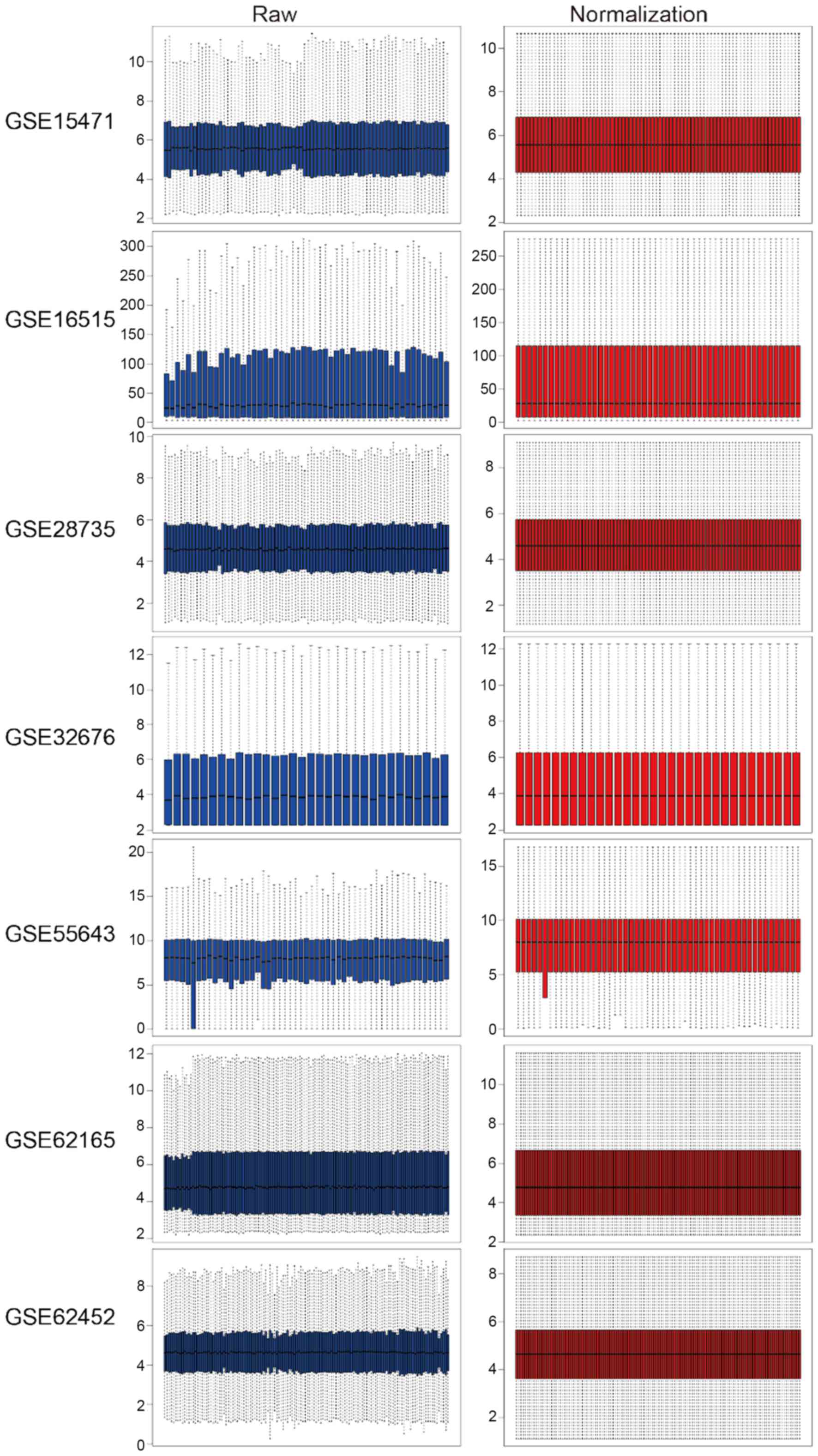
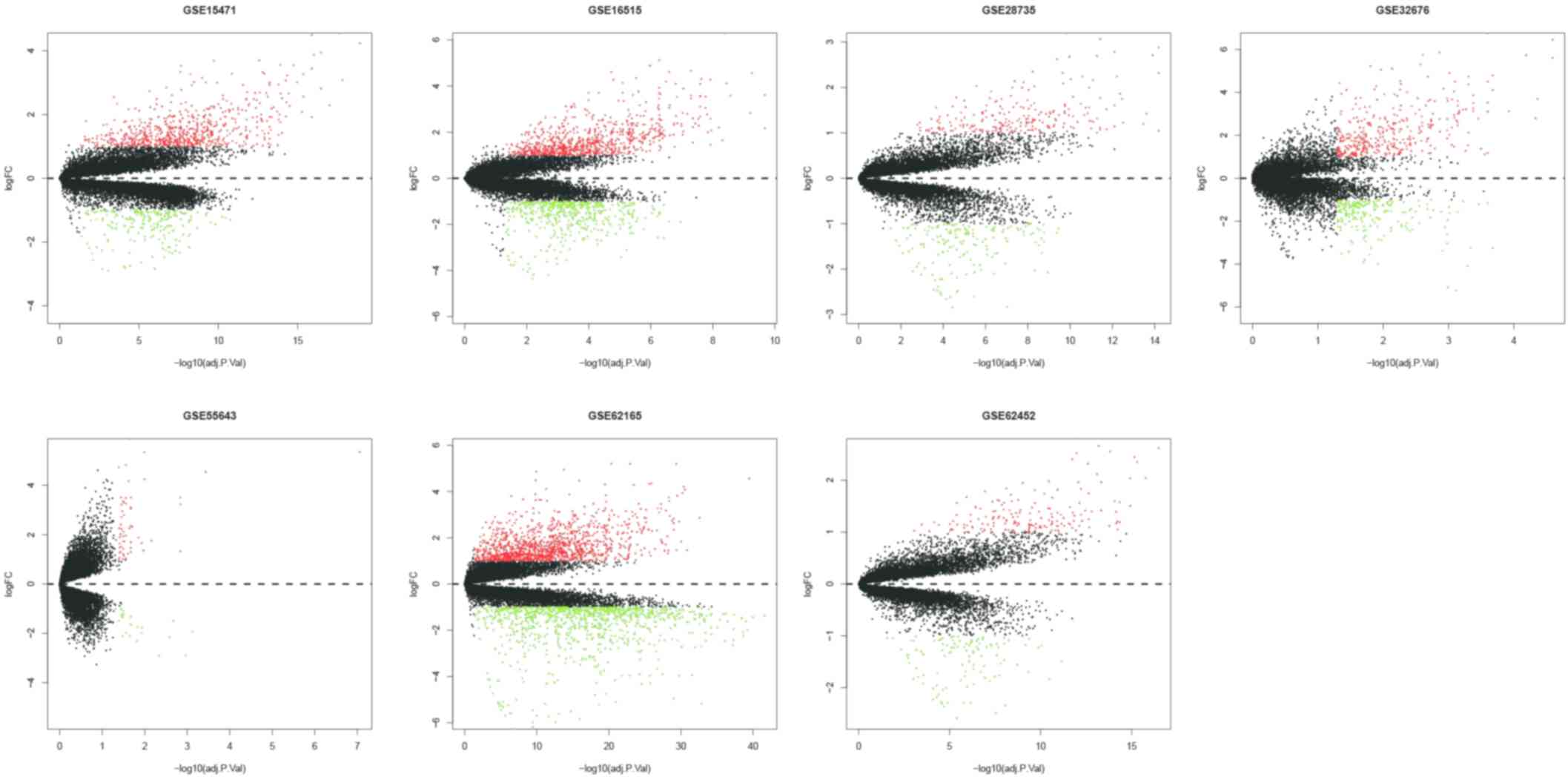
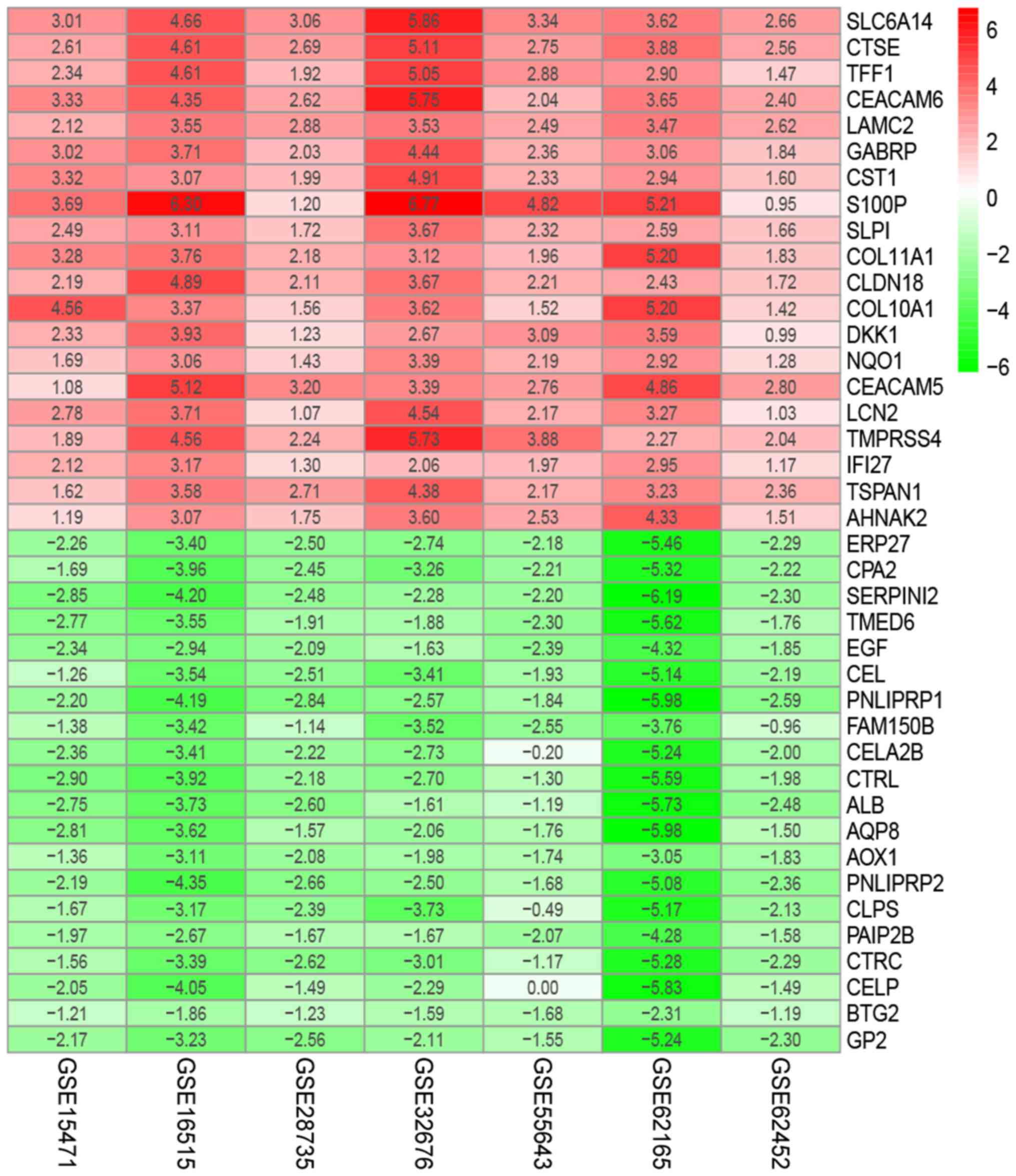
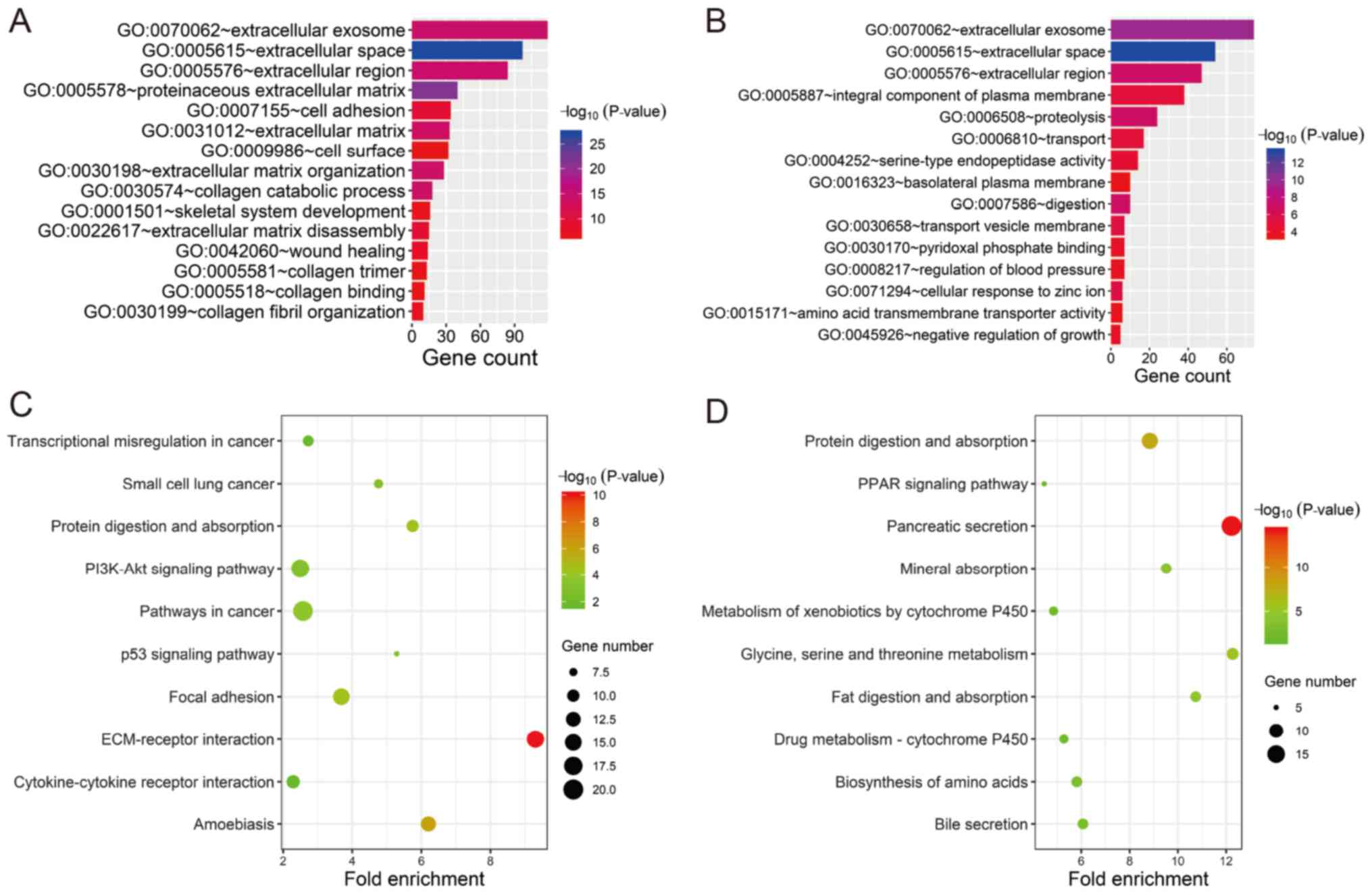
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gordon-Dseagu VL, Devesa SS, Goggins M and
Stolzenberg-Solomon R: Pancreatic cancer incidence trends: Evidence
from the surveillance, epidemiology and end results (SEER)
population-based data. Int J Epidemiol. 47:427–439. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Golan T, Sella T, Margalit O, Amit U,
Halpern N, Aderka D, Shacham-Shmueli E, Urban D and Lawrence YR:
Short- and long-term survival in metastatic pancreatic
adenocarcinoma, 1993-2013. J Natl Compr Canc Netw. 15:1022–1027.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mackay MT, van Erning FN, van der Geest
