Introduction
Breast carcinoma is a primary cause of
cancer-associated mortality in women aged 20–59 years. Statistical
studies have demonstrated that the incidence of breast carcinoma is
increasing annually and accounts for 30% of new cancer diagnoses in
women alone in USA in 2019 (1). The
therapeutic modalities that are currently applied are selected
primarily according to the most extensively studied biomarkers:
Estrogen receptor, progesterone receptor and human epidermal growth
factor receptor 2 (2). However,
previous studies have demonstrated that the occurrence,
tumorigenesis and metastasis of breast cancer are controlled by
complex signaling networks (3–5). Thus, a
complete understanding of the molecular mechanism of breast
carcinogenesis is required to eliminate obstacles in the early
detection and treatment of breast cancer.
Aberrant protein phosphorylation is one of most
typical characteristics of tumor cells. Protein tyrosine
phosphatases (PTPs) are critical enzymes that modulate the
phosphorylation status of intracellular signaling molecules
(6–8). It is well-established that PTPs
negatively or positively regulate cancer-associated signaling
pathways in breast cancer (8–10). PTP1B
overexpression promotes proliferation and migration by regulating
the phosphorylation of steroid receptor coactivator (11). PTPδ has been predicted to be an
enhancer of tumorigenicity and its high expression has been tested
in clinical breast cancer samples (12). Furthermore, PTP receptor type (PTPR)
K potentially serves a negative role in breast cancer and a low
PTPRK transcript level is associated with poor prognosis and low
survival rates (13). Furthermore,
tumor function inhibition via PTPN12 expression alteration
suppresses breast cancer development and metastasis in vivo
(14). Additionally, treatment of
MCF-7 cells with c-Jun N-terminal kinase or extracellular
signal-regulated kinase inhibitors partially rescue the effects of
PTPRM knockdown on cell migration, indicating that PTPRM inhibits
tumor metastasis by decreasing the activity of oncogenic protein
tyrosine kinases (13,15).
Similar to other PTPs, PTPRA is closely associated
with the tumorigenic phenotype of breast cancer via its control of
the balance between PTKs and PTPs (16). A significant increase in PTPRA the
transcription and translation levels has been confirmed in the
majority of primary breast cancer types (16–18).
Nonetheless, the role of PTPRA in breast cancer remains
controversial. Ardini et al demonstrated that PTPTA is an
inhibitor of breast cancer cell proliferation and significantly
delays cancer cell migration and invasion in vivo and in
vitro (10), while other in
vivo studies indicate that PTPRA enhances malignant activities,
such as migration and invasion of tumor cells (16,17).
Mechanistically, PTPs, including PTPRA, are
primarily physiological upstream activators of oncogenic SRC that
act by dephosphorylating key signaling factors (19). Certain PTPs also directly interact
with cell adhesion molecules, such as E-cadherin and β-catenin, to
regulate cancer cell transformation (6). Furthermore, PTPRA has been reported to
respond to different stimuli, such as insulin-like growth factor
(IGF)-1, and activate IGF-1-medidated downstream signaling pathways
that are critical in tumorigenesis and metastasis (20). Therefore, the present study concluded
that further work concerning the precise molecular and cellular
mechanisms is still essential to elucidate the role of PTPRA in
breast cancer.
The present study clarified the oncogenic role of
PTPRA and its underlying mechanism in breast cancer using loss and
gain of function analyses, demonstrating the effect of PTPRA on the
proliferation, colony formation and migration of MCF-7 cells.
Furthermore, a luciferase reporter gene assay was used to screen
for the possible PTPRA-mediated signaling pathway. Overall, the
present study may provide new insight for breast cancer diagnosis
and therapy.
Materials and methods
Reagents
Human recombinant TNF-α was obtained from T&L
Biological Technology. Transcription factor E2F (E2F), p53, NF-κB,
eukaryotic initiation factor 2 α kinase 1 (EIK1), transforming
growth factor (TGF)-β), JNK, myc proto-oncogene protein (c-MYC),
PI3K/AKT, Wnt, protein giant-lens (Gil), Notch, STAT3 and ETS
transcription factor (Elk1) luciferase reporter plasmids and the
pHAGE puro vector were gifted by the School of life sciences, Wuhan
University, Wuhan, China. These luciferase reporter constructs
contain the DNA-binding motifs of transcription factor in these
signaling pathways.
Analyzed datasets
PTPRA mRNA data in patients with the breast cancer
gene (BRCA) were analyzed using the Gene Expression Profiling
Interactive Analysis database (gepia.cancer-pku.cn) with Kaplan
Meier analysis and log-rank tests as previously described (15). The expression data of 1,085 tumors
and 291 adjacent normal tissues were obtained from The Cancer
Genome Atlas (TCGA; www.tcga.org).
Group cut-off was at 50% and based on this, patients were divided
into the low PTPRA or the high PTPRA group. The overall survival
was calculated in low and high PTPRA groups for 250 months
according to previous reports (21–23)
using the GEPIA database to display the relevance of PTPRA mRNA in
patients with breast cancer.
Cell culture
MCF-7 and HEK293T cells were purchased from the
American Type Culture Collection. MCF-7 cells were cultured in
DMEM/nutrient mixture F12 (1:1) (Gibco; Thermo Fisher Scientific,
Inc.) medium supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin. HEK293T cells were cultured in DMEM medium
supplemented with 10% FBS. All cells were cultured in a 5%
CO2 chamber at 37°C.
Construction of the FLAG-PTPRA-pGEM-T
plasmid and transfection
Total RNA was extracted out using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) from MCF-7
cells and reverse transcribed into cDNA using a high capacity cDNA
Reverse Transcription kit (Thermo Fisher Scientific, Inc.) at 4°C
according to the manufacturer's protocols. cDNA encoding PTPRA was
amplified with PCR with the following primers: Forward:
5′-GATCCGCCACCAUGGATGGATTCCTGGTTCATTCTTGTTC-3′ and reverse:
5′-TCGAGCTTGAAGTTGGCATAATCTG-3′. The PTPRA fragment was
gel-purified via BamH I and EcoRI and rejoined to BamH I-EcoRI
digested pHAGE puro plasmid. A total of 10 µg Flag-PTPRA expression
plasmid were introduced to MCF-7 cells for exogenous overexpression
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. As
the control, the empty pHAGE puro plasmid was introduced to
MCF-7.
Construction of the PTPRA-deficient
cell line
PTPRA knockout plasmids were constructed using the
clustered regularly interspaced short palindromic repeat (CRISPR)
knockout method (24,25). Briefly, the gRNA targeting sequence
was obtained and inserted into CRISPR/Cas9 Plasmid using the
Precision X Multiplex gRNA kit (SBI http://systembio.com/products/crispr-cas9-systems/)
according to the manufacturer's protocol. The non-specific binding
targets of the CRISPR/Cas9 plasmid served as the negative control.
In total, 2.5×103 HEK293T cells per well at 80–90%
confluence were transfected with 400 ng CRISPR/Cas9-PTPRA plasmid
or the control plasmid for 2 days. Next, MCF-7 cells were
transfected with 10 µl lentiviral particles for 72 h at 37°C. The
infected MCF-7 cells were screened in the presence of 1 µg/ml
puromycin for 7 days. The puromycin-resistant cells were subjected
to further confirmation by agarose gel electrophoresis and western
blotting. The cells carrying the non-specific binding targets
CRISPR/Cas9 plasmid were used as a control.
Western blot analysis
Collected cells were lysed with RIPA buffer
(BioVision, Inc.) and the supernatant was extracted for SDS-PAGE
analysis. After quantification using the Bradford assay, protein
lysates (10 µg/lane) were separated by 8% SDS-PAGE, transferred to
PVDF membranes and blocked with TBS containing 5% non-fat milk at
room temperature for 1 h. The membranes were probed with mouse
monoclonal anti-Flag antibody (1:3,000; cat. no. 81069; ProteinTech
Group, Inc.) or PTPRA antibody (1:2,000; cat. no. 13079–1-AP;
ProteinTech Group, Inc.), β-actin antibody (1:2,000; cat. no.
Ag27042; ProteinTech Group, Inc.) for 1–2 h at room temperature.
The blots were then incubated and reprobed with horseradish
peroxidase-conjugated secondary antibody (1:2,000; cat. no.
SA00001-1; ProteinTech Group, Inc.) for 1 h at room temperature.
Band signals were visualized using an ECL system (GE Healthcare).
The following antibodies were diluted in TBS and used for
immunoblotting: Anti-FLAG, anti-β-actin and anti-PTPRA.
Colony formation, cell proliferation
and Transwell cell migration assay
Overexpressed-PTPRA MCF-7 cells or two
PTPRA-knockout independent clones (PTPRA−/−1# and
PTPRA−/−2#) and their corresponding control MCF-7 cells
were harvested and prepared in single-cell suspension
(1×104 cells/ml) for the subsequent cell assays.
A colony formation experiment was conducted to
estimate the clonogenic activity of breast cancer cells. Prepared
cells were seeded in 6-well plates and cultured in an incubator at
37°C. Following 8-day culture, the colonies were fixed using 100%
methanol for 15 min and stained using 0.5% crystal violet for 20
min at room temperature before quantification under an inverted
light microscope (ECLIPSE TE2000-S; Nikon) at 200× magnification.
The indicated colony formation units were recorded in 5 random
fields for every replicate and plotted.
In the Cell Counting Kit (CCK)-8 assay, the
aforementioned cells were cultured in 96-well plates
(1×103 cells/well). Following incubation for 1, 3, 5 and
7 days, diluted CCK-8 solution (Dojindo Molecular Technologies,
Inc.) was supplemented into each well according to the
manufacturer's manual. After incubation for another 1–2 h at 37°C,
cell proliferation was evaluated spectrophotometrically at a
wavelength of 450 nm with an Automated Enzyme Immunoassay Analyzer
(Shanghai Dongcao Biotechnology Co., Ltd, Tosoh Corporation;
http://www.biomart.cn/infosupply/57204830.htm).
For Transwell migration assay, Transwell inserts
(Corning Inc.) with porous polycarbonate membranes were firstly
placed in 24-well plates. The lower compartment was filled with 2.6
ml DMEM containing 40% FBS. MCF-7 cells (1×104) were
added to the upper compartment and cultured in Transwell plates at
37°C for 2 days. Cell debris that did not migrate through the
membrane were removed with a cotton swab. The migratory cells were
fixed with 5% glutaraldehyde for 10 min and stained 1% crystal
violet in 2% ethanol for 20 min before images were captured and
quantification under an inverted light microscope (ECLIPSE
TE2000-S; Nikon). All comparison experiments were performed in
triplicate.
Luciferase reporter gene assay
In order to screen the target signaling pathways of
PTPRA, a series of luciferase reporter gene assays were performed
to determine the effect of PTPRA on the transcriptional activity of
several documented tumor signaling markers: E2F, p53, NF-κB, EIK1,
TGF-β, JnK, c-MYC, Wnt, Gil, Notch, STAT3 and Elk1.
A total of 1×104 HEK293T cells were
cultured in 24-well plates overnight at 37°C and transfected with
the aid of Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.). Each transfection contained the indicated
luciferase reporter vector (200 ng/well) and prl-tk (10 ng/well)
empty control plasmid or PTPRA expression plasmid or control
vector. Following 36 h incubation at 37°C, the released cells were
treated with trypsin and luciferase activity was measured using a
Dual-Glo Luciferase Assay kit (Promega Corporation), according to
the manufacturer's protocol.
Furthermore, HEK293T cells were transfected with
pNFKB-luc and PTPRA plasmids at various diluted concentrations
(100, 200 and 400 ng/well) to further confirm the effect of PTPRA
on NF-κB transcriptional activity. Luciferase activity was
normalized using the Renilla luciferase activity.
RNA isolation and reverse
transcription-quantitative PCR
Total RNA from cells was extracted from lysed cells
using TRIzol® (Thermo Fisher Scientific, Inc.). Reverse
transcription was performed using oligo dT primers using RT kit
(Invitrogen) according to the manufacturer's protocol. mRNA of IKBα
(one of classic downstream molecules of the NF-κB signaling pathway
(26) in and two PTPRA deficient
MCF-7 cells(PTPRA−/−1# and PTPRA-/−2#) cells
and their corresponding control MCF-7 cells (PTPRA+/+)
was assessed upon TNF-α stimulation or not. The relative expression
was quantified by the 2−ΔΔCq method (27).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software (version 6.0; GraphPad Software). Log-rank test was
carried out to calculate significance of PTPRA in predicting
overall survival of breast cancer patients using GEPIA according to
the creator of this website (28).
For continuous variables, measured data are
presented as the mean ± SEM. Unpaired two-tailed Student's t-test
was used to compare differences between two groups. One-way ANOVA
was used to evaluate the statistical significance among multiple
groups followed by Tukey's post hoc corrective test. P<0.05 was
considered to indicate a statistically significant difference. All
experiments except the analysis from GEPIA were performed at least
three times.
Results
PTPRA expression and prognostic value
in breast cancer
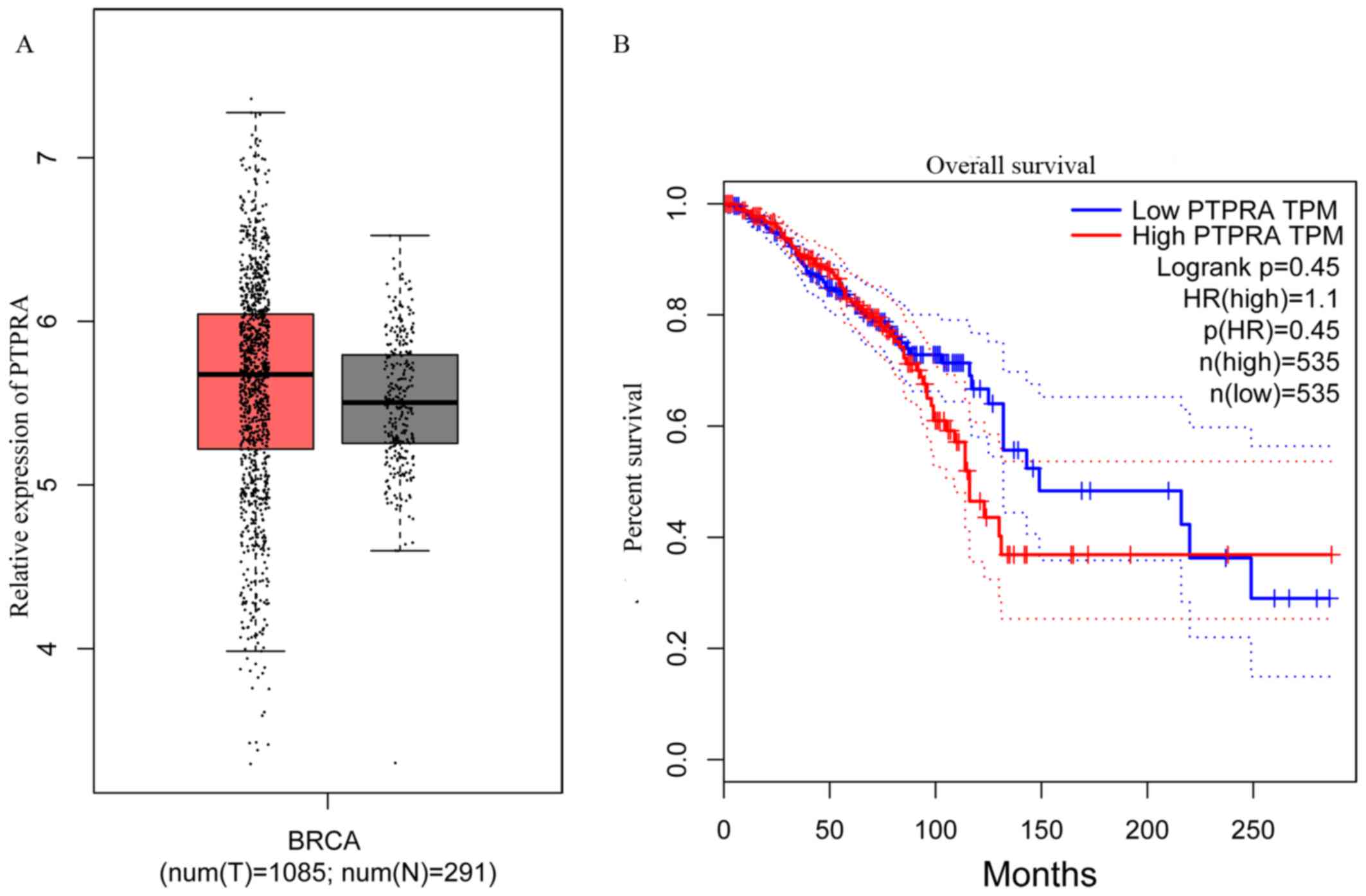
The BRCA database from GEPIA was analyzed and used
to compare the expression of PTPRA in breast cancer and normal
tissues in order to determine the prognostic value of PTPRA in
breast cancer. PTPRA expression was significantly increased in
breast cancer tissues compared with normal tissues (Fig. 1A). As presented in Fig. 1B, patients with high PTPRA
demonstrated worse clinical outcomes compared with patients with
PTPRA, while there was no significant difference between groups
(P=0.45). These results indicated that PTPRA expression level may
serve a putative role in breast cancer malignancy.
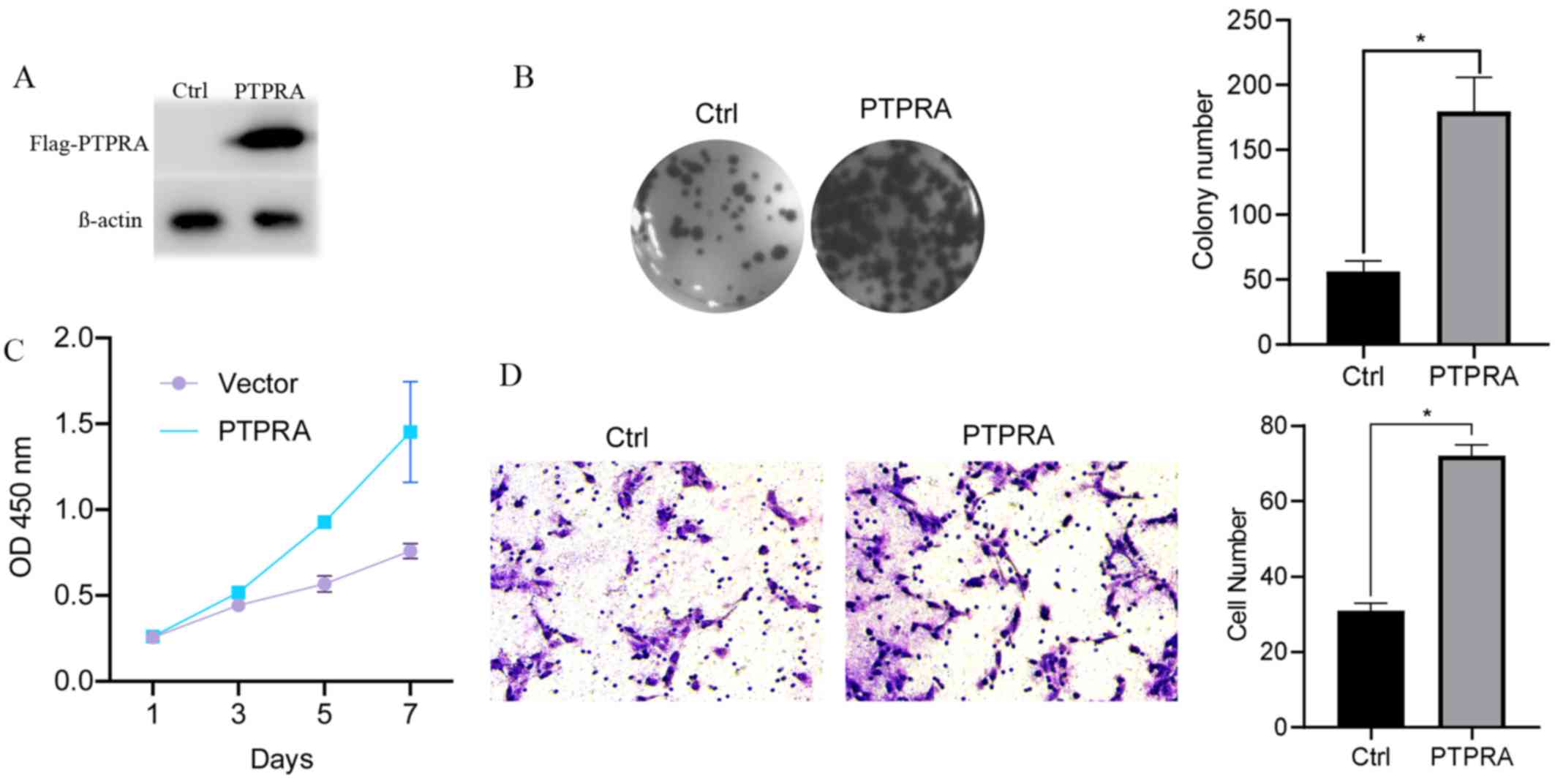
PTPRA overexpression promotes
proliferation, colony formation and migration of MCF-7 cells
A vector containing Flag-PTPRA was constructed and
transfected into MCF-7 cells using Lipofectamine 2000 to verify the
specific biological function of PTPRA. PTPRA overexpression was
confirmed using anti-Flag antibodies via western blotting (Fig. 2A). Furthermore, PTPRA overexpression
significantly promoted the colony formation ability of MCF cells
(Fig. 2B). A growth curve analysis
demonstrated that PTPRA overexpression dramatically enhanced
proliferation compared with the control cells (Fig. 2C). Additionally, the Transwell assay
demonstrated that the number of migratory cells in the
overexpression group was significantly increased compared with the
control group (Fig. 2D). These
results indicated that PTPRA increased the tumorigenic properties
of MCF7 cells in vivo.
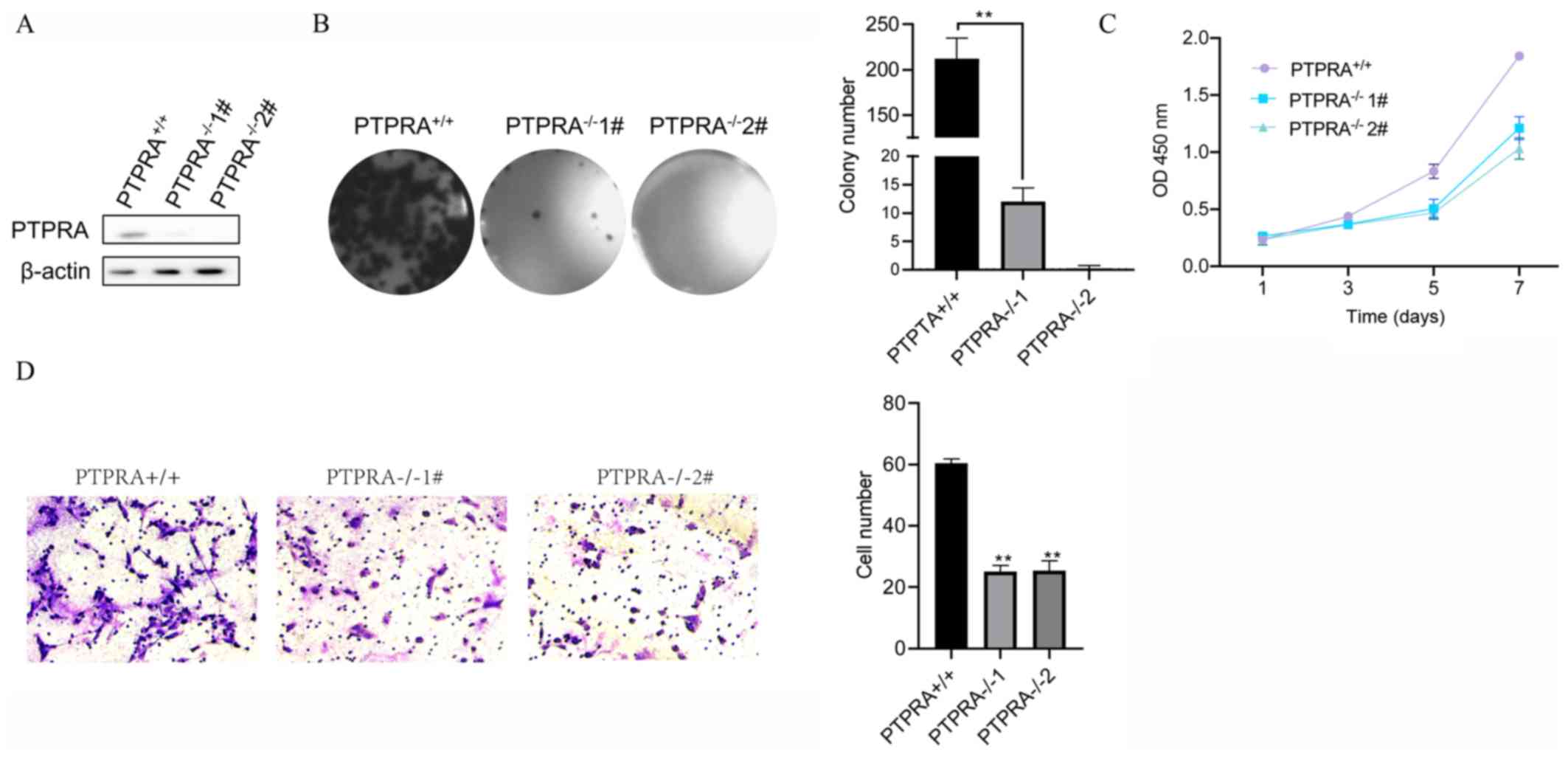
Knockout of PTPRA suppresses the
proliferation and migration ability of MCF-7 cells in vitro
PTPRA was further depleted in MCF-7 cells using
CRISPR to confirm the effect of PTPRA on cell behaviors. Western
blotting revealed that PTPRA was almost completely silenced
(Fig. 3A). Furthermore, the colony
formation ability and proliferation of PTPRA-deficient MCF-7 cells
were significantly decreased compared with that of control MCF-7
cells (PTPRA+/+) (Fig. 3B and
C, respectively). Consistently, MCF7 cells deficient in PTPRA
had fewer migratory cells than the control cells
(PTPRA+/+) (Fig. 3D).
These results suggested that PTPRA deficiency decreased cell colony
formation ability and inhibited tumor cell migration ability.
Signaling pathway is regulated by
PTPRA in breast cancer
Oncogenesis, the development and prognosis of
tumors, involves complicated pathway networks that are implicated
in numerous signal pathways, such as microtubule-associated protein
kinase, NF-κB and signal transducer and activator of transcription
factor 3 (29). The results of
luciferase reporter gene assays demonstrated that NF-κB
transcriptional activity was markedly increased compared with
controls (Fig. 4A). The luciferase
reporter gene assay did not demonstrate any obvious alterations in
the expression of the other signaling molecules. NF-κB
transcriptional activity was also demonstrated to be increased in a
dose-dependent manner (Fig. 4B).
These results indicated that PTPRA overexpression in HEK293T cells
stimulated NF-κB transcriptional activity.
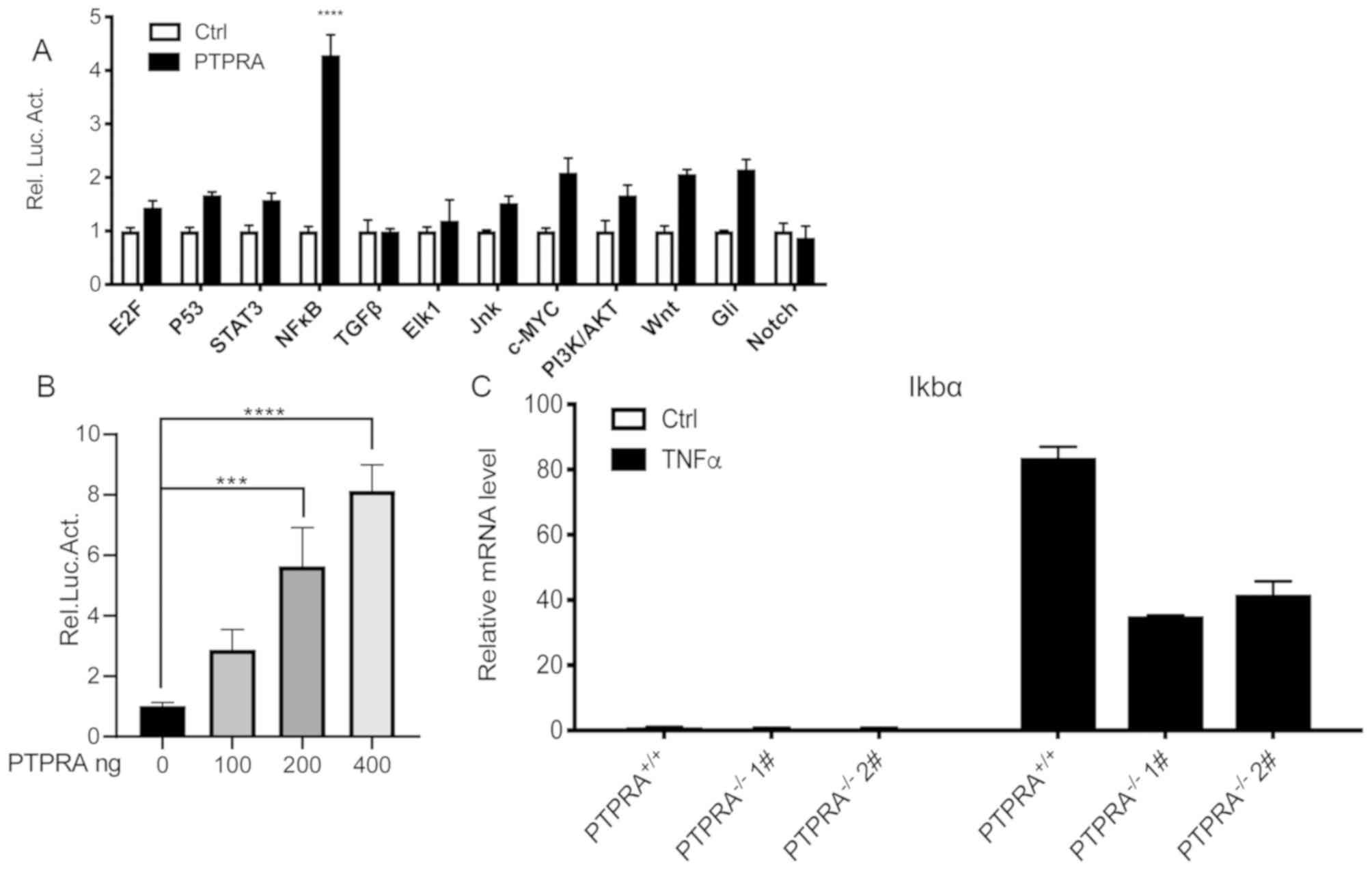 | Figure 4.Overexpression of PTPRA contributes
to an enhanced inflammatory response in MCF-7 breast cancer and
HEK293T cells. (A) Screening of human cancer pathways demonstrated
that PTPRA markedly enhanced NF-κB transcriptional activity in
HEK293T cells. (B) NF-κB activity was increased in a
PTPRA-dependent manner as detected by luciferase assays in MCF-7
cells. (C) Reverse transcription-quantitative PCR analysis was
performed to examine the expression level of NF-κB-targeted gene
followed by TNF-α stimulation. ***P<0.001, ****P<0.0001 vs.
PTPRA+/+ MCF-7 cells. PTPRA, protein tyrosine
phosphatase receptor type A; NF-κB, nuclear factor
κ-light-chain-enhancer of activated B cells; TNF-α, tumor necrosis
factor α; Rel. Luc. Act., relative luciferase activity; STAT3,
signal transducer and activator of transcription 3; TGF-β,
transforming growth factor β; Jnk, c-Jun N-terminal kinase;
PI3K/AKT, phosphoinositide 3-kinase/protein kinase B; E2F,
transcription factor E2F; c-MYC, myc proto-oncogene protein; Gil,
protein giant-lens. |
TNFα-activates NF-κB in MCF-7
cells
RT-qPCR was utilized to quantify one of the classic
downstream molecules of the NF-κB signaling pathway, IKBα in MCF-7
cells. No transcription of the IKBα gene was detected in
PTPRA−/− and PTPRA+/+ MCF-7 cells without
TNF-α treatment (Fig. 4C). However,
the TNF-α stimulus changed these outcomes. IKBα gene transcription
in PTPRA+/+ MCF-7 cells was increased compared with that
in PTPRA-deficient MCF-7 cells. These outcomes indicated that
TNF-α-mediated PTPRA stimulated the activation of NF-κB and
promoted the tumor phenotype of breast cancer cells.
Discussion
PTPRA is closely associated with neoplastic
transformation through its effects on proliferation and migration
in breast cancer cells (30).
However, the oncogenic characteristics of PTPRA remain elusive
in vitro. The present study demonstrated the significance of
PTPRA on the migration and metastatic potential of MCF-7 breast
cancer cells. Additionally, to the best of our knowledge, these
results are the first to reveal that PTPRA may act as a
proto-oncogene in the TNF-α-dependent inflammatory responses by
directly binding to NF-κB in vitro.
The results of the present study demonstrated that
PTPRA expression was significantly increased in the tumor tissues
of patients with breast cancer compared with normal tissues via
analysis of TCGA data from GEPIA. The GEPIA dataset also suggested
that patients with breast cancer exhibiting high expression levels
of PTPRA and slightly worse clinical outcomes when compared with
low-PTPRA patients, though the difference was not statistically
significant. These results revealed that PTPRA acts as an enhancer
of tumorigenicity and increases the malignancy of breast cancer
types. In the present study, clonogenic and migratory behaviors in
PTPRA-overexpressing or PTPRA-deficient breast cancer cell lines
were investigated. The results were consistent with a recent study
that demonstrated that PTPRA accumulation in MCF-7 cells
facilitates focal adhesion formation and cell migration in
vitro (17), indicating that
PTPRA may be a pro-migratory factor. Furthermore, a retrospective
cohort analysis demonstrated PTPRA overexpression in squamous cell
lung cancer (19). PTPRA
overexpression promotes lung cancer cell proliferation and is
associated with worse overall survival, suggesting that PTPRA
overexpression may be an effective predictive or prognostic marker
for squamous cell lung cancer (19).
Protein phosphatases are critical modulators of cell
signaling. Their functional roles in aberrant signaling are
critical for tumor pathogenesis. PTPRA, one of the classic PTPs,
executes its signaling functions primarily through directly
dephosphorylating key signaling molecules or activating the
oncogenic focal adhesion kinase-Src complex in breast cancer cells
(6,31). Furthermore, a previous study using an
animal model of pulmonary fibrosis has revealed that PTPRA directly
interacts with mothers against decapentaplegic homolog (Smad)
protein and increases Smad transcriptional activity in response to
TGF-β stimuli, indicating that PTPRA has a profound effect on the
genesis of inflammatory pulmonary fibrosis (32), which lead to the present study
investigating the detailed information regarding the oncogenic
action of PTPRA. The present study utilized a series of luciferase
pathway screening assays and the results revealed that alterations
in the inflammatory NF-κB signaling pathway were largest compared
with those of other oncogenic signaling pathways. Furthermore, the
NF-κB inflammation signaling pathway was activated by TNF-α
stimulus, an extensively used approach to assess the mediation of
target protein to certain signaling pathways, such as PI3K/AKT
signaling (33–37), in order to further validate the
regulatory function of PTPRA. These results indicated an oncogenic
role of PTPRA in the TNF-α-induced inflammatory pathway, which has
also been linked to the inflammogenesis of breast cancer (38). Ghandadi et al (39) reported similar results by
demonstrating that the treatment of MCF-7 cells with TNF-α
triggered activation of NF-κB, ultimately leading to
receptor-interacting serine/threonine-protein kinase 1
ubiquitination and non-apoptotic death.
Activation of NF-κB is a crucial event which
supports chronic inflammation and cancer progression (40). Previous studies have demonstrated
that the NF-κB pathway may exert a number of roles in different
settings or cellular contexts. In enterocytes, NF-κB has been
implicated in tumorigenesis; however, it has not been implicated in
cancer progression or growth (41).
These results were supported by Ardini et al (10), indicating that PTPRA is positively
correlated with low tumor grade. The present study also supports
the hypothesis that PTPRA is a downstream target of TNF-α and
triggers the genesis of breast tumors (42). In the present study, TNF-α stimuli
contributed to a significant PTPRA upregulation in
PTPRA+/+ MCF-7 cells compared with PTPRA−/−
MCF-7 cells, indicating that crosstalk between PTPRA and TNF-α
activates downstream signaling (43). Hence, it is essential to determine to
what extent PTPRA influences breast cancer by TNF-α-induced NF-κB
activation.
To date, there has been compelling evidence that
PTPRA is responsible for Src tyrosine 530 dephosphorylation, which
leads to cellular transformation (19,44,45). For
instance, Lai et al (44)
confirmed that PTPRA overexpression activated pp60c-src kinase
in vitro and in vivo, which contributed to cellular
transformation and induced lung tumorigenesis in vivo. In
the present study, the results demonstrated that PTPRA directly
bound to an NF-κB promoter and enhanced its transcriptional
activity, promoting the clonogenic and migratory tumor phenotype of
breast cancer. We also noticed that PTPRA can increased the
expression level of IKBα:one of classic downstream molecules of the
NF-κB signaling pathway. Whether c-Src is one of the signaling
checkpoints in this signaling pathway is yet to be determined. Li
et al (46) reported that
TNF-α triggered two parallel, but independent, signaling pathways
(Src and TNF receptor 1/NF-κB) to regulate neuroses in the mouse
fibrosarcoma L929 cell line. Another previous study supported the
notion that a cloned osteoclastic protein-tyrosine phosphatase
(PPT-oc) enhances osteoclast activity partially via the
PPT-oc/c-Src/NF-κB signaling pathway (47). These diverse signaling networks may
explain the dual roles that PTPRA serves in breast cancer. In
future studies, whether c-Src is a crucial participant in these
regulatory mechanisms will be investigated.
There are limitations in the current study that need
to be noted. The main limitation is that a single breast cancer
cell line MCF-7 was used. Future studies should focus on more
breast cancer cell lines, which will further elucidate the
underlying mechanisms of PTPRA in breast cancer. Another limitation
is that only the level of IKBα mRNA in MCF-7 cell lines was
assessed following the screening of underlying oncogenic signaling
pathways in HEK293T cells, further systematic approaches, including
chromatin immunoprecipitation, mutational experiments and in
vivo assays, should be performed to further validate results.
Additionally, vectors that overexpressed multiple genes were
generated in our lab, therefore flag antibody was used to screen
the proposed-gene overexpressing cells. But only PTPRA
overexpression was performed in this study.
In conclusion, the present study demonstrated that
PTPRA is upregulated in patients with breast cancer. The oncogenic
gene PTPRA is mediated by TNF-α and may partially activate
the NF-κB inflammation signaling pathway in MCF-7 breast cancer
cells. These results further elucidate the function of PTPRA in
breast cancer and indicate that PTPRA may be an effective
diagnostic curative target for breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was funded by Shantou Scientific
Research Project (grant no. 181203164010454).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article and GEPIA repository
(gepia.cancer-pku.cn).
Authors' contributions
FZ designed the study. CL contributed to analysis
and manuscript preparation. SX performed data analysis and wrote
the manuscript. XH provided assistance for data acquisition, data
analysis and statistical analysis and constitutive analysis. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lin SX, Chen J, Mazumdar M, Poirier D,
Wang C, Azzi A and Zhou M: Molecular therapy of breast cancer:
Progress and future directions. Nat Rev Endocrinol. 6:485–493.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ruvolo PP: Role of protein phosphatases in
the cancer microenvironment. Biochim Biophys Acta Mol Cell Res.
1866:144–152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Merino Bonilla JA, Torres Tabanera M and
Ros Mendoza LH: Breast cancer in the 21st century: From early
detection to new therapies. Radiologia. 59:368–379. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ahmed M, Carrascosa LG, Ibn Sina AA,
Zarate EM, Korbie D, Ru KL, Shiddiky MJA, Mainwaring P and Trau M:
Detection of aberrant protein phosphorylation in cancer using
direct gold-protein affinity interactions. Biosens Bioelectron.
91:8–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bollu LR, Mazumdar A, Savage MI and Brown
PH: Molecular pathways: Targeting protein tyrosine phosphatases in
cancer. Clin Cancer Res. 23:2136–2142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim M, Morales LD, Jang IS, Cho YY and Kim
DJ: Protein tyrosine phosphatases as potential regulators of STAT3
signaling. Int J Mol Sci. 19:27082018. View Article : Google Scholar
|
|
8
|
Nunes-Xavier CE, Mingo J, López JI and
Pulido R: The role of protein tyrosine phosphatases in prostate
cancer biology. Biochim Biophys Acta Mol Cell Res. 1866:102–113.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yao Z, Darowski K, St-Denis N, Wong V,
Offensperger F, Villedieu A, Amin S, Malty R, Aoki H, Guo H, et al:
A global analysis of the receptor tyrosine kinase-protein
phosphatase interactome. Mol Cell. 65:347–360. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ardini E, Agresti R, Tagliabue E, Greco M,
Aiello P, Yang LT, Ménard S and Sap J: Expression of protein
tyrosine phosphatase alpha (RPTPalpha) in human breast cancer
correlates with low tumor grade, and inhibits tumor cell growth in
vitro and in vivo. Oncogene. 19:4979–4987. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mei W, Wang K, Huang J and Zheng X: Cell
transformation by PTP1B truncated mutants found in human colon and
thyroid tumors. PLoS One. 11:e01665382016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chaudhary F, Lucito R and Tonks NK:
Missing-in-metastasis regulates cell motility and invasion via
PTPdelta-mediated changes in SRC activity. Biochem J. 465:89–101.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun PH, Ye L, Mason MD and Jiang WG:
Protein tyrosine phosphatase kappa (PTPRK) is a negative regulator
of adhesion and invasion of breast cancer cells, and associates
with poor prognosis of breast cancer. J Cancer Res Clin Oncol.
139:1129–1139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Davidson D, Martins Souza C, Zhong
MC, Wu N, Park M, Muller WJ and Veillette A: Loss of PTPN12
stimulates progression of ErbB2-dependent breast cancer by
enhancing cell survival, migration, and epithelial-to-mesenchymal
transition. Mol Cell Biol. 35:4069–4082. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun PH, Ye L, Mason MD and Jiang WG:
Protein tyrosine phosphatase µ (PTP µ or PTPRM), a negative
regulator of proliferation and invasion of breast cancer cells, is
associated with disease prognosis. PLoS One. 7:e501832012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meyer DS, Aceto N, Sausgruber N, Brinkhaus
H, Müller U, Pallen CJ and Bentires-Alj M: Tyrosine phosphatase
PTPα contributes to HER2-evoked breast tumor initiation and
maintenance. Oncogene. 33:398–402. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao J, Gao Y, Yang F, Wang C, Xu Y, Chang
R, Zha X and Wang L: β1,6 GlcNAc branches-modified protein tyrosine
phosphatase alpha enhances its stability and promotes focal
adhesion formation in MCF-7 cells. Biochem Biophys Res Commun.
482:1455–1461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Boivin B, Chaudhary F, Dickinson BC, Haque
A, Pero SC, Chang CJ and Tonks NK: Receptor protein-tyrosine
phosphatase alpha regulates focal adhesion kinase phosphorylation
and ErbB2 oncoprotein-mediated mammary epithelial cell motility. J
Biol Chem. 288:36926–36935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gu Z, Fang X, Li C, Chen C, Liang G, Zheng
X and Fan Q: Increased PTPRA expression leads to poor prognosis
through c-Src activation and G1 phase progression in squamous cell
lung cancer. Int J Oncol. 51:489–497. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen SC, Khanna RS, Bessette DC,
Samayawardhena LA and Pallen CJ: Protein tyrosine phosphatase-alpha
complexes with the IGF-I receptor and undergoes IGF-I-stimulated
tyrosine phosphorylation that mediates cell migration. Am J Physiol
Cell Physiol. 297:C133–C139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hou GX, Liu P, Yang J and Wen S: Mining
expression and prognosis of topoisomerase isoforms in
non-small-cell lung cancer by using oncomine and kaplan-meier
plotter. PLoS One. 12:e01745152017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lou W, Chen J, Ding B, Chen D, Zheng H,
Jiang D, Xu L, Bao C, Cao G and Fan W: Identification of
invasion-metastasis-associated microRNAs in hepatocellular
carcinoma based on bioinformatic analysis and experimental
validation. J Transl Med. 16:2662018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lou W, Liu J, Ding B, Chen D, Xu L, Ding
J, Jiang D, Zhou L, Zheng S and Fan W: Identification of potential
miRNA-mRNA regulatory network contributing to pathogenesis of
HBV-related HCC. J Transl Med. 17:72019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li SZ, Zeng F, Li J, Shu QP, Zhang HH, Xu
J, Ren JW, Zhang XD, Song XM and Du RL: Nemo-like kinase (NLK)
primes colorectal cancer progression by releasing the E2F1 complex
from HDAC1. Cancer Lett. 431:43–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ran FA, Hsu PD, Lin CY, Gootenberg JS,
Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y and
Zhang F: Double nicking by RNA-guided CRISPR Cas9 for enhanced
genome editing specificity. Cell. 154:1380–1389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen J, Cheng J, Zhu S, Zhao J, Ye Q, Xu
Y, Dong H and Zheng X: Regulating effect of baicalin on
IKK/IKB/NF-kB signaling pathway and apoptosis-related proteins in
rats with ulcerative colitis. Int Immunopharmacol. 73:193–200.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li L, Tang P, Li S, Qin X, Yang H, Wu C
and Liu Y: Notch signaling pathway networks in cancer metastasis: A
new target for cancer therapy. Med Oncol. 34:1802017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liang J, Shi J, Wang N, Zhao H and Sun J:
Tuning the protein phosphorylation by receptor type protein
tyrosine phosphatase epsilon (PTPRE) in normal and cancer cells. J
Cancer. 10:105–111. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tremper-Wells B, Resnick RJ, Zheng X,
Holsinger LJ and Shalloway D: Extracellular domain dependence of
PTPalpha transforming activity. Genes Cells. 15:711–724. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
!!! INVALID CITATION !!!
|
|
33
|
Nakano N, Itoh S, Watanabe Y, Maeyama K,
Itoh F and Kato M: Requirement of TCF7L2 for TGF-beta-dependent
transcriptional activation of the TMEPAI gene. J Biol Chem.
285:38023–38033. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singha PK, Pandeswara S, Geng H, Lan R,
Venkatachalam MA and Saikumar P: TGF-β induced TMEPAI/PMEPA1
inhibits canonical Smad signaling through R-Smad sequestration and
promotes non-canonical PI3K/Akt signaling by reducing PTEN in
triple negative breast cancer. Genes Cancer. 5:320–336.
2014.PubMed/NCBI
|
|
35
|
Zhang L, Wang X, Lai C, Zhang H and Lai M:
PMEPA1 induces EMT via a non-canonical TGF-β signalling in
colorectal cancer. J Cell Mol Med. 23:3603–3615. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang MH, Zhang HH, Du XH, Gao J, Li C,
Shi HR and Li SZ: UCHL3 promotes ovarian cancer progression by
stabilizing TRAF2 to activate the NF-kB pathway. Oncogene.
39:322–333. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao X, Luo G, Fan Y, Ma X, Zhou J and
Jiang H: ILEI is an important intermediate participating in the
formation of TGF-β1-induced renal tubular EMT. Cell Biochem Funct.
36:46–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zahid H, Simpson ER and Brown KA:
Inflammation, dysregulated metabolism and aromatase in obesity and
breast cancer. Curr Opin Pharmacol. 31:90–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ghandadi M, Behravan J, Abnous K, Ehtesham
Gharaee M and Mosaffa F: TNF-α exerts cytotoxic effects on
multidrug resistant breast cancer MCF-7/MX cells via a
non-apoptotic death pathway. Cytokine. 97:167–174. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bhatelia K, Singh K and Singh R: TLRs:
Linking inflammation and breast cancer. Cell Signal. 26:2350–2357.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Luo C and Zhang H: The role of
proinflammatory pathways in the pathogenesis of colitis-associated
colorectal cancer. Mediators Inflamm. 2017:51260482017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Herrera Abreu MT, Penton PC, Kwok V,
Vachon E, Shalloway D, Vidali L, Lee W, McCulloch CA and Downey GP:
Tyrosine phosphatase PTPalpha regulates focal adhesion remodeling
through Rac1 activation. Am J Physiol Cell Physiol. 294:C931–C944.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Stanford SM, Svensson MN, Sacchetti C,
Pilo CA, Wu DJ, Kiosses WB, Hellvard A, Bergum B, Muench GRA, Elly
C, et al: Receptor protein tyrosine phosphatase α-mediated
enhancement of rheumatoid synovial fibroblast signaling and
promotion of arthritis in mice. Arthritis Rheumatol. 68:359–369.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lai X, Chen Q, Zhu C, Deng R, Zhao X, Chen
C, Wang Y, Yu J and Huang J: Regulation of RPTPα-c-Src signalling
pathway by miR-218. FEBS J. 282:2722–2734. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yuan BY, Chen YH, Wu ZF, Zhuang Y, Chen
GW, Zhang L, Zhang HG, Cheng JCH, Lin Q and Zeng ZC:
MicroRNA-146a-5p attenuates fibrosis-related molecules in
irradiated and TGF-beta1-treated human hepatic stellate cells by
regulating PTPRA-SRC signaling. Radiat Res. 192:621–629. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li L, Chen W, Liang Y, Ma H, Li W, Zhou Z,
Li J, Ding Y, Ren J, Lin J, et al: The Gβg-Src signaling pathway
regulates TNF-induced necroptosis via control of necrosome
translocation. Cell Res. 24:417–432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Amoui M, Sheng MH, Chen ST, Baylink DJ and
Lau KH: A transmembrane osteoclastic protein-tyrosine phosphatase
regulates osteoclast activity in part by promoting osteoclast
survival through c-Src-dependent activation of NFkappaB and JNK2.
Arch Biochem Biophys. 463:47–59. 2007. View Article : Google Scholar : PubMed/NCBI
|