Introduction
Cervical cancer (CC) represents the fourth highest
cause of mortality in the female population worldwide, with
~569,847 newly diagnosed CC cases and ~311,365 CC-associated deaths
in 2018 (1). The most prevalent
histological subtypes are squamous cell carcinoma (SCC) and
adenocarcinoma (AC), with SCC representing ~70% of CC cases
(2). Persistent infection associated
with high-risk human papillomavirus (HR-HPV) is considered to be a
key risk factor in the development of CC, particularly HPV types 16
and 18 (3). Additionally, the
integration of HR-HPV DNA in fragile host-genome sites, favors the
overexpression of oncoproteins E6 and E7. This, as a result,
promotes the progression and transformation of malignant cells,
inducing genetic and epigenetic instability (4).
From the cytological point of view, the Bethesda
system classifies precursor lesions of CC as low-grade squamous
intraepithelial lesions (LSIL) and high-grade lesions (HSIL). LSIL
are characterized by the presence of cells with karyomegaly,
perinuclear halo and binucleation (koilocytes), while HSIL is
associated with the presence of intense binucleation and
dyskaryosis in basal and parabasal cells with little cytoplasm,
hyperchromatic and big nuclei. Furthermore, SCC is characterized by
large undifferentiated and multinucleated cells lacking cytoplasm,
pleomorphic nuclei and irregular distribution of chromatin
(5).
Integration of HR-HPV, in addition to altering
significant transcription patterns and regulating the expression of
E6 and E7, also regulates the expression of host genes in fragile
integration sites (6). It is
important to note that a number of these genes have an oncogenic
function, for example as oncogenic microRNAs (miRNAs/miRs)
(7). miRNAs suppress genetic
translation by binding to the 3′-untranslated region of their
target genes (8). Previous studies
have shown that numerous miRNAs are implicated in CC, such as
miR-16-1 (9), miR-21 (10), miR-22-3p (11) and miR-486-5p (12). These miRNAs contribute to a number of
cellular processes, such as cell proliferation (9–12),
invasion (9,10,12),
migration (12) and apoptosis
(11).
miR-16-1 is a regulator of gene expression at the
post-transcriptional level. Studies have reported that miRNAs are
often increased in certain cancer types, including breast cancer
(13), hepatocellular carcinoma
(14) and CC (9,15–20).
miR-15a and miR-16-1 form part of a cluster in an intron region of
the deleted in lymphocytic leukemia 2 (DLEU2) transcript on
chromosome 13q14.3, which is frequently deleted in chronic
lymphocytic leukemia. Both are implicated in cellular invasion,
survival and proliferation (21).
It has been reported that the expression of miR-16-1
is decreased in CC cell lines that are positive for HPV16 and HPV18
following transfection with a siRNA for oncoprotein E7, suggesting
that the increase in miR-16-1 expression may be due to the
E7/E2F/miR-16-1 pathway (22).
Furthermore, other studies have indicated that miR-16-1 possesses
an oncomiR function in CC (15–18,20). The
overexpression of miR-16-1 is associated with activation of genes
implicated in the cell cycle, including CDK6, CDC27, CARD10,
C10orf46 (23) and CCNE1 (9), which promote the proliferation of
cancerous cells. The aim of the present study was to investigate
the association of the miR-16-1 expression level with squamous
intraepithelial lesions (SIL) and with the integration of HR-HPV
DNA.
Materials and methods
Participants and sample
collection
The present study included 80 liquid-based cytology
samples obtained from the squamous-column transformation zone (TZ)
of the uterine cervix of female patients aged 18–71 years, who
resided within the State of Guerrero, Mexico. The mean age was 42
years. Between March 2012 and September 2018, the patients
presented at the Integral Diagnostic Service for the Timely
Detection of Cervical Cancer and HPV of the Autonomous University
of Guerrero (Chilpancingo, Mexico), to the Dysplasia Clinic of the
General Hospital ‘Raymundo Abarca Alarcón’ (Chilpancingo, Mexico)
and to the State Institute of Cancerology ‘Arturo Beltrán Ortega’
(Acapulco, Mexico).
The study was approved by the Ethics Committee of
the Autonomous University of Guerrero, Guerrero, Mexico. All
patients signed an informed consent for the use of their cervical
samples and clinical information. This study was also performed
according to the ethical guidelines of the Declaration of Helsinki
2008 (24).
Cytological examinations, HPV genotyping and
measurements of miR-16-1 by reverse transcription-quantitative PCR
(RT-qPCR) were performed in the present study. Three ectocervical
and three endocervical samples were obtained from each patient,
utilizing an Ayre spatula (ectocervix) and cytobrush (endocervix),
ensuring cytological material was from the TZ of the uterine
cervix. The cytological diagnosis was performed by a
cytotechnologist (LdCA-R) who was accredited and certified by the
Mexican Council of Technicians in Pathobiology, A.C., and the
Mexican Council of Anatomopathological Physicians, A.C., with 29
years of experience, utilizing the criteria of the Bethesda System
(5). The colposcopic diagnosis was
performed by the colposcopist Dr Raúl Peralta-Catalán responsible
for the Dysplasia Clinic of the General Hospital ‘Raymundo Abarca
Alarcón’ (Chilpancingo, Mexico). The histopathological diagnosis,
for confirmation of SIL and SCC, was performed by the pathologist
Marco Antonio Jiménez-López at the State Institute of Cancerology
‘Arturo Beltrán Ortega’ (Acapulco, Mexico) (25).
Cytological examination
Slides with the cytological smears of the TZ for
conventional cytology examination were fixed in ethanol for 10 min.
The slides were then stained using the Papanicolaou kit (cat. no.
64294; Hycel, Chemical Reagents). Briefly, the slides were hydrated
in a descending alcohol series and then incubated at room
temperature for 45 sec with Harris hematoxylin to stain the nuclei.
Additionally, Orange G colorant was added and incubated at room
temperature for 80 sec, followed by EA-50 incubated at room
temperature for 3 min, which stained the eosinophils and basophils
cells, respectively. The slides were then cleared with Xylol
reagent prior to microscopic observation (DM1000 LED; Leica
Microsystems, Inc.; magnification, ×10-x20).
Alternatively, the samples for liquid-based cytology
were processed according to the manufacturer's protocol of
liquid-PREP™ (LGM International, Inc.). Briefly, a clearing
solution was added to each sample and then the samples were
centrifuged at 1,000 × g for 5 min at room temperature. The
supernatant was discarded after the addition of the cell base
solution, which conserved the pellet. The samples were mixed and 10
µl was added to a slide, which was fixed at room temperature with
ethanol for 10 min, following by staining using Papanicolaou kit
and microscopic observation (DM1000 LED; Leica Microsystems, Inc.;
magnification, ×10-x20).