LG, Koerkamp BG, van Laarhoven MH, Bonsing BA, Wilmink JW, van
Santvoort HC, de Vos-Geelen Jd, van Eijck CH, et al: Association of
the location of pancreatic ductal adenocarcinoma (head, body, tail)
with tumor stage, treatment, and survival: A population-based
analysis. Pancreatology. 18 (Suppl):S1322018. View Article : Google Scholar
|
|
5
|
Latenstein AEJ, van der Geest LGM, Bonsing
BA, Groot Koerkamp B, Haj Mohammad N, de Hingh IHJT, de Meijer VE,
Molenaar IQ, van Santvoort HC, van Tienhoven G, et al: Nationwide
trends in incidence, treatment and survival of pancreatic ductal
adenocarcinoma. Eur J Cancer. 125:83–93. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mokdad AA, Minter RM, Yopp AC, Porembka
MR, Wang SC, Zhu H, Augustine MM, Mansour JC, Choti MA and Polanco
PM: Comparison of overall survival between preoperative
chemotherapy and chemoradiotherapy for resectable pancreatic
adenocarcinoma. J Natl Compr Canc Netw. 16:1468–1475. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moore MJ and Stathis A: Advanced
pancreatic carcinoma: Current treatment and future challenges. Nat
Rev Clin Oncol. 7:163–172. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jun I, Park HS, Piao H, Han JW, An MJ, Yun
BG, Zhang X, Cha YH, Shin YK, Yook JI, et al: ANO9/TMEM16J promotes
tumourigenesis via EGFR and is a novel therapeutic target for
pancreatic cancer. Br J Cancer. 117:1798–1809. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chio IIC, Jafarnejad SM, Ponz-Sarvise M,
Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Öhlund D, et
al: NRF2 promotes tumor maintenance by modulating mrna translation
in pancreatic cancer. Cell. 166:963–976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jain A, Brown SZ, Thomsett HL, Londin E
and Brody JR: Evaluation of post-transcriptional gene regulation in
pancreatic cancer cells: Studying RNA binding proteins and their
mRNA targets. Methods Mol Biol. 1882:239–252. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Uchida S, Kinoh H, Ishii T, Matsui A,
Tockary TA, Takeda KM, Uchida H, Osada K, Itaka K and Kataoka K:
Systemic delivery of messenger RNA for the treatment of pancreatic
cancer using polyplex nanomicelles with a cholesterol moiety.
Biomaterials. 82:221–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hutter C and Zenklusen JC: The cancer
genome atlas: Creating lasting value beyond its data. Cell.
173:283–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ho J, Li X, Zhang L, Liang Y, Hu W, Yau
JC, Chan H, Gin T, Chan MT, Tse G and Wu WK: Translational genomics
in pancreatic ductal adenocarcinoma: A review with re-analysis of
TCGA dataset. Semin Cancer Biol. 55:70–77. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barrett T: NCBI GEO: Mining millions of
expression profiles-database and tools. Nucleic Acids Res.
33:D562–D566. 2004. View Article : Google Scholar
|
|
15
|
Li C, Zeng X, Yu H, Gu Y and Zhang W:
Identification of hub genes with diagnostic values in pancreatic
cancer by bioinformatics analyses and supervised learning methods.
