Introduction
Lung cancer represents one of the leading causes of
cancer-associated mortality, both globally as well as within China
(1,2). Non-small cell lung cancer (NSCLC)
accounts for 80–85% of lung cancer cases, the majority of which are
lung adenocarcinoma (LUAD) (3).
Despite advances in the prevention, diagnosis and treatment of
LUAD, the 5-year overall survival (OS) rate is <15% (4). There have been numerous studies
investigating LUAD prognosis-associated factors, including age,
sex, tumor stage and certain genetic mutations [such as epidermal
growth factor receptor (EGFR)], which may help to predict the OS;
however, effective monitoring of certain genetic mutations is yet
to be achieved, as they are either invasive or costly (5,6).
Therefore, the identification of more economical and convenient
biomarkers able to predict OS time in patients with LUAD is
imperative to facilitate efficient management and treatment of the
disease.
The inhibitor of DNA binding (ID) proteins is a
class of helix-loop-helix (HLH) transcription regulatory factors,
including ID1, ID2, ID3 and ID4 (7).
These proteins have been demonstrated to serve crucial roles in a
wide range of tumor-associated processes, such as tumorigenesis,
cell differentiation, cell cycle, migration and invasion,
angiogenesis and metastasis (8,9).
Aberrant expression of the ID proteins has been revealed in a
variety of human malignancies, including LUAD (10) and breast cancer (11,12).
However, the association between the expression of IDs and LUAD
prognosis is yet to be elucidated. In the present study, compared
with other malignancies, IDs, including ID1, ID2, ID3 and ID4, were
universally downregulated in LUAD samples. Notably, cigarette smoke
contains a complex mixture of >6000 chemicals, thus may alter
gene expression and contribute to smoking-associated disease such
as lung cancer. Thus, the association between IDs expression and
smoking status was also analyzed in LUAD here.
Notably, to the best of our knowledge, the
prognostic value of IDs expression in predicting OS time in
patients with LUAD has not yet been investigated. Therefore, the
purpose of the present study was to summarize and update the
currently available evidence from openly accessible databases
regarding the association between the expression of various IDs and
OS time in patients with LUAD.
Materials and methods
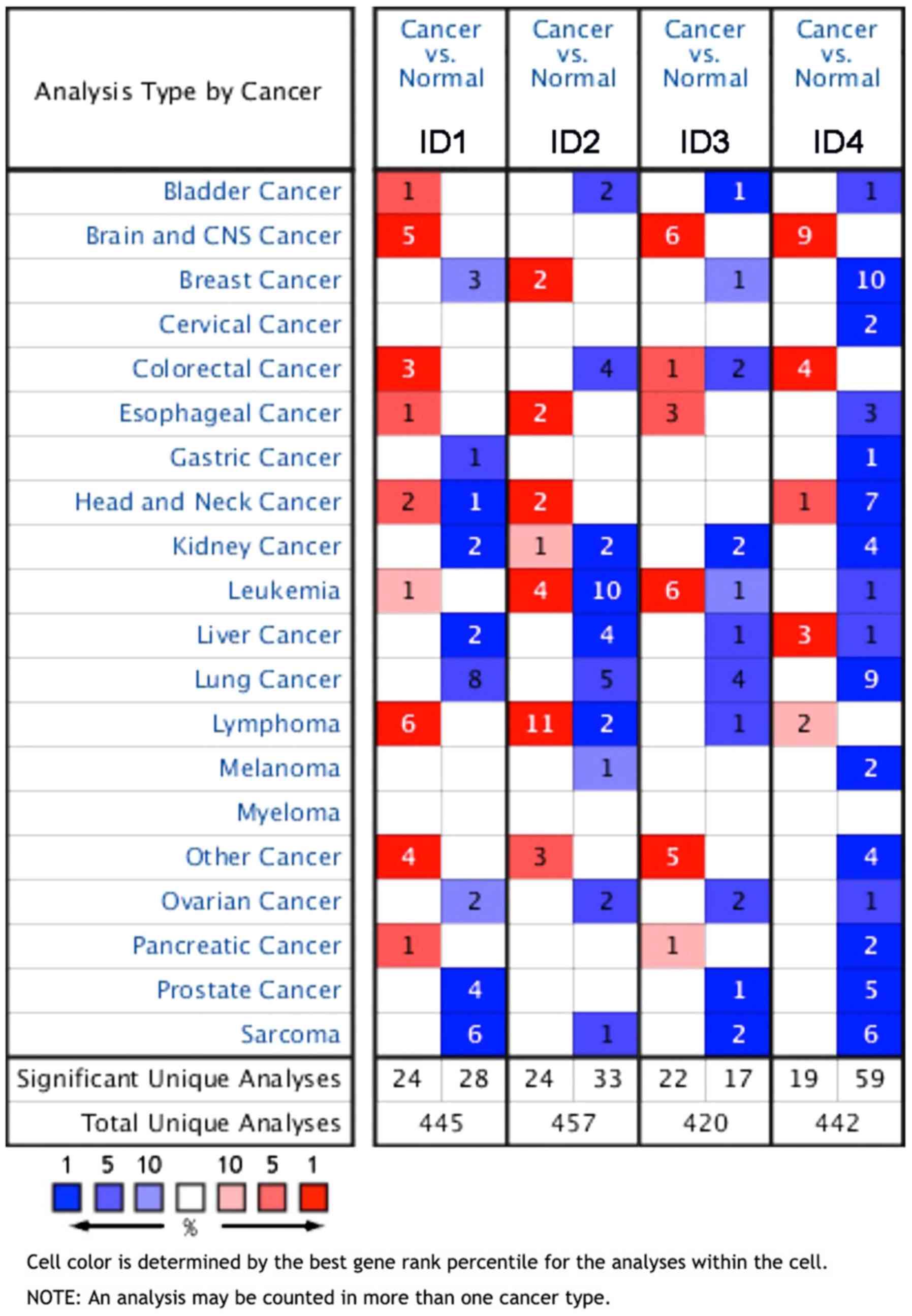
Oncomine analysis
The expression level of all ID proteins was analyzed
in different types of human malignancies using a free-access public
online database, Oncomine (www.oncomine.org), which contains a large amount of
cancer microarray information. The paired Student's t-test was used
to compare cancer vs. normal tissues and a fold-change of 2 with a
P-value of <0.01 was considered to represent a clinically
significance difference in expression, as in a previous study
(13). Gene rank was analyzed by the
percentile of the target gene in the top of all genes(top 10%)
measured in each study. The cell color was determined by the gene
rank percentile for the analyses within the cell as follow: Dark
blue, 100%; blue, 50%; light blue 10%; dark red, 100%, red, 50%;
and light red, 10%.
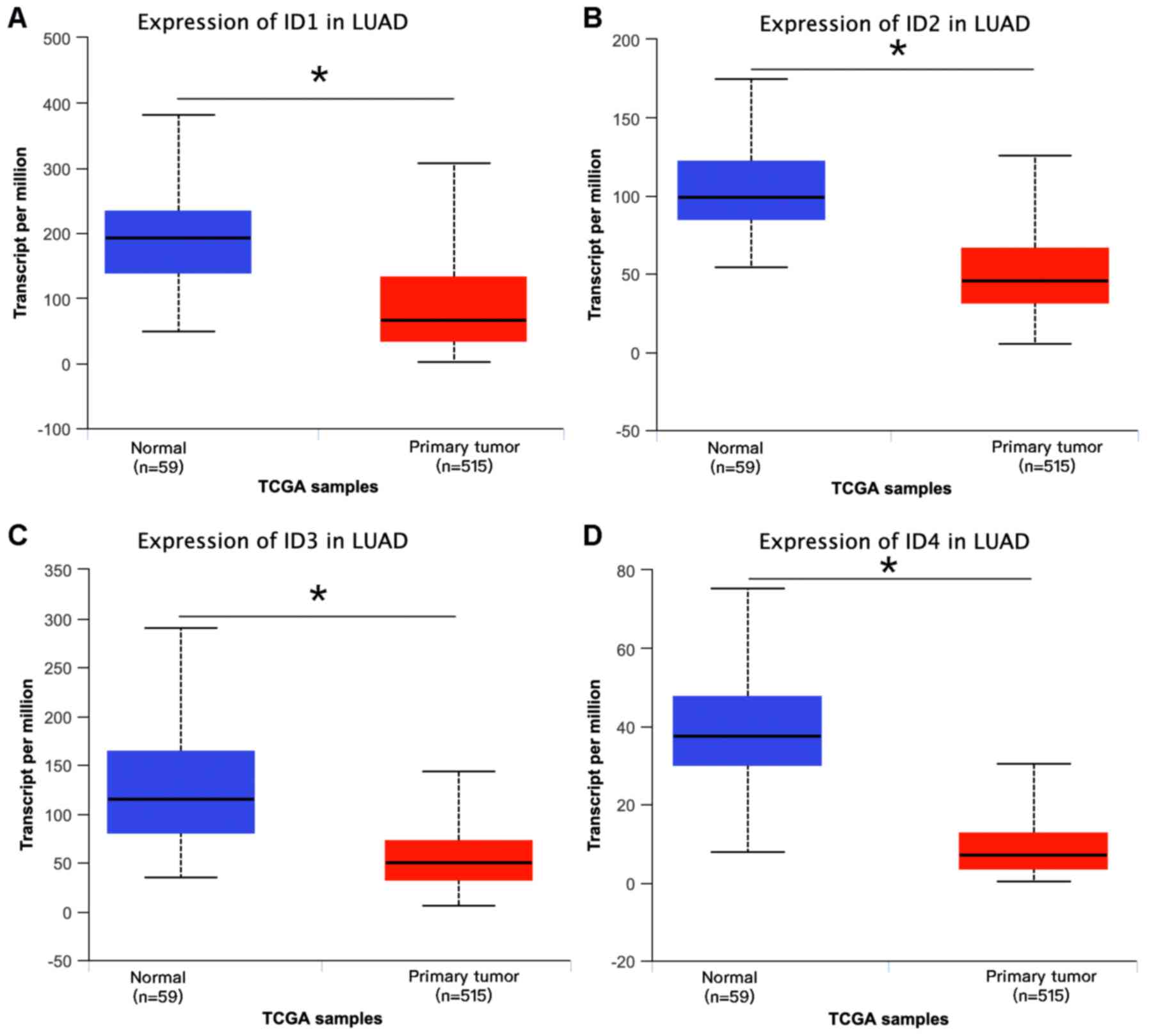
UCLAN database analysis
The open access database UCLAN (http://ualcan.path.uab.edu/analysis.html) was used to
analyze the association between the expression levels of various
IDs and Tumor-Node-Metastasis (TNM) staging and smoking status of
patients with LUAD (14). This
database provides easy access to the publicly available cancer
OMICS information from The Cancer Genome Atlas (TCGA) and the
MET500 (a database comprised of whole-exome and -transcriptome
sequencing of 500 adult patients with metastatic solid tumors of
diverse lineage and biopsy sites), allowing users to identify
biomarkers or perform in silico validation of potential
genes of interest. The median expression value was used as cut-off
and significant difference between groups