Genotyping and the physical state of
HR-HPV
Using cervical cytology samples in PBS (pH 7.0), DNA
was extracted by the standard method of phenol-chloroform
extraction (26). For HPV
genotyping, the reverse INNO-LiPA Genotyping Extra assay
(Invitrogen; Thermo Fisher Scientific, Inc.) was used according to
the manufacturer's instructions. This method allowed the
simultaneous identification of 28 different HPV genotypes. Briefly,
the L1 region of HPV was PCR-amplified with the SPF10 primers. The
biotinylated amplicons were denatured and hybridized with specific
and immobile oligonucleotides anchored to a membrane, and then
Streptavidin conjugated with alkaline phosphatase was added,
followed by Chromogen BCIP/NBT to reveal the reaction. The HLA-DPB1
gene was employed as a control for DNA amplification, and the L1
region of HPV6 was utilized as a positive control.
The determination of the physical state of viral DNA
was performed by means of the Dako GenPoint™ Tyramide Signal
Amplification System for Biotinylated Probes (Agilent Technologies,
Inc.), according to the manufacturer's protocol. Briefly,
liquid-based cytology smears were submitted to permeabilization for
30 min at 120°C, followed by enzymatic digestion for 5 sec with K
proteinase (1:1,000). The samples were then added to 1 µl test
reagent (Dako GentPoint™ HPV DNA Probe Cocktail, Biotinylated) for
13 HR-HPV genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59
and 68; Agilent Technologies, Inc.). The slides were subjected to
DNA denaturation for 10 min at 95°C and hybridization for 20 h at
37°C on a Dako Hybridizer (Agilent Technologies, Inc.).
Subsequently, the slides were placed in an astringent solution
(1:20) for 20 min at 55°C, followed by the addition of 30 µl
primary streptavidin-HRP conjugate (1:50) for 1 h and incubation in
a humidified chamber at room temperature. Subsequently, 30 µl
biotinyl tyramide was added for 40 min and 30 µl secondary
streptavidin-HRP conjugate was added for 1 h in a humidified
chamber at room temperature. Diaminobenzidine (1:20) was then added
for 10 sec, followed by counterstaining with Harris hematoxylin
(Merck KGaA) for 10 sec, both incubations at room temperature. The
positive reaction with the nucleus was identified by a brown color,
which was classified as diffuse (episomal state), punctate
(integrated state) or mixed (episomal and integrated state). As a
positive control, the SiHa cervical cancer cell line (catalog no.
HTB-35; American Type Culture Collection) was used and, as negative
control, SiHa cells were used without the test reagent. The cell
line was cultivated in DMEM (Invitrogen; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Merck KGaA), 50 µg/ml
penicillin/streptomycin (Merck KGaA), 2 mM L-glutamine (Merck KGaA)
and 250 ng/ml fungizone (Merck KGaA), and placed at 37°C in a
humidified incubator containing 5% CO2.
Analysis of miR-16-1 expression
Using the cervical cytology samples, total RNA was
extracted using TRIzol® Reagent (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Briefly, 1 ml TRIzol® Reagent was added to each sample
and incubated for 5 min to permit the dissociation of the
nucleus-protein complex. Subsequently, 200 µl chloroform for every
1 ml TRIzol® Reagent was added, followed by vigorous
shaking for 15 sec and incubation for 3 min. Following incubation,
the sample was centrifuged at 12,000 × g at 4°C for 5 min. The
aqueous phase, which contains the RNA, was transferred into a new
tube. Next, 500 µl isopropanol per 1 ml TRIzol® Reagent
was used for lysis for 10 min at −70°C, followed by centrifugation
at 14,000 x g at 4°C for 15 min. The supernatant was then discarded
and the sediment was re-suspended in 20 µl RNase-free water. The
concentration of RNA was evaluated by UV absorbance at 260 nm
(A260) using a Thermo Scientific NanoDrop 200c (Thermo Fisher
Scientific, Inc.). Synthesis of the complementary DNA (cDNA) was
carried out using the TaqMan MicroRNA Reverse Transcription kit
(Thermo Fisher Scientific, Inc.), which possesses a high capacity
for synthesizing cDNA from miRNA. In total, 5 ng total RNA was
utilized for synthesizing the cDNA of the miR-16-1. Briefly,
inverse transcription assays were prepared with 3 µl 5X RT primer,
5 µl RNA sample and 7 µl Master mix [100 mM dNTPs (with dTTP) (0.15
µl), MultiScribe™ Reverse Transcriptase (50 U/µl; 1 µl), 10X
Reverse Transcription Buffer (1.50 µl), RNAse Inhibitor (20 U/µl;
0.19 µl) and Nuclease-Free water (4.16 µl) in a total volume of 15
µl]. The reactions were incubated in an Eppendorf thermocycler
(Eppendorf Mastercycler EP Gradient Model 5341) for 30 min at 16°C,
30 min at 42°C, and 5 min at 85°C, followed by storage at 4°C until
later use.
The PCR reaction was conducted at 95°C for 10 min
followed by 40 cycles at 95°C for 10 sec and at 60°C for 60 sec
using TaqMan™ MicroRNA Assays (Thermo Fisher Scientific, Inc.).
Specific primers (Thermo Fisher Scientific, Inc.) were used for
hsa-miR-16-1 (5′-UAGCAGCACGUAAAUAUUGGCG-3′) and RNU44
(5′-CCTGGATGATGATAGCAAATGCTGACTGAACATGAAGGTCTTAATTAGCTCTAACTGACT-3′),
which was used for normalization. The reaction was incubated in PCR
tubes and caps, RNase-free, 0.2 ml (catalog no. AM12230; Thermo
Fisher Scientific, Inc.) in the CFX96 Touch™ Real-Time PCR
Detection system supplied with analytical software (Bio-Rad
Laboratories, Inc.). Each reaction was performed in triplicate. The
2−ΔΔCT method (27) was
employed to evaluate the relative abundance of miR-16-1 compared
with the expression of RNU44, which is a small nuclear RNA and is
one of the 18 human endogenous controls identified as the most
abundant in all tissues, based on CT averages (22-28.9),
good linearity test (R2>0.96) and its relatively
stable expression (28).
Statistical analysis
Data are presented as frequencies for the
qualitative variables and as the mean ± standard error for
quantitative variables. One-way analysis of variance followed by
Bonferroni's post hoc test was used to compare the expression level
of miR-16-1 between study groups. The association of cytological
diagnosis or the physical state of the HPV with the expression
level of miR-16-1 was evaluated through linear regression models.
This obtained the regression coefficients (β), as the average
change in the expression of miR-16-1 by cytological diagnosis or
physical state of the HPV, in comparison with the reference
category (NSIL). P<0.05 was considered to indicate a
statistically significant difference. Statistical analysis was
performed using STATA V.13 statistical software (StataCorp
LLC).
Results
Genotypes and physical state of the
HPV
Once the cytological results were obtained, the
following cytological samples of women were selected as follows: 20
samples with negative diagnosis for SIL and without HPV infection;
20 samples with LSIL; 20 samples with HSIL (of which 45% were
diagnosed with carcinoma in situ); and 20 samples with SCC.
Women with SIL or SCC had HR-HPV infection. From the findings of
the present study, eight types of HR-HPV could be identified; the
most frequent being 16, 18, 31, 33, 45, 51, 52 and 58. The
frequency of HPV16, in relation to the cytological diagnosis, was
20% in LSIL, 30% in HSIL and 40% in SCC. Notably, 15% of LSIL, 20%
of HSIL, and 40% of SCC presented with multiple infection (MI) with
the genotypes of HR-HPV, including HPV16 (data not shown). On the
other hand, it is important to note that when analyzing the
physical state of the HR-HPV DNA, an integrated state was
identified in 40% of women with LSIL, in 35% of women with HSIL and
in 90% of women with SCC (Table I).
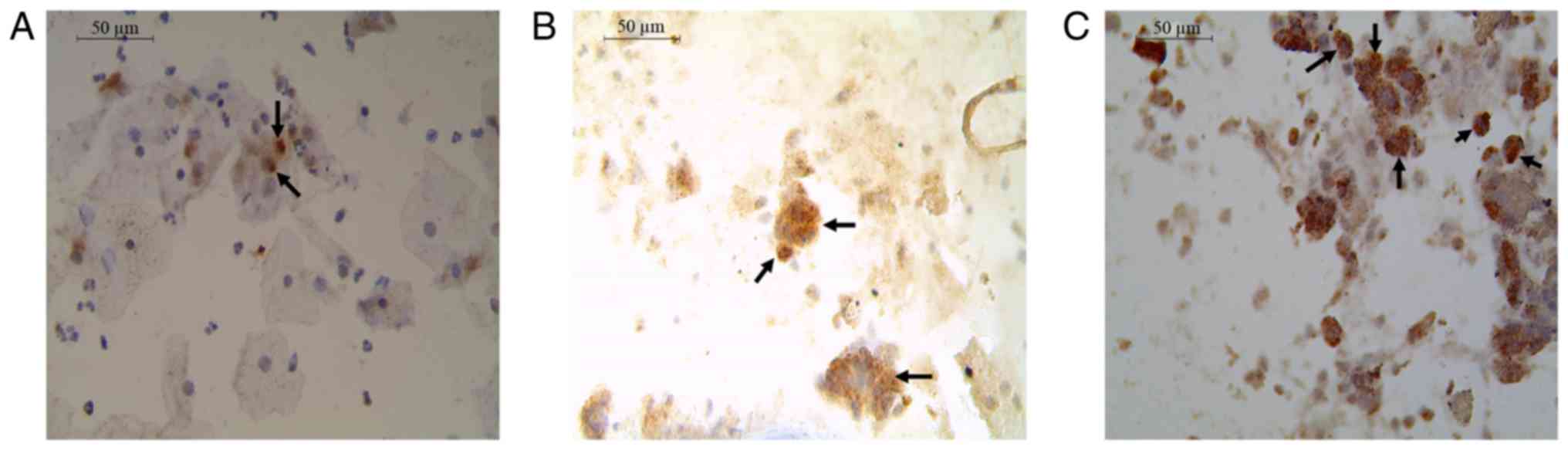
Cytologically in the LSIL cases, some intermediate cells with
karyomegaly, binucleation, perinuclear halo and hyperchromatic
nuclei were observed. These are considered to be characteristics of
the HPV infection. The intermediate cells with karyomegaly
presented 1–2 viral copies integrated (Fig. 1A). While in cytologies with HSIL,
small groups of cells with moderate to intense dyskaryosis and
binucleation were observed, in immature basal and parabasal cells
with little cytoplasm, big and hyperchromatic nuclei. These cells
presented multiple integrated copies (Fig. 1B). Finally, in the cases of SCC,
multiple integrated copies were observed in large undifferentiated
cells, multinucleated, devoid of cytoplasm, pleomorphic nuclei and
irregular distribution of chromatin (Fig. 1C).
 | Table I.HR-HPV state according to cytological
diagnosis. |
Table I.
HR-HPV state according to cytological
diagnosis.
|
| Diagnosis |
|---|
|
|
|
|---|
| HR-HPV state | LSIL, n (%) | HSIL, n (%) | SCC, n (%) |
|---|
| Integrated | 8 (40) | 7 (35) | 18 (90) |
| Mixed | 12 (60) | 13 (65) | 2 (10) |
| Total | 20 (100) | 20 (100) | 20 (100) |
Expression of miR-16-1
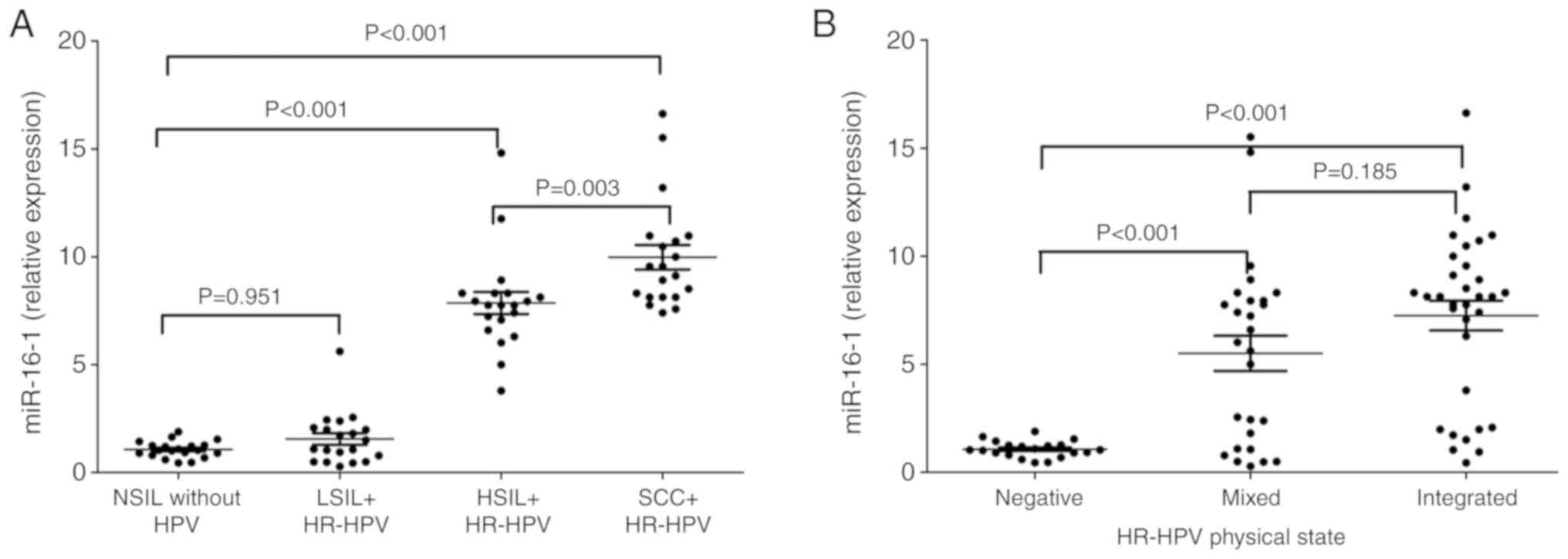
It was demonstrated that the mean expression level
of miR-16-1 was increased significantly in women with HSIL
(7.9±0.5) and SCC (10±0.6), in comparison with women with NSIL or
LSIL (P<0.001). Although a small increase was identified in the
expression of miR-16-1 in patients with LSIL (1.6±0.3) in
comparison with women with NSIL (1.1±0.1), this was not
statistically significant (P=0.95; Fig.
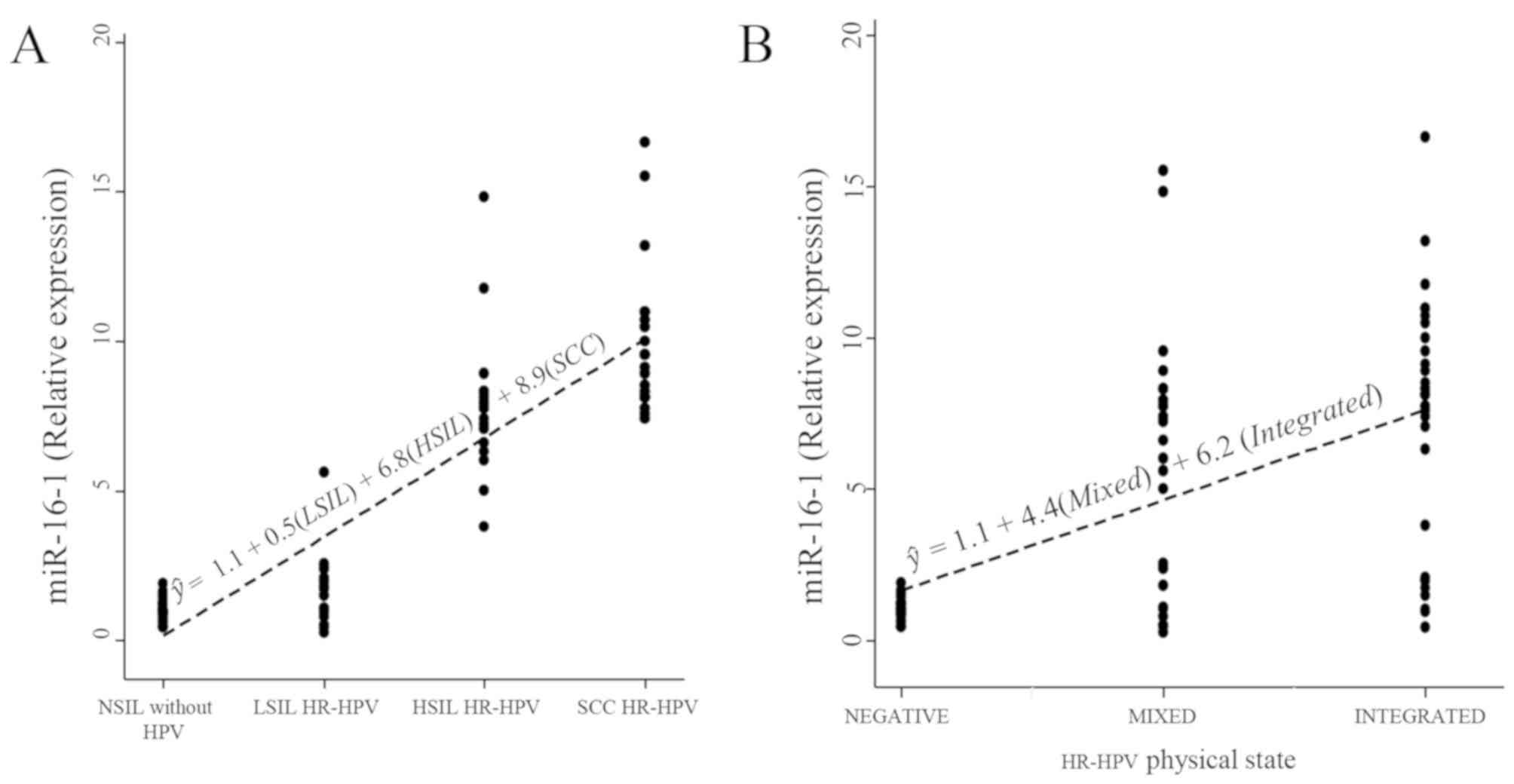
2A). Linear regression analysis revealed a significant increase
in the expression of miR-16-1 in women with HSIL (β=6.8;
P<0.001) and in women with SCC (β=8.9; P<0.001) in comparison
with women NSIL, with an explanation percentage of 83% (Table II).
Additionally, significant differences were
identified for the expression level of miR-16-1 in the samples with
a mixed or integrated HPV physical state compared with the samples
that did not present with HPV infection (P<0.001; Fig. 2B). In addition, a significant
influence of the mixed (β=4.4; P<0.001) or integrated (β=6.2;
P<0.001) state on the expression level of miR-16-1 (P<0.001)
was identified, compared with the samples negative for HPV
infection; with an explanation percentage of 33%. It is also
important to comment that any relevant changes were not identified
in the level of expression of miR-16-1 between the integrated
physical state and the mixed state (β=1.7; P=0.105; Table II; Fig.
3).
Discussion
CC represents a serious public-health problem.
Despite it being a preventable disease, it has high rates of
incidence and mortality in developing countries, and in Mexico, CC
is the third most common cancer in women with 7,689 new cases
reported in 2018 (1). In Mexico, a
nationwide cytology-based cervical cancer screening program was
implemented in 1974, but the subsequent decrease in incidence and
mortality have been modest (29). In
Guerrero, Mexico, from the 2000 to 2013, CC represented the second
most common cancer in women, which therefore makes CC the fifth
highest contributor to mortality rates nationally (30). In Guerrero, Mexico, HPV16 has been
identified as the most frequent genotype in CC, followed by HPV18
(31). It is important to note that
our group has previously reported that, in the state of Guerrero,
there are five circulating variants of E6 of HPV16 (E-G350, AA-a,
AA-c, E-C188/G350 and E-A176/G350). These variants have been
associated with the development of SCC, and the AA-a variant has
the greatest association with the development of CC (odds ratio,
69.01; confidence interval, 7.57-628.96) in comparison with the
E-prototype variant (32).
In the present study, women diagnosed with LSIL, as
well as women diagnosed with HSIL, presented with HPV16 with
greater frequency in unique infection or MI compared with other
types of HR-HPV. It has been noted that genotypes 16 and 18 are
present in >70% of SCC cases (3).
A number of studies have reported that MI with HR-HPV can increase
the risk of cervical intraepithelial neoplasia (CIN) progressing to
SCC (33–35). However, other reports that HPV16, in
itself, can increase the risk of developing a HSIL should also be
considered (36).
Integration of viral DNA occurs because of
chromosomal instability, which is induced by the aberrant
expression of oncoproteins E6 and E7 (37). The biotinyl-tyramide-based in
situ hybridization (ISH) amplification method has the advantage
of allowing the in situ examination of the physical state of
HPV DNA, preserving the morphology of the cells or tissues, as well
as it being optimized to enable the reproducible detection of one
to two integrated copies of the HPV-16 (38–40). It
has been reported that in cervical scrapes or biopsy samples
positive for HPV16 or 18 from 187 female patients without SIL,
LSIL, HSIL and CC, ISH has a high concordance (96.1%) with qPCR to
determine the physical state of HPV. These results suggest that ISH
has good concordance with qPCR with regards to the detection of HPV
integration. Therefore, this method can be used for determining the
physical status of HPV (41). The
present study identified that women with LSIL (60%), with HSIL
(65%) and with SCC (10%) had principally a mixed state. In this
regard, we reported previously that the mixed state of HR-HPV can
be found in cytologies with LSIL with HR-HPV (42), while other investigators have
reported it in HSIL (43). On the
other hand, with respect to the integration of viral DNA, it was
identified that 40% of LSIL, 35% of HSIL and 90% of women with SCC
had viral integration. In our work group and with regards to
previous studies, it was identified that in women with LSIL with
HR-HPV, 10% had viral DNA integration (42), while some studies have reported that
viral integration is an indicator of HSIL (44), and a predictive indicator that is
markedly unfavorable for the survival of patients with primary CC,
in comparison with mixed forms of HR-HPV (45). One possible reason why the percentage
of women with viral integration was similar between LSIL and HSIL
is that in both study groups, HPV16 and 18 were present in single
or multiple infection with other HR-HPV (data not shown). However,
differences in the number of copies integrated between both groups
were observed (Fig. 1). It is
important to note that in LSIL the number of altered cells is lower
compared with HSIL. In addition, it has been reported that viral
integration in SIL and CC is more frequently related to HPV16, 18
and 58 genotypes (41,46).
It must be considered that previous reports have
demonstrated that viral integration is an early event in the
progression of the disease (47,48). In
addition, it has been reported that populations of cells with
integrated HPV16 possess a selective advance in growth, compared
with cells that maintain episomal HPV16 genomes (49). The present study identified that the
number of cells and the integrated copies in them increased in
cytologies with HSIL and SCC compared with cytologies with LSIL
(Fig. 1A-C). These results are
important since it has been reported that those cells with multiple
integrated copies of HPV16 has an increase in methylation patterns
in the upstream regulatory region (URR) region of the viral genome,
compared with those with only 1–2 integrated copies or those that
present only episomal copies. This suggests that methylation in E2
binding sites, in the URR region of HPV16, can lead to deregulation
of E6 and E7 expression in early stages of cell transformation
induced by HR-HPV (50).
Furthermore, a wide range of studies have reported
that miRNAs serve an important role in the regulation of gene
expression, and the deregulation of miRNAs plays an important role
in the development of human cancers (51). The expression of miR-16-1 is of great
interest for further analysis, as it has been reported to be
increased in a variety of human cancers, in which its function has
been described as an oncomiR (9,15–20,22).
The present study did not identify significant differences in the
expression of miR-16-1 between women who presented with NSIL and
without HPV with those who presented with LSIL with HR-HPV
(P=0.951). One limitation of the present study that must be
considered when examining the results was the small sample size
used to identify significant differences between these two groups.
However, we found that HSIL, SCC or HPV physical state had an
effect on the increase in miR-16-1 expression. The patients with
HSIL and SCC with HR-HPV exhibited a significant increase in the
expression of miR-16-1 in comparison with women with NSIL without
HPV (P<0.001; Table II). There
are studies that have evaluated this miRNA in cell lines and
tissues with SCC, for example. miR-16-1 was increased in 19 SCC
tissue samples, 7 adenocarcinomas, 2 adenosquamous cell carcinomas
and 2 small-cell carcinomas, all of these with HR-HPV, in
comparison with normal tissue (15).
Furthermore, an increase was found in the expression of miR-16-1 in
ten tissues with invasive SCC in comparison with normal tissue
according to RT-qPCR (16). Through
Northern blot analysis, it has also been demonstrated that the
expression of miR-16-1 is increased in cell lines with HPV and in
CC tissues in comparison with normal tissue (17). Through microarrays, the expression of
diverse miRNAs has been studied, and results demonstrated that the
expression of miR-16-1 increased according to the grade of CIN,
with higher expression observed in cases with CIN III and SCC, in
comparison with CIN I and normal tissue (18). By contrast, through RT-qPCR, it was
identified that the expression of miR-16-1 was lower in ten normal
tissues was compared with 18 cases of CIN II and CIN III, 9 cases
of adenocarcinoma and 10 cases of SCC, in which the expression
increased according to the grade of CIN (19). Similar results were confirmed in a
study in which the expression of miR-16-1 was higher in CIN I, CIN
II, CIN III and CC in comparison with normal tissue (20).
In the present study, a basal expression of
miRNA-16-1 was found in samples without SIL that were negative for
HPV. It has been reported that the normal function of miR-16-1 is
to negatively regulate the progression of the cell cycle, by
regulating cell targets, such as CDK1, CDK2, CDK6, cyclin D1,
cyclin D3 and cyclin E1 (52). In
addition, during the progression of the normal cell cycle, the
endogenous inactivation of E2F leads to an increase in the basal
expression of miR-16-1 and miR-15, and consequently the arrest of
the cell cycle occurs in the G1 phase (53). The expression of miR-16-1 is
increased in tissues of CIN I, CIN II, CIN III and CC compared with
in normal tissue (20). Further
studies have shown that when HR-HPV infection is present, the
oncoprotein E7 dissociates the RB/E2F complex, resulting in the
endogenous activation of E2F (54,55). By
in silico analysis, it has been reported that the promoter
of the human gene DLEU2 contains a binding site conserved for E2F
in the position −4 to +4; therefore, the expression of miR-16-1 is
endogenously regulated by E2F (53).
These findings are important because cyclin E1 plays a crucial role
in the transition of the G1/S phase, and it is known that this
cyclin is transcriptionally regulated by E2F (56) and post-transcriptionally regulated by
miR-16-1 (9,57). These two molecular alterations can
cooperate during tumor development, maintaining an increased
proliferation of transformed cells.
It is noteworthy that the increased expression of
miR-16-1 was mainly related to HSIL and SCC in 6.8 and 8.9 relative
expression units, respectively, compared with NSIL (Table II). In this regard, the viral genome
is replicated as episomal DNA during productive infections, while
viral integration in the host chromosome by the HR-HPV has been
associated with the progression of SIL to SCC (58). Deletion of the E2 gene results in the
loss of negative regulation of the transcription of oncogenes E6
and E7, favoring dyscontrolled cellular proliferation and
immortalization (59). Having found
an increased expression in LSIL (1.6±0.3), in comparison with
normal samples (1.1±0.1), it can be suggested that these cells
possess a high proliferative capacity and that this could
facilitate the detection of cells with potential transformation
into an HSIL. These conclusions allow us to consider the importance
of strict follow-up of patients with LSIL with HR-HPV alone or with
MI, which also would allow the evaluation of the prognostic value
of miR-16-1.
Furthermore, the present study identified that the
mixed or integrated HPV states exhibited a significant effect on
the expression level of miR-16-1, in comparison with samples
negative for HPV (P<0.001); however, the variability in the
expression level of miR-16-1 has a greater explanation by the
changes induced by SIL and SCC (83%) than by the HPV physical state
(33%). In addition, when comparing women who presented with the
mixed state and those with the integrated state, no significant
difference was observed (P=0.105). A limitation of the present
study was that patients who presented only with the HPV episomal
physical state were not included, which could have provided
additional information on the expression level of miR-16-1 compared
with those with an integrated physical state. Notably, to the best
of our knowledge, no studies have analyzed this relationship
before. However, it has been suggested that the increased
expression of miR-16-1 in CC could be due to the molecular
mechanism induced by the interaction of E7 of the HPV16 and E2F
(53). To demonstrate whether E7 is
directly associated with the increase in the expression of miR-16-1
but not E6, a study was performed with tissue samples derived from
human keratinocytes, with and without HPV16 and HPV18. The results
demonstrated that on inducing the expression of E6, E7 and E6/E7,
the increase in the expression level of miR-16-1 was only observed
in the presence of E7. This suggests that E7 was responsible for
the overexpression of miR-16-1 in CC cells. In addition, by
silencing the expression of E7 by small interfering RNA in CaSki
(HPV16) and HeLa (HPV18) cell lines, it was demonstrated that
E7-knockdown decreased the expression of miR-16-1 in comparison
with the control cells (22).
It has been reported that HPV16 possesses an
integration site in chromosome 13q14 (60), where the DLEU2 gene is localized, and
this could activate the transcription of miR-16-1 (61). It has also been reported that HPV18
contains an integration site on chromosome 8q23-24, near the c-Myc
gene (62,63), which is known to be able to activate
the DLEU2 gene and induce the transcription of miR-16-1 (64). The overexpression of miR-16-1 has
been found to be associated with the activation of genes implicated
in cellular proliferation, such as CDK6, CDC27, CARD10, C10orf46
(23), CDC7 (21) and CCNE1 (9). Additionally, E6, on degrading into p53,
inhibits the expression of kinase-inhibitor proteins (55), generating an uncontrolled environment
for the proliferation and immortalization of cancerous cells.
In conclusion, the present results demonstrated that
the increased expression of miR-16-1 was associated with increased
cellular proliferation of HSIL and SCC in the presence of the
integrated state of the HR-HPV DNA alone or in MI. This suggests
that the expression level of miR-16-1 could serve as an additional
tool in the diagnosis of HSIL that exhibits potential progression
to SCC. Therefore, follow-up studies on a larger scale are required
in order to examine the clinical usefulness of the expression of
miR-16-1 as a prognostic biomarker of SIL, particularly in women
with a diagnosis of LSIL and the integrated state of the HR-HPV,
which can later progress to HSIL.
Acknowledgements
The authors would like to thank the pathologist Dr
Marco Antonio Jiménez-López (State Institute of Cancerology,
‘Arturo Beltrán Ortega’, Acapulco, Mexico) for contributing to the
histopathological diagnoses of squamous cell carcinoma. The authors
would also like to thank colposcopist gynecologist Dr Raúl
Peralta-Catalán (Dysplasia Clinic, General Hospital, ‘Raymundo
Abarca Alarcón’, Chilpancingo, Mexico) for contributing to the
diagnoses of low-grade squamous intraepithelial lesions.
Funding
This study was financially supported by grant from
CONACyT of the Sectoral Fund for Research in Health and Social
Security (grant no. 201579), Mexico.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LDCAR and EILBP designed and supervised the
research. LDCAR performed the cytological diagnosis. BIA carried
out the molecular diagnosis of the HPV. YCC and HJW collected the
samples and the survey data. MIZG conducted the extraction of the
RNA for reverse transcription-quantitative PCR. KIGP carried out
the liquid-based cytology for in situ hybridization with
amplification with tyramide. EFA performed the statistical
analysis. MIZG, LDCAR and EFA wrote and revised the manuscript. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
All patients signed an informed consent for the use
of their cervical samples and clinical information, and this study
was approved by the Bioethics Committee at the Autonomous
University of Guerrero, Guerrero, Mexico (approval no.
CB-003/2018).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bulk S, Berkhof J, Bulkmans NWJ, Zielinski
GD, Rozendaal L, Van Kemenade FJ, Snijders PJF and Meijer CJLM:
Preferential risk of HPV16 for squamous cell carcinoma and of HPV18
for adenocarcinoma of the cervix compared to women with normal
cytology in The Netherlands. Br J Cancer. 94:171–175. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Serrano B, de Sanjosé S, Tous S, Quiros B,
Muñoz N, Bosch X and Alemany L: Human papillomavirus genotype
attribution for HPVs 6, 11, 16, 18, 31, 33, 45, 52 and 58 in female
anogenital lesions. Eur J Cancer. 51:1732–1741. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiménez-Wences H, Peralta-Zaragoza O and
Fernández-Tilapa G: Human papillomavirus, DNA methylation and
microRNA expression in cervical cancer (Review). Oncol Rep.
31:2467–2476. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nayar R and Wilbur DC: The Bethesda system
for reporting cervical cytology. Definitions, criteria, and
explanatory notes. Springer International Publishing. (3rd).
(Switzerland). 2015.
|
|
6
|
Ferber MJ, Thorland EC, Brink AA, Rapp AK,
Phillips LA, McGovern R, Gostout BS, Cheung TH, Chung TKH, Fu WY
and Smith DI: Preferential integration of human papillomavirus type
18 near the c-myc locus in cervical carcinoma. Oncogene.
22:7233–7242. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmitz M, Driesch C, Jansen L, Runnebaum
IB and Dürst M: Non-random integration of the HPV genome in
cervical cancer. PLoS One. 7:e396322012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Medina PP and Slack FJ: MicroRNAs and
cancer: An overview. Cell Cycle. 7:2485–2492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zubillaga-Guerrero MI, Alarcón-Romero LC,
Illades-Aguiar B, Flores-Alfaro E, Bermúdez-Morales VH, Deas J and
Peralta-Zaragoza O: MicroRNA miR-16-1 regulates CCNE1 (cyclin E1)
gene expression in human cervical cancer cells. Int J Clin Exp Med.
8:15999–16006. 2015.PubMed/NCBI
|
|
10
|
Park S, Eom K, Kim J, Bang H, Wang HY, Ahn
S, Kim G, Jang H, Kim S, Lee D, et al: MiR-9, miR-21, and miR-155
as potential biomarkers for HPV positive and negative cervical
cancer. BMC Cancer. 17:6582017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lv KT, Liu Z, Feng J, Zhao W, Hao T, Ding
WY, Chu JP and Gao LJ: miR-22-3p regulates cell proliferation and
inhibits cell apoptosis through targeting the eIF4EBP3 gene in
human cervical cancer squamous carcinoma cells. Int J Med Sci.
15:142–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li C, Zheng X, Li W, Bai F, Lyu J and Meng
QH: Serum miR-486-5p as a diagnostic marker in cervical cancer:
With investigation of potential mechanisms. BMC Cancer. 18:612018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Z, Peng Z, Gu S, Zheng J, Feng D, Qin Q
and He J: Global analysis of miRNA-mRNA interaction network in
breast cancer with brain metastasis. Anticancer Res. 37:4455–4468.
2017.PubMed/NCBI
|
|
14
|
Mourad L, El-Ahwany E, Zoheiry M,
Abu-Taleb H, Hassan M, Ouf A, Rahim AA, Hassanien M and Zada S:
Expression analysis of liver-specific circulating microRNAs in
HCV-induced hepatocellular carcinoma in Egyptian patients. Cancer
Biol Ther. 19:400–406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lui WO, Pourmand N, Patterson BK and Fire
A: Patterns of known and novel small RNAs in human cervical cancer.
Cancer Res. 67:6031–6043. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ,
Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, et al: Altered microRNA
expression in cervical carcinomas. Clin Cancer Res. 14:2535–2542.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and tumor
suppressive microRNAs in cervical cancer is required for cancer
cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pereira PM, Marques JP, Soares AR, Carreto
L and Santos MAS: microRNA expression variability in human cervical
tissues. PLoS One. 5:e117802010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wilting SM, Snijders PJ, Verlaat W,
Jaspers A, van de Wiel MA, van Wieringen WN, Meijer GA, Kenter GG,
Yi Y, le Sage C, et al: Altered microRNA expression associated with
chromosomal changes contributes to cervical carcinogenesis.
Oncogene. 32:106–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Wang HK, Li Y, Hafner M, Banerjee
NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, et al: microRNAs
are biomarkers of oncogenic human papillomavirus infections. Proc
Natl Acad Sci USA. 111:4262–4267. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Calin GA, Cimmino A, Fabbri M, Ferracin M,
Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzón R, Aqeilan RI,
et al: miR-15a and miR-16-1 cluster functions in human leukemia.
Proc Natl Acad Sci USA. 105:5166–5171. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng ZM and Wang X: Regulation of
cellular miRNA expression by human papillomaviruses. Biochim
Biophys Acta. 1809:668–677. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Linsley PS, Schelter J, Burchard J,
Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond
CK, Dai H, et al: Transcripts targeted by the microRNA-16 family
cooperatively regulate cell cycle progression. Mol Cell Biol.
27:2240–2252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Williams JR: The declaration of Helsinki
and public health. Bull World Health Organ. 86:650–652. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benedet JL, Bender H, Jones H III, Ngan HY
and Pecorelli S: FIGO staging classifications and clinical practice
guidelines in the management of gynecologic cancers. FIGO Committee
on Gynecologic Oncology. Int J Gynaecol Obstet. 70:209–262. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Leonard DG, Michael KW and James BF:
Basics methods in Molecular Biology. McGraw-Hill Professional
(2nd). Appleton and Lange. (Norwalk, CT, USA). 1994.
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong L, Lee K, Russell I and Chen C:
Endogenous controls for real-time quantitation of miRNA Using
TaqMan® MicroRNA Assays. Application note
TaqMan® MicroRNA Assays. Applied Biosystems. 2007.
|
|
29
|
Palacio-Mejía LS, Lazcano-Ponce E,
Allen-Leigh B and Hernández-Avila M: Regional differences in breast
and cervical cancer mortality in Mexico between 1979–2006. Salud
Publica Mex. 51 (Suppl 2):S208–S219. 2009.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Secretaría de Salud: Información
estadística. Estadísticas de cáncer cervicouterino. 22–03.
2020
|
|
31
|
Illades-Aguiar B, Alarcón-Romero Ldel C,
Antonio-Véjar V, Zamudio-López N, Sales-Linares N, Flores-Alfaro E,
Fernández-Tilapa G, Vénces-Velázquez A, Muñoz-Valle JF and
Leyva-Vázquez MA: Prevalence and distribution of human
papillomavirus types in cervical cancer, squamous intraepithelial
lesions, and with no intraepithelial lesions in women from Southern
Mexico. Gynecol Oncol. 117:291–296. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ortiz-Ortiz J, Alarcón-Romero Ldel C,
Jiménez-López MA, Garzón-Barrientos VH, Calleja-Macías I,
Barrera-Saldaña HA, Leyva-Vázquez MA and Illades-Aguiar B:
Association of human papillomavirus 16 E6 variants with cervical
carcinoma and precursor lesions in women from Southern Mexico.
Virol J. 12:292015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen Y, Gong JM, Li YQ, Gong YM, Lei DM,
Cheng GM and Li XF: Epidemiology and genotype distribution of human
papillomavirus (HPV) in women of Henan Province, China. Clin Chim
Acta. 415:297–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lacobone AD, Bottari F, Radice D, Preti
EP, Franchi D, Vidal-Urbinati AM, Boveri S, Passerini R and Sandri
MT: Distribution of high-risk human papillomavirus genotypes and
multiple infections in preneoplastic and neoplastic cervical
lesions of unvaccinated women: A cross-sectional study. J Low Genit
Tract Dis. 23:259–264. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li M, Du X, Lu M, Zhang W, Sun Z, Li L, Ye
M, Fan W, Jiang S, Liu A, et al: Prevalence characteristics of
single and multiple HPV infections in women with cervical cancer
and precancerous lesions in Beijing, China. J Med Virol.
91:473–481. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chaturvedi AK, Katki HA, Hildesheim A,
Rodríguez AC, Quint W, Schiffman M, Van Doorn LJ, Porras C,
Wacholder S, González P, et al: Human papillomavirus infection with
multiple types: Pattern of coinfection and risk of cervical
disease. J Infect Dis. 203:910–920. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vinokurova S, Wentzensen N, Kraus I, Klaes
R, Driesch C, Melsheimer P, Kisseljov F, Dürst M, Schneider A and
von Knebel Doeberitz M: Type dependent integration frequency of
human papillomavirus genomes in cervical lesions. Cancer Res.
68:307–313. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Evans MF, Mount SL, Beatty BG, Cooper K
and Phil D: Biotinyl-tyramide-based in situ hybridization signal
patterns distinguish human papillomavirus type and grade of
cervical intraepithelial neoplasia. Mod Pathol. 15:1339–1347. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vega-Peña A, Illades-Aguiar B,
Flores-Alfaro E, López-Bayghen E, Leyva-Vázquez MA,
Castañeda-Saucedo E and Alarcón-Romero LC: Risk of progression of
early cervical lesions is associated with integration and
persistence of HPV-16 and expression of E6, Ki-67, and telomerase.
J Cytol. 30:226–232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tagle DK, Sotelo DH, Illades-Aguiar B,
Leyva-Vazquez MA, Alfaro EF, Coronel YC, Hernández Odel M and
Romero Ldel C: Expression of E6, p53 and p21 proteins and physical
state of HPV16 in cervical cytologies with and without low grade
lesions. Int J Clin Exp Med. 7:186–193. 2014.PubMed/NCBI
|
|
41
|
Torres-Rojas FI, Alarcón-Romero LCD,
Leyva-Vázquez MA, Ortiz-Ortiz J, Mendoza-Catalán MA,
Hernández-Sotelo D, Del Moral-Hernández O, Rodríguez-Ruiz HA,
Leyva-Illades D, Flores-Alfaro E and Illades-Aguiar B: Methylation
of the L1 gene and integration of human papillomavirus 16 and 18 in
cervical carcinoma and premalignant lesions. Oncol Lett.
15:2278–2286. 2018.PubMed/NCBI
|
|
42
|
Zubillaga-Guerrero MI, Illades-Aguiar B,
Leyva-Vazquez MA, Flores-Alfaro E, Castañeda-Saucedo E, Muñoz-Valle
JF and Alarcón-Romero LC: The integration of HR-HPV increases e
expression of cyclins A and E in cytologies with and without
low-grade lesions. J Cytol. 30:1–7. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cricca M, Venturoli S, Leo E, Costa S,
Musiani M and Zerbini M: Molecular analysis of HPV 16 E6I/E6II
spliced mRNAs and correlation with the viral physical state and the
grade of the cervical lesion. J Med Virol. 81:1276–1282. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Saunier M, Monnier-Benoit S, Mauny F,
Dalstein V, Briolat J, Riethmuller D, Kantelip B, Schwarz E, Mougin
C and Prétet JL: Analysis of human papillomavirus type 16 (HPV16)
DNA load and physical state for identification of HPV16-infected
women with high-grade lesions or cervical carcinoma. J Clin
Microbiol. 46:3678–3685. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ibragimova M, Tsyganov M, Shpileva O,
Churuksaeva O, Bychkov V, Kolomiets L and Litviakov N: HPV status
and its genomic integration affect survival of patients with
cervical cancer. Neoplasma. 65:441–448. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim J, Kim K, Jeon DS, Lee CH, Roh JW, Kim
Y and Park SY: Type-specific viral load and physical state of HPV
type 16, 18, and 58 as diagnostic biomarkers for high-grade
squamous intraepitelial lesions or cervical cancer. Cancer Res
Treat. 52:396–405. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gallo G, Bibbo M, Bagella L, Zamparelli A,
Sanseverino F, Giovagnoli MR, Vecchione A and Giordano A: Study of
viral integration of HPV-16 in young patients with LSIL. J Clin
Pathol. 56:532–536. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kulmala SMA, Syrjänen SM, Gyllensten UB,
Shabalova IP, Petrovichev N, Tosi P, Syrjänen KJ and Johansson BC:
Early integration of high copy HPV16 detectable in women with
normal and low grade cervical cytology and histology. J Clin
Pathol. 59:513–517. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Peitsaro P, Johansson B and Syrjänen S:
Integrated human papillomavirus type 16 is frequently found in
cervical cancer precursors as demonstrated by a novel quantitative
real-time PCR technique. J Clin Microbiol. 40:886–891. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chaiwongkot A, Vinokurova S, Pientong C,
Ekalaksananan T, Kongyingyoes B, Kleebkaow P, Chumworathayi B,
Patarapadungkit N, Reuschenbach M and von Knebel Doeberitz M:
Differential methylation of E2 binding sites in episomal and
integrated HPV 16 genomes in preinvasive and invasive cervical
lesions. Int J Cancer. 132:2087–2094. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Schickel R, Boyerinas B, Park SM and Peter
ME: MicroRNAs: Key players in e immune system, differentiation,
tumorigenesis and cell death. Oncogene. 27:5959–5974. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bueno MJ and Malumbres M: MicroRNAs and
the cell cycle. Biochim Biophys Acta. 1812:592–601. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ofir M, Hacohen D and Ginsberg D: MiR-15
and miR-16 are direct transcriptional targets of E2F1 at limit
E2F-induced proliferation by targeting cyclin E. Mol Cancer Res.
9:440–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gonzalez SL, Stremlau M, Basile JR and
Münger K: Degradation of the retinoblastoma tumor suppressor by the
human papillomavirus type 16 E7 oncoprotein is important for
functional inactivation and is separable from proteasomal
degradation of E7. J Virol. 75:7583–7591. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nguyen CL and Münger K: Direct association
of the HPV16 E7 oncoprotein with cyclin A/CDK2 and cyclin E/CDK2
complexes. Virology. 380:21–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ohtani K, DeGregori J and Nevins JR:
Regulation of the cyclin E gene by transcription factor E2F1. Proc
Natl Acad Sci USA. 92:12146–12150. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang F, Fu XD, Zhou Y and Zhang Y:
Down-regulation of the cyclin E1 oncogene expression by
microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB
Rep. 42:725–730. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pett M and Coleman N: Integration of
high-risk human papillomavirus: A key event in cervical
carcinogenesis? J Pathol. 212:356–367. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kalantari M, Blennow E, Hagmar B and
Johansson B: Physical state of HPV16 and chromosomal mapping of the
integrated form in cervical carcinomas. Diagn Mol Pathol. 10:46–54.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Thorland EC, Myers SL, Gostout BS and
Smith DI: Common fragile sites are preferential targets for HPV16
integrations in cervical tumors. Oncogene. 22:1225–1237. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Popescu NC, DiPaolo JA and Amsbaugh SC:
Integration sites of human papillomavirus 18 DNA sequences on HeLa
cell chromosomes. Cytogenet Cell Genet. 44:58–62. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chang TC, Yu D, Lee YS, Wentzel EA, Arking
DE, West KM, Dang CV, Thomas-Tikhonenko A and Mendell JT:
Widespread microRNA repression by Myc contributes to tumorigenesis.
Nat Genet. 40:43–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lerner M, Harada M, Loven J, Castro J,
Davis Z, Oscier D, Henriksson M, Sangfelt O, Grander D and Corcoran
MM: DLEU2, frequently deleted in malignancy, functions as a
critical host gene of the cell cycle inhibitory microRNAs miR-15a
and miR-16-1. Exp Cell Res. 315:2941–2952. 2009. View Article : Google Scholar : PubMed/NCBI
|