World J Surg Oncol. 16:2232018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang H, Wei P, Chang P, Li Y, Yan D, Liu
C, Hassan M and Li D: Genetic polymorphisms associated with
pancreatic cancer survival: A genome-wide association study. Int J
Cancer. 141:678–686. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sivakumar S, de Santiago I, Chlon L and
Markowetz F: Master regulators of oncogenic KRAS response in
pancreatic cancer: An integrative network biology analysis. PLoS
Med. 14:e10022232017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Muzumdar MD, Chen PY, Dorans KJ, Chung KM,
Bhutkar A, Hong E, Noll EM, Sprick MR, Trumpp A and Jacks T:
Survival of pancreatic cancer cells lacking KRAS function. Nat
Commun. 8:10902017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wolfgang CL, Herman JM, Laheru DA, Klein
AP, Erdek MA, Fishman EK and Hruban RH: Recent progress in
pancreatic cancer. CA Cancer J Clin. 63:318–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
21
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang G, Schetter A, He P, Funamizu N,
Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, et al:
DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity
and predicts clinical outcome in pancreatic ductal adenocarcinoma.
PLoS One. 7:e315072012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Donahue TR, Tran LM, Hill R, Li Y,
Kovochich A, Calvopina JH, Patel SG, Wu N, Hindoyan A, Farrell JJ,
et al: Integrative survival-based molecular profiling of human
pancreatic cancer. Clin Cancer Res. 18:1352–1363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lunardi S, Jamieson NB, Lim SY, Griffiths
KL, Carvalho-Gaspar M, Al-Assar O, Yameen S, Carter RC, McKay CJ,
Spoletini G, et al: IP-10/CXCL10 induction in human pancreatic
cancer stroma influences lymphocytes recruitment and correlates
with poor survival. Oncotarget. 5:11064–11080. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Janky R, Binda MM, Allemeersch J, Van den
Broeck A, Govaere O, Swinnen JV, Roskams T, Aerts S and Topal B:
Prognostic relevance of molecular subtypes and master regulators in
pancreatic ductal adenocarcinoma. BMC Cancer. 16:6322016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang S, He P, Wang J, Schetter A, Tang W,
Funamizu N, Yanaga K, Uwagawa T, Satoskar AR, Gaedcke J, et al: A
novel MIF signaling pathway drives the malignant character of
pancreatic cancer by targeting NR3C2. Cancer Res. 76:3838–3850.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nie K, Shi L, Wen Y, Pan J, Li P, Zheng Z
and Liu F: Identification of hub genes correlated with the
pathogenesis and prognosis of gastric cancer via bioinformatics
methods. Minerva Med. 111:213–225. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu L, Lin J and He H: Identification of
potential crucial genes associated with the pathogenesis and
prognosis of endometrial cancer. Front Genet. 10:3732019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Prevo R, Pirovano G, Puliyadi R, Herbert
KJ, Rodriguez-Berriguete G, O'Docherty A, Greaves W, McKenna WG and
Higgins GS: CDK1 inhibition sensitizes normal cells to DNA damage
in a cell cycle dependent manner. Cell Cycle. 17:1513–1523. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei D, Parsels LA, Karnak D, Davis MA,
Parsels JD, Marsh AC, Zhao L, Maybaum J, Lawrence TS, Sun Y and
Morgan MA: Inhibition of protein phosphatase 2A radiosensitizes
pancreatic cancers by modulating CDC25C/CDK1 and homologous
recombination repair. Clin Cancer Res. 19:4422–4432. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Z, Fan M, Candas D, Zhang TQ, Qin L,
Eldridge A, Wachsmann-Hogiu S, Ahmed KM, Chromy BA, Nantajit D, et
al: Cyclin B1/Cdk1 coordinates mitochondrial respiration for
cell-cycle G2/M progression. Dev Cell. 29:217–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Levasseur MD, Thomas C, Davies OR, Higgins
JM and Madgwick S: Aneuploidy in oocytes is prevented by sustained
CDK1 activity through degron masking in cyclin B1. Dev Cell.
48:672–684. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu H: Cdc20: A WD40 activator for a cell
cycle degradation machine. Mol Cell. 27:3–16. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang DZ, Ma Y, Ji B, Liu Y, Hwu P,
Abbruzzese JL, Logsdon C and Wang H: Increased CDC20 expression is
associated with pancreatic ductal adenocarcinoma differentiation
and progression. J Hematol Oncol. 5:152012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang WY, Hsu CC, Wang TY, Li CR, Hou YC,
Chu JM, Lee CT, Liu MS, Su JJ, Jian KY, et al: A gene expression
signature of epithelial tubulogenesis and a role for ASPM in
pancreatic tumor progression. Gastroenterology. 145:1110–1120.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hsu CC, Liao WY, Chan TS, Chen WY, Lee CT,
Shan YS, Huang PJ, Hou YC, Li CR and Tsai KK: The differential
distributions of ASPM isoforms and their roles in Wnt signaling,
cell cycle progression, and pancreatic cancer prognosis. J Pathol.
249:498–508. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu G, Zhao J, Pan B, Ma G and Liu L:
UBE2C overexpression in melanoma and its essential role in G2/M
transition. J Cancer. 10:2176–2184. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jin D, Guo J, Wu Y, Du J, Wang X, An J, Hu
B, Kong L, Di W and Wang W: UBE2C, directly targeted by
miR-548e-5p, increases the cellular growth and invasive abilities
of cancer cells interacting with the EMT marker protein zinc finger
E-box binding homeobox 1/2 in NSCLC. Theranostics. 9:2036–2055.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Yin L, Yang L, Zheng Y, Liu S,
Yang J, Cui H and Wang H: Silencing ubiquitin-conjugating enzyme 2C
inhibits proliferation and epithelial-mesenchymal transition in
pancreatic ductal adenocarcinoma. FEBS J. 286:4889–4909. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gomes-Filho SM, Dos Santos EO, Bertoldi
ER, Scalabrini LC, Heidrich V, Dazzani B, Levantini E, Reis EM and
Bassères DS: Aurora A kinase and its activator TPX2 are potential
therapeutic targets in KRAS-induced pancreatic cancer. Cell Oncol
(Dordr). 43:445–460. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Warner SL, Stephens BJ, Nwokenkwo S,
Hostetter G, Sugeng A, Hidalgo M, Trent JM, Han H and Von Hoff DD:
Validation of TPX2 as a potential therapeutic target in pancreatic
cancer cells. Clin Cancer Res. 15:6519–6528. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Heestand GM, Schwaederle M, Gatalica Z,
Arguello D and Kurzrock R: Topoisomerase expression and
amplification in solid tumours: Analysis of 24,262 patients. Eur J
Cancer. 83:80–87. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pei YF, Yin XM and Liu XQ: TOP2A induces
malignant character of pancreatic cancer through activating
β-catenin signaling pathway. Biochim Biophys Acta Mol Basis Dis.
1864:197–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mills CA, Suzuki A, Arceci A, Mo JY,
Duncan A, Salmon ED and Emanuele MJ: Nucleolar and
spindle-associated protein 1 (NUSAP1) interacts with a SUMO E3
ligase complex during chromosome segregation. J Biol Chem.
292:17178–17189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Garrido-Rodríguez M, Ortea I, Calzado MA,
Muñoz E and García V: SWATH proteomic profiling of prostate cancer
cells identifies NUSAP1 as a potential molecular target for
galiellalactone. J Proteomics. 193:217–229. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Taniuchi K, Furihata M and Saibara T:
KIF20A-mediated RNA granule transport system promotes the
invasiveness of pancreatic cancer cells. Neoplasia. 16:1082–1093.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Imai K, Hirata S, Irie A, Senju S, Ikuta
Y, Yokomine K, Harao M, Inoue M, Tomita Y, Tsunoda T, et al:
Identification of HLA-A2-restricted CTL epitopes of a novel
tumour-associated antigen, KIF20A, overexpressed in pancreatic
cancer. Br J Cancer. 104:300–307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Asahara S, Takeda K, Yamao K, Maguchi H
and Yamaue H: Phase I/II clinical trial using HLA-A24-restricted
peptide vaccine derived from KIF20A for patients with advanced
pancreatic cancer. J Transl Med. 11:2912013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hewit K, Sandilands E, Martinez RS, James
D, Leung HY, Bryant DM, Shanks E and Markert EK: A functional
genomics screen reveals a strong synergistic effect between
docetaxel and the mitotic gene DLGAP5 that is mediated by the
androgen receptor. Cell Death Dis. 19:10692018. View Article : Google Scholar
|
|
50
|
Mello SS, Valente LJ, Raj N, Seoane JA,
Flowers BM, McClendon J, Bieging-Rolett KT, Lee J, Ivanochko D,
Kozak MM, et al: A p53 super-tumor suppressor reveals a tumor
suppressive p53-Ptpn14-yap axis in pancreatic cancer. Cancer Cell.
32:460–473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Connor AA, Denroche RE, Jang GH, Lemire M,
Zhang A, Chan-Seng-Yue M, Wilson G, Grant RC, Merico D, Lungu I, et
al: Integration of genomic and transcriptional features in
pancreatic cancer reveals increased cell cycle progression in
metastases. Cancer Cell. 35:267–282. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jiang W, Zhao S, Jiang X, Zhang E, Hu G,
Hu B, Zheng P, Xiao J, Lu Z, Lu Y, et al: The circadian clock gene
Bmal1 acts as a potential anti-oncogene in pancreatic cancer by
activating the p53 tumor suppressor pathway. Cancer Lett.
371:314–325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang H, Zhang X, Li X, Meng WB, Bai ZT,
Rui SZ, Wang ZF, Zhou WC and Jin XD: Effect of CCNB1 silencing on
cell cycle, senescence, and apoptosis through the p53 signaling
pathway in pancreatic cancer. J Cell Physiol. 234:619–631. 2019.
View Article : Google Scholar
|
|
54
|
Fraser SP, Diss JK, Chioni A, Mycielska
ME, Pan H, Yamaci RF, Pani F, Siwy Z, Krasowska M, Grzywna Z, et
al: Voltage-Gated sodium channel expression and potentiation of
human breast cancer metastasis. Clin Cancer Res. 11:5381–5389.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Diss JK, Archer SN, Hirano J, Fraser SP
and Djamgoz MB: Expression profiles of voltage-gated Na(+) channel
alpha-subunit genes in rat and human prostate cancer cell lines.
Prostate. 48:165–178. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Stellas D and Patsavoudi E: Inhibiting
matrix metalloproteinases, an old story with new potentials for
cancer treatment. Anticancer Agents Med Chem. 12:707–717. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Fujisawa T, Rubin B, Suzuki A, Patel PS,
Gahl WA, Joshi BH and Puri RK: Cysteamine suppresses invasion,
metastasis and prolongs survival by inhibiting matrix
metalloproteinases in a mouse model of human pancreatic cancer.
PLoS One. 7:e344372012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tian J, Zhang R, Piao H, Li X, Sheng W,
Zhou J, Dong M, Zhang X, Yan X, Shang W, et al: Silencing Tspan1
inhibits migration and invasion, and induces the apoptosis of human
pancreatic cancer cells. Mol Med Rep. 18:3280–3288. 2018.PubMed/NCBI
|
|
59
|
Hou FQ, Lei XF, Yao JL, Wang YJ and Zhang
W: Tetraspanin 1 is involved in survival, proliferation and
carcinogenesis of pancreatic cancer. Oncol Rep. 34:3068–3076. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lupski JR, Gonzaga-Jauregui C, Yang Y,
Bainbridge MN, Jhangiani S, Buhay CJ, Kovar CL, Wang M, Hawes AC,
Reid JG, et al: Exome sequencing resolves apparent incidental
findings and reveals further complexity of SH3TC2 variant alleles
causing charcot-marie-tooth neuropathy. Genome Med. 5:572013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Stendel C, Roos A, Kleine H, Arnaud E,
Özçelik M, Sidiropoulos PN, Zenker J, Schüpfer F, Lehmann U, Sobota
RM, et al: SH3TC2, a protein mutant in charcot-marie-tooth
neuropathy, links peripheral nerve myelination to endosomal
recycling. Brain. 133:2462–2474. 2010. View Article : Google Scholar : PubMed/NCBIPubMed/NCBI
|