individual MAGE-A genes were identified by observing the
fragment pattern. For patient 17 (Fig.
3A), the fragments of 787 bp was observed after BclI
digestion, 720 bp was observed after SphI digestion, 747 bp
was observed after EcoRI digestion, and 438 bp was observed
after AflIII digestion, indicating the presence of
MAGE-A1, -A2, -A3 and -A6 in the PCR product. The
expression pattern of MAGE-As in patient 45 is presented in
Fig. 3B; the expression of
MAGE-A1 and -A6 was not observed, but the 720 bp band
was observed after SphI digestion, and the fragment of 747
bp was observed after EcoRI digestion, indicating the
presence of MAGE-A2 and -A3 in the PCR product.
Association between MAGE-A gene
expression in the peripheral blood and the clinicopathological
characteristics of patients with LSCC
The association between MAGE-A gene
expression in the peripheral blood and the clinicopathological
characteristics of patients with LSCC was evaluated (Table IV). The expression of MAGE-A
genes (MAGE-As, -A1, -A2, -A3, -A4, -A6, -A9 and
-A11) were not associated with age, smoking history, tumor
size and location, but was positively associated with lymph node
metastasis (P=0.001, P=0.022, P<0.001, P=0.001, P=0.016,
P<0.001, P=0.001 and P=0.016, respectively). High expression
levels of MAGE-As in LSCC were associated with the
histological degree (P=0.007). Among individual MAGE-As,
positive MAGE-A1, -A3, -A4 and -A6 expression was
more frequent in patients with histological grade G3 compared with
those with histological grades G1/G2 (P=0.005, P=0.013, P=0.001 and
P=0.001, respectively). In addition, more frequent positive
expression of MAGE-A3, -A6, -A9 and -A11 was observed
in patients with high clinical stage compared with that in patients
with low clinical stage (P=0.044, P=0.048, P=0.01 and P=0.045,
respectively). No associations were observed between each
individual MAGE-A genes expression and other
clinicopathological factors.
 | Table IV.Clinicopathological characteristics
and MAGE-A gene expression in the peripheral blood from 104
patients with laryngeal squamous cell carcinoma. |
Table IV.
Clinicopathological characteristics
and MAGE-A gene expression in the peripheral blood from 104
patients with laryngeal squamous cell carcinoma.
| Variable | N | MAGE-As, n (%) | P | MAGE-A1, n (%) | P | MAGE-A2, n (%) | P | MAGE-A3, n (%) | P | MAGE-A4, n (%) | P | MAGE-A6, n (%) | P | MAGE-A9, n (%) | P | MAGE-A11, n
(%) | P |
|---|
| Age, years |
|
<60 | 45 | 12 (26.7) | 0.541 | 6 (13.3) | 0.255 | 6 (13.3) | 0.128 | 9 (20.0) | 0.966 | 6 (13.3) | 0.613 | 4 (8.9) | 0.460 | 9 (20.0) | 0.226 | 9 (20.0) | 0.117 |
|
≥60 | 59 | 19 (32.2) |
| 13 (22.0) |
| 15 (25.4) |
| 12 (20.3) |
| 10 (16.9) |
| 8 (13.6) |
| 18 (30.5) |
| 20 (33.9) |
|
| Tumor location |
|
Glottic | 65 | 17 (26.2) | 0.293 | 12 (18.5) | 0.948 | 10 (15.4) | 0.115 | 10 (15.4) | 0.115 | 11 (16.9) | 0.575 | 8 (12.3) | 0.751 | 13 (20.0) | 0.073 | 18 (27.7) | 0.955 |
|
Supraglottic | 39 | 14 (35.9) |
| 7 (17.9) |
| 11 (28.2) |
| 11 (28.2) |
| 5 (12.8) |
| 4 (10.3) |
| 14 (35.9) |
| 11 (28.2) |
|
| Smoking index |
|
<400 | 41 | 9 (22.0) | 0.158 | 5 (12.2) | 0.196 | 6 (14.6) | 0.255 | 5 (12.2) | 0.101 | 3 (7.3) | 0.066 | 5 (12.2) | 0.866 | 7 (17.1) | 0.095 | 8 (19.5) | 0.125 |
|
≥400 | 63 | 22 (34.9) |
| 14 (22.2) |
| 15 (23.8) |
| 16 (25.4) |
| 13 (20.6) |
| 7 (11.1) |
| 20 (31.7) |
| 21 (33.3) |
|
| Tumor size, cm |
|
<2 | 43 | 13 (30.2) | 0.937 | 7 (16.3) | 0.659 | 7 (16.3) | 0.404 | 10 (23.3) | 0.513 | 9 (20.9) | 0.188 | 4 (9.3) | 0.549 | 8 (18.6) | 0.151 | 10 (23.3) | 0.377 |
| ≥2 | 61 | 18 (29.5) |
| 12 (19.7) |
| 14 (23.0) |
| 11 (18.0) |
| 7 (11.5) |
| 8 (13.1) |
| 19 (31.1) |
| 19 (31.1) |
|
| Lymph node
metastasis |
| N | 32 | 17 (53.1) | 0.001 | 10 (31.3) | 0.022 | 14 (43.8) | <0.001 | 13 (40.6) | 0.001 | 9 (28.1) | 0.016 | 9 (28.1) | <0.001 | 15 (46.9) | 0.001 | 14 (43.8) | 0.016 |
| N0 | 72 | 14
(43.8)a |
| 9
(12.5)a |
| 7
(9.7)a |
| 8
(11.1)a |
| 7
(9.7)a |
| 3
(4.2)a |
| 12
(16.7)a |
| 15
(20.8)a |
|
| Histological
grade |
|
G1/G2 | 91 | 23 (25.3) | 0.007 | 13 (14.3) | 0.005 | 17 (18.7) | 0.292 | 15 (16.5) | 0.013 | 10 (11.0) | 0.001 | 7 (7.7) | 0.001 | 22 (24.2) | 0.315 | 25 (27.5) | 0.752 |
| G3 | 13 | 8
(61.5)a |
| 6
(46.2)a |
| 4 (30.8) |
| 6
(46.2)a |
| 6
(46.2)a |
| 5
(38.5)a |
| 5 (38.5) |
| 4 (30.8) |
|
| Clinical stage |
|
I/II | 45 | 10 (22.2) | 0.140 | 7 (15.6) | 0.532 | 7 (15.6) | 0.304 | 5 (11.1) | 0.044 | 5 (11.1) | 0.291 | 2 (4.4) | 0.048 | 6 (13.3) | 0.010 | 8 (17.8) | 0.045 |
|
III/IV | 59 | 21 (35.6) |
| 12 (20.3) |
| 14 (23.7) |
| 16
(27.1)a |
| 11 (18.6) |
| 10
(16.9)a |
| 21
(35.6)a |
| 21
(35.6)a |
|
Association between MAGE-A gene
expression in the peripheral blood and the overall survival of
patients with LSCC
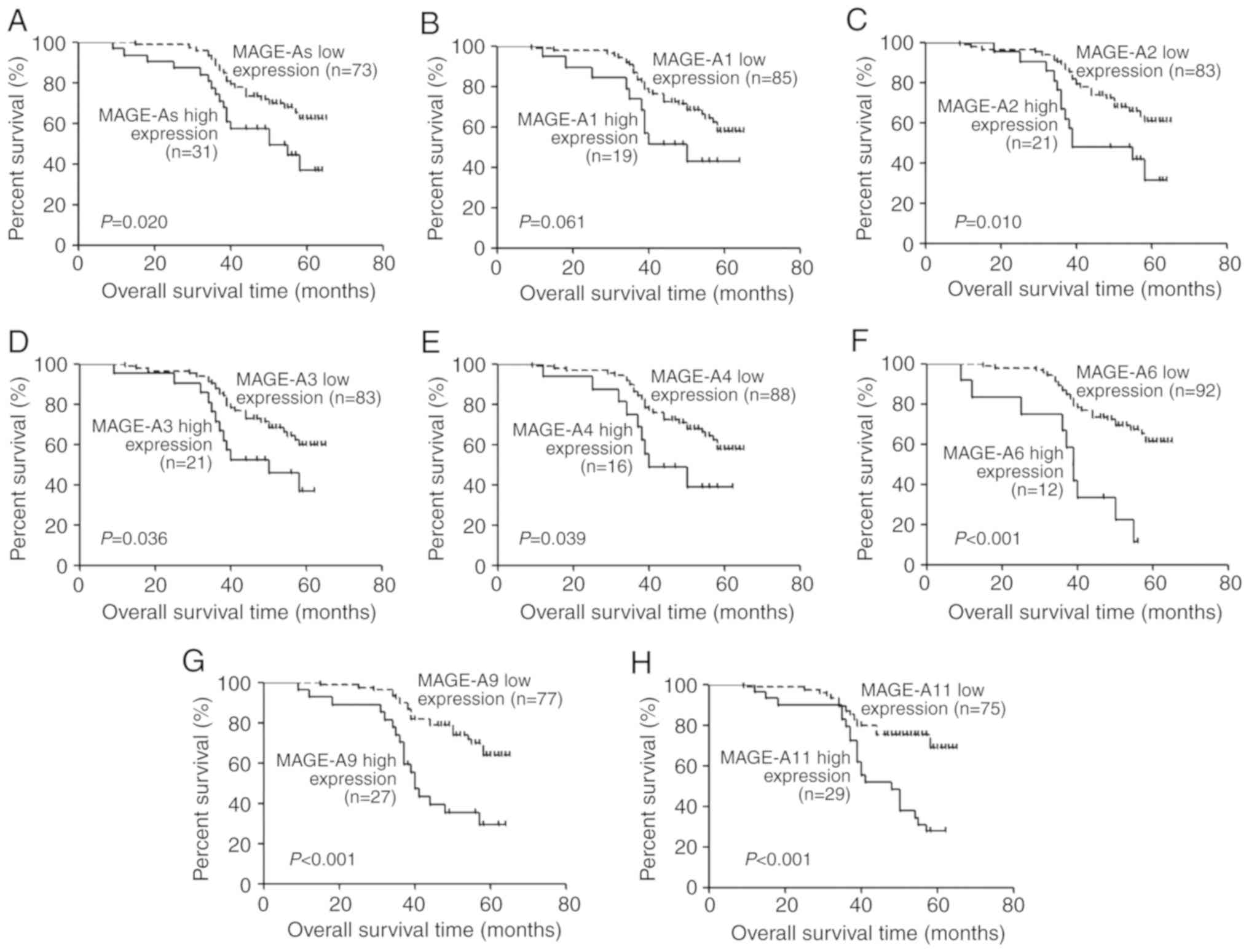
All 104 patients with LSCC were followed up for
18–65 months. Kaplan-Meier analysis was performed to determine the
association between MAGE-A gene (MAGE-A1, -A2, -A3, -A4,
-A6, -A9 and -A11) expression levels and the overall
survival of patients with LSCC. Overall survival of patients with
high MAGE-A gene expression in the peripheral blood was
significantly lower compared with those with low expression
(P=0.020, P=0.061, P=0.010, P=0.036, P=0.039, P<0.001,
P<0.001 and P<0.001, respectively; Fig. 4). To further evaluate the prognostic
significance of MAGE-A expression, univariate analysis of
clinicopathological factors and overall survival was performed.
High expression of MAGE-As (P=0.024), -A2 (P=0.014),
-A3 (P=0.041), -A4 (P=0.046), -A6
(P<0.001), -A9 (P<0.001) and -A11 (P<0.001),
as well as lymph node metastasis (P<0.01), low clinical stage
(P=0.013) and high histological grade (P=0.028) were demonstrated
to be predictors of poor overall survival (Table V). clinical stage, which includes
tumor size and lymph node metastasis, and MAGE-As, which
includes MAGE-A1, -A2, -A3, -A4 and -A6, were not
considered as independent prognostic factors. Lymph node
metastasis, histological grade and the expression of MAGE-A2,
-A3, -A4, -A6, -A9 and -A11 were further analyzed by
multivariate Cox regression analysis. As demonstrated in Table V, high expression of MAGE-A11
(P=0.032) and lymph node metastasis (P<0.01) were determined to
be independent prognostic factors for poor prognosis of patients
with LSCC.
 | Table V.Univariate and multivariate analyses
of prognostic factors for overall survival of patients with
laryngeal squamous cell carcinoma. |
Table V.
Univariate and multivariate analyses
of prognostic factors for overall survival of patients with
laryngeal squamous cell carcinoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | P-value | 95% CI | HR | P-value | 95% CI |
|---|
| Expression of
MAGE-As, high vs. low | 2.048 | 0.024a | 1.099-3.816 |
|
|
|
| Expression of
MAGE-A1, high vs. low | 1.948 | 0.068 | 0.951-3.990 |
|
|
|
| Expression of
MAGE-A2, high vs. low | 2.297 | 0.014a | 1.187-4.443 | 2.137 | 0.050 | 1.001-4.561 |
| Expression of
MAGE-A3, high vs. low | 2.016 | 0.041a | 1.028-3.954 | 1.207 | 0.731 | 0.412-3.540 |
| Expression of
MAGE-A4, high vs. low | 2.131 | 0.046a | 1.013-4.482 | 2.339 | 0.174 | 0.686-7.970 |
| Expression of
MAGE-A6, high vs. low | 4.050 |
<0.001a | 1.958-8.376 | 1.410 | 0.480 | 0.543-3.665 |
| Expression of
MAGE-A9, high vs. low | 3.204 |
<0.001a | 1.722-5.961 | 2.082 | 0.065 | 0.954-4.543 |
| Expression of
MAGE-A11, high vs. low | 3.019 |
<0.001a | 1.636-5.573 | 2.438 | 0.032a | 1.080-5.504 |
| Age, years, <60
vs. ≥60 | 1.369 | 0.327 | 0.730-2.566 |
|
|
|
| Tumor location,
supraglottic vs. glottic | 1.839 | 0.053 | 0.992-3.409 |
|
|
|
| Smoking index,
<400 vs. ≥400 | 1.335 | 0.381 | 0.700-2.547 |
|
|
|
| Tumor size, cm,
<2 vs. ≥2 | 1.924 | 0.057 | 0.981-3.777 |
|
|
|
| Histological grade,
G1/G2 vs. G3 | 2.013 | 0.028a | 1.078-3.760 | 1.290 | 0.544 | 0.568-2.931 |
| Clinical stage
(AJCC), I/II vs. III/IV | 2.345 | 0.013a | 1.194-4.604 |
|
|
|
| Metastatic state of
lymph node, N vs. N0 | 10.788 |
<0.001a | 5.036-23.110 | 21.112 |
<0.001a | 7.927-56.226 |
Discussion
Clinical imaging is often the first to identify the
tumor and is important for the diagnosis of recurrence and
metastasis of the auxiliary technology (2). Failure to identify the primary or
metastatic tumors by imaging means that the best time for diagnosis
and treatment is missed, and late metastasis of the tumor has no
effective treatment and the prognosis is poor (2). The process by which tumor cells invade
the body remains unclear and may involve the active invasion of
cells via epithelial-to-mesenchymal transition (16) and passively shed individual cells or
clusters of tumor cells from impaired tumor blood vessels (17,18). The
presence of CTCs in the peripheral blood is considered to be the
key factor of tumor metastasis (19). A number of studies have evaluated the
clinical application value of CTCs for metastatic breast (20,21),
prostate (22), colon (23) and lung (24,25)
cancer. Using the FDA-approved Cell Search system (Veridex), the
peripheral blood CTC count test and analysis of the prognosis of
patients suggest that CTCs can be used as an independent prognostic
indicator (26,27). The technology detects the biomarkers
on the cell surface to identify the CTCs (26,27). The
downregulation or absence of epithelial markers should have an
effect on detecting CTCs (19).
Thus, the discovery of new tumor markers may help identify CTCs in
the peripheral blood of patients with malignant tumors.
RT-PCR, which has a wide range of applications, is
considered the most sensitive method for detecting CTCs in the
peripheral blood (28). However, the
following issues may exist: i) Large amounts of water in the
peripheral blood may dilute the normal mRNA and cause false
negative results (29); ii)
contamination of the target gene in the peripheral blood, leading
to DNA amplification, may result in false positive results
(30), and iii) low levels of
abnormal transcription and amplification of target genes in the
peripheral blood tumor cells may also cause false-positive results
(30). Improving the detection
technology, designing more suitable primers and selecting accurate
tumor markers may help avoid the aforementioned issues. Previous
studies have used RT-nested PCR to detect the mRNA expression of
CTCs in the peripheral blood of non-cancerous and colorectal cancer
cells, and demonstrated high sensitivity and specificity (31,32). It
has been demonstrated that the expression of MAGE-A genes
can be used as a biomarker of CTCs in colorectal, breast and
gastric cancer (33). The results of
the previous study demonstrated that the MAGE-A antigen is a
tumor-associated antigen of LSCC (34). Therefore, MAGE-A mRNA may be
used to detect CTCs in the peripheral blood of patients with LSCC
as a specific tumor marker, which may guide the diagnosis and
prognosis of LSCC. In the present study, fresh blood was used for
further analysis of MAGE-A gene expression; fresh blood is
easy to collect and observe, allowing for timely guidance on
clinical diagnosis and treatment.
Due to the high homology, it is difficult to design
specific primers for a single MAGE-A gene. Therefore, a pair
of primer sequences containing two forward primers and two reverse
primers were designed in the present study for the amplification of
a mixture of MAGE-A1, -A2, -A3, -A4 and -A6 genes by
multi-RT-nested PCR. The expression of MAGE-A9 and
-A11 mRNA was detected by RT-PCR. MAGE-A mRNA was expressed
in different degrees in peripheral blood of patients with LSCC,
while MAGE-A expression was not detected in the healthy
donors. These are consistent with previous studies (35,36). A
total of 18 patients were positive for only one MAGE-A gene,
12 patients were positive for two genes, seven patients were
positive for three genes, eight patients were positive for four
genes, eight patients were positive for five genes, and two
patients were positive for six genes. Since the MAGE-A gene only
exists in tumor cells (9), if MAGE-A
gene expression is detected in peripheral blood, they must be
expressed in tumor cells. In view of the definition of CTC, the
results of the present study indicated that MAGE-A is expressed in
CTCs. The results of the present study suggested that MAGE-A
mRNA was only expressed in tumor cells and may be detected only in
the presence of tumor cells in the peripheral blood of patients
with LSCC. Therefore, the expression of MAGE-A genes may be
used as a specific marker for detecting CTCs in the peripheral
blood of patients with LSCC.
The association between MAGE-A gene
expression in the peripheral blood and the clinicopathological
features of patients with LSCC was statistically analyzed in the
present study. The expression of MAGE-A1, -A3, -A4 and
-A6 were more frequently detected in patients with
histological grade G3 tumors compared with patients with tumor
grades G1/G2. A previous study has demonstrated that histological
grade and tumor prognosis are significantly associated in breast
cancer (37). Therefore, the
expression of MAGE-A1, -A3, -A4 and -A6 genes may be
an important indicator of the prognosis of LSCC. In the present
study, the expression of MAGE-A3, -A6, -A9 and -A11
was observed in patients with a high clinical stage more frequently
compared with that in patients with a low clinical stage. Patients
with late clinical staging usually have a poor prognosis (37). In addition, the expression of
MAGE-As in the peripheral blood of patients with LSCC was
positively associated with the lymph node metastasis status in the
present study. For each individual MAGE-A gene, including
MAGE-A1, -A2, -A3, -A4, -A6, -A9 and -A11, the
expression frequency in patients with lymph node metastasis was
significantly higher compared with that in patients without lymph
node metastasis. Previous studies have demonstrated that CTCs are
commonly present in advanced metastatic malignancies, with more
CTCs in the peripheral blood of distantly metastatic breast cancer
compared with early-stage breast cancer (38), and similar observations in ovarian
cancer (39). In the present
univariate analysis, individual MAGE-A expression,
metastatic state of the lymph nodes, clinical stage and
histological grade qualified to enter the regression model;
however, as clinical stage includes the metastatic state of the
lymph nodes and distant metastasis, only the metastatic state of
the lymph nodes was entered into the regression model. The results
demonstrated that multiple MAGE-As expression, the
metastatic state of the lymph nodes and distant metastasis were
risk factors for the 5-year survival of patients with LSCC.
To date, serological hallmark and high-resolution
imaging technology still cannot identify micrometastasis and
reflect the efficacy of treatments. CTCs detected in the peripheral
blood suggest the possibility of early occult micrometastasis
(26,27). CTCs cannot only be used to study the
biological characteristics of malignant tumors, but they overcome
the disadvantage of traditional tissue biopsy and allow real-time
dynamic monitoring of the changes in the tumor (40). Previous studies have demonstrated
that the changes in the number of CTCs can reflect the efficacy of
treatments and provide the basis for individual treatment. Qiao
et al (41), reported the
quantitative variation of CTCs in a patient with ESCC before and
after surgery and during a 5-year follow-up period; the results
demonstrated that the number of CTCs before and the initial period
after surgery remained high. By contrast, following combined
treatment, the number of CTCs deceased, and after 117 weeks, the
number gradually stabilized at a low level (41). Thus, a change in the number of
peripheral blood CTCs may be used to monitor disease status and
treatment efficacy. Another previous study reported that clinically
acquired drug resistance did not become resistant at the cellular
level during the course of treatment, but clinically acquired drug
resistance was the selective reaction of heterogeneous cancer cells
to target cells (42). Through the
dynamic supervision of certain molecular indicators of CTCs, the
endpoint for therapy can be determined (43). CTCs in the peripheral blood of
patients with LSCC can be examined to dynamically monitor the
changes and development of the tumor in order to achieve an
improved understanding of personalized therapy. CTCs in the
peripheral blood of patients with LSCC may be monitored by
detecting the MAGE-A genes to guide clinical treatment and
the judgement of prognosis.
In conclusion, the results of the present study
identified the expression patterns of multiple MAGE-A genes
in the peripheral blood of patients with LSCC. MAGE-A gene
expression in the peripheral blood may therefore be used as a
molecular marker for guiding the treatment and monitoring the
prognosis of patients with LSCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Financial Supporting
Program of Hebei Province (grant no. 2014]1257), the Financial
Department of Hebei Province (grant no. 2016]361006) and the Hebei
Health Department (grant no. 20150786).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
BS and MS contributed to the experimental design and
fundraising. YZ and YX performed some of the expriments. RZ and LG
assisted in the data analysis. SL analyzed the data and drafted the
initial manuscript.
Ethics approval and consent to
participate
This study was approved by the Medical Ethics
Committee of The Fourth Hospital of Hebei Medical University
(Shijiazhuang, China; approval no. 2011KY112). Written informed
consent was provided by all participants before enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang DS, Pan CC, Lai HC and Huang JM:
Expression of HMGA1 and ezrin in laryngeal squamous cell carcinoma.
Acta Otolaryngol. 133:626–632. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Steuer CE, El-Deiry M, Parks JR, Higgins
KA and Saba NF: An update on larynx cancer. CA Cancer J Clin.
67:31–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li Y, Liu J, Hu W, Zhang Y, Sang J, Li H,
Ma T, Bo Y, Bai T, Guo H, et al: miR-424-5p promotes proliferation,
migration and invasion of laryngeal squamous cell carcinoma.
OncoTargets Ther. 12:10441–10453. 2019. View Article : Google Scholar
|
|
4
|
Yang Q, Sun J, Ma Y, Zhao C and Song J:
LncRNA DLX6-AS1 promotes laryngeal squamous cell carcinoma growth
and invasion through regulating miR-376c. Am J Transl Res.
11:7009–7017. 2019.PubMed/NCBI
|
|
5
|
Bonner J, Giralt J, Harari P, Spencer S,
Schulten J, Hossain A, Chang SC, Chin S and Baselga J: Cetuximab
and radiotherapy in laryngeal preservation for cancers of the
larynx and hypopharynx: A secondary analysis of a randomized
clinical trial. JAMA Otolaryngol Head Neck Surg. 142:842–849. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nishimura G, Sano D, Arai Y, Hatano T,
Takahashi H, Tanabe T, Wada T, Morishita D and Oridate N: A
prospective clinical trial of the second-look procedure for
transoral surgery in patients with T1 and T2 laryngeal,
oropharyngeal, and hypopharyngeal cancer. Cancer Med. 8:7197–7206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nishimura G, Sano D, Yabuki K, Arai Y,
Chiba Y, Tanabe T and Oridate N: The second-look procedure for
transoral videolaryngoscopic surgery for T1 and T2 laryngeal,
oropharyngeal, and hypopharyngeal cancer patients: Protocol for a
nonrandomized clinical trial. JMIR Res Protoc. 6:e2352017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Strieth S, Ernst BP, Both I, Hirth D,
Pfisterer LN, Künzel J and Eder K: Randomized controlled
single-blinded clinical trial of functional voice outcome after
vascular targeting KTP laser microsurgery of early laryngeal
cancer. Head Neck. 41:899–907. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van der Bruggen P, Traversari C, Chomez P,
Lurquin C, De Plaen E, van den Eynde B, Knuth A and Boon T: A gene
encoding an antigen recognized by cytolytic T lymphocytes on a
human melanoma. Science. 254:1643–1647. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meek DW and Marcar L: MAGE-A antigens as
targets in tumour therapy. Cancer Lett. 324:126–132. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sang M, Lian Y, Zhou X and Shan B: MAGE-A
family: Attractive targets for cancer immunotherapy. Vaccine.
29:8496–8500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sang M, Wang L, Ding C, Zhou X, Wang B,
Wang L, Lian Y and Shan B: Melanoma-associated antigen genes - an
update. Cancer Lett. 302:85–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De Plaen E, Arden K, Traversari C, Gaforio
JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethé B,
Lurquin C, et al: Structure, chromosomal localization, and
expression of 12 genes of the MAGE family. Immunogenetics.
40:360–369. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lian Y, Sang M, Ding C, Zhou X, Fan X, Xu
Y, Lü W and Shan B: Expressions of MAGE-A10 and MAGE-A11 in breast
cancers and their prognostic significance: A retrospective clinical
study. J Cancer Res Clin Oncol. 138:519–527. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lydiatt WM, Patel SG, O'Sullivan B,
Brandwein MS, Ridge JA, Migliacci JC, Loomis AM and Shah JP: Head
and neck cancers-major changes in the American Joint Committee on
Cancer eighth edition cancer staging manual. CA Cancer J Clin.
67:122–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fidler IJ: The relationship of embolic
homogeneity, number, size and viability to the incidence of
experimental metastasis. Eur J Cancer. 9:223–227. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liotta LA, Saidel MG and Kleinerman J: The
significance of hematogenous tumor cell clumps in the metastatic
process. Cancer Res. 36:889–894. 1976.PubMed/NCBI
|
|
19
|
Batth IS, Mitra A, Rood S, Kopetz S,
Menter D and Li S: CTC analysis: An update on technological
progress. Transl Res. 212:14–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, Riethdorf S, Wu G, Wang T, Yang
K, Peng G, Liu J and Pantel K: Meta-analysis of the prognostic
value of circulating tumor cells in breast cancer. Clin Cancer Res.
18:5701–5710. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bidard FC, Peeters DJ, Fehm T, Nolé F,
Gisbert-Criado R, Mavroudis D, Grisanti S, Generali D, Garcia-Saenz
JA, Stebbing J, et al: Clinical validity of circulating tumour
cells in patients with metastatic breast cancer: A pooled analysis
of individual patient data. Lancet Oncol. 15:406–414. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Bono JS, Scher HI, Montgomery RB,
Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ
and Raghavan D: Circulating tumor cells predict survival benefit
from treatment in metastatic castration-resistant prostate cancer.
Clin Cancer Res. 14:6302–6309. 2008. View Article : Google Scholar
|
|
23
|
Cohen SJ, Punt CJ, Iannotti N, Saidman BH,
Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et
al: Relationship of circulating tumor cells to tumor response,
progression-free survival, and overall survival in patients with
metastatic colorectal cancer. J Clin Oncol. 26:3213–3221. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krebs MG, Sloane R, Priest L, Lancashire
L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G,
et al: Evaluation and prognostic significance of circulating tumor
cells in patients with non-small-cell lung cancer. J Clin Oncol.
29:1556–1563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hou JM, Krebs MG, Lancashire L, Sloane R,
Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, et
al: Clinical significance and molecular characteristics of
circulating tumor cells and circulating tumor microemboli in
patients with small-cell lung cancer. J Clin Oncol. 30:525–532.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Balic M, Lin H, Williams A, Datar RH and
Cote RJ: Progress in circulating tumor cell capture and analysis:
Implications for cancer management. Expert Rev Mol Diagn.
12:303–312. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van de Stolpe A, Pantel K, Sleijfer S,
Terstappen LW and den Toonder JM: Circulating tumor cell isolation
and diagnostics: Toward routine clinical use. Cancer Res.
71:5955–5960. 2011. View Article : Google Scholar
|
|
28
|
Savino M, Garrubba M, Parrella P, Baorda
F, Copetti M, Murgo R, Zelante L, Carella M, Valori VM and Santini
SA: Development of real-time quantitative reverse transcription-PCR
for Her2 detection in peripheral blood from patients with breast
cancer. Clin Chim Acta. 384:52–56. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stutterheim J, Ichou FA, den Ouden E,
Versteeg R, Caron HN, Tytgat GA and van der Schoot CE: Methylated
RASSF1a is the first specific DNA marker for minimal residual
disease testing in neuroblastoma. Clin Cancer Res. 18:808–814.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Wang F, Ning N, Chen Q, Yang Z,
Guo Y, Xu D, Zhang D, Zhan T and Cui W: Patterns of circulating
tumor cells identified by CEP8, CK and CD45 in pancreatic cancer.
Int J Cancer. 136:1228–1233. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yoon SO, Kim YT, Jung KC, Jeon YK, Kim BH
and Kim CW: TTF-1 mRNA-positive circulating tumor cells in the
peripheral blood predict poor prognosis in surgically resected
non-small cell lung cancer patients. Lung Cancer. 71:209–216. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gervasoni A, Sandri MT, Nascimbeni R,
Zorzino L, Cassatella MC, Baglioni L, Panigara S, Gervasi M, Di
Lorenzo D and Parolini O: Comparison of three distinct methods for
the detection of circulating tumor cells in colorectal cancer
patients. Oncol Rep. 25:1669–1703. 2011.PubMed/NCBI
|
|
33
|
Kim DD, Yang CS, Chae HD, Kwak SG and Jeon
CH: Melanoma antigen-encoding gene family member A1-6 and hTERT in
the detection of circulating tumor cells following CD45- depletion
and RNA extraction. Oncol Lett. 14:837–843. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu S, Sang M, Xu Y, Gu L, Liu F and Shan
B: Expression of MAGE-A1, -A9, -A11 in laryngeal squamous cell
carcinoma and their prognostic significance: A retrospective
clinical study. Acta Otolaryngol. 136:506–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Trippel A, Halling F, Heymann P, Ayna M,
Al-Nawas B and Ziebart T: The expression of melanoma-associated
antigen A (MAGE-A) in oral squamous cell carcinoma: An evaluation
of the significance for tumor prognosis. Oral Maxillofac Surg.
23:343–352. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Poojary M, Jishnu PV and Kabekkodu SP:
Prognostic value of melanoma-associated antigen-A (MAGE-A) gene
expression in various human cancers: A systematic review and
meta-analysis of 7428 patients and 44 studies. Mol Diagn Ther.
doi.org/10.1007/s40291-020-00476-5. PubMed/NCBI
|
|
37
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer. I. The value of histological
grade in breast cancer: Experience from a large study with
long-term follow-up. Histopathology. 19:403–410. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stemke-Hale K, Hennessy B, Mills GB and
Mitra R: Molecular screening for breast cancer prevention, early
detection, and treatment planning: Combining biomarkers from DNA,
RNA, and protein. Curr Oncol Rep. 8:484–491. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sang M, Wu X, Fan X, Sang M, Zhou X and
Zhou N: Multiple MAGE-A genes as surveillance marker for the
detection of circulating tumor cells in patients with ovarian
cancer. Biomarkers. 19:34–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hart CD, Galardi F, Luca FD, Pestrin M and
Di Leo A: Circulating tumour cells as liquid biopsy in breast
cancer-advancing from prognostic to predictive potential. Curr
Breast Cancer Rep. 7:53–58. 2015. View Article : Google Scholar
|
|
41
|
Qiao YY, Lin KX, Zhang Z, Zhang DJ, Shi
CH, Xiong M, Qu XH and Zhao XH: Monitoring disease progression and
treatment efficacy with circulating tumor cells in esophageal
squamous cell carcinoma: A case report. World J Gastroenterol.
21:7921–7928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Marusyk A, Almendro V and Polyak K:
Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev
Cancer. 12:323–334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Krawczyk N, Meier-Stiegen F, Banys M,
Neubauer H, Ruckhaeberle E and Fehm T: Expression of stem cell and
epithelial-mesenchymal transition markers in circulating tumor
cells of breast cancer patients. BioMed Res Int.
2014:415721–415731. 2014. View Article : Google Scholar : PubMed/NCBI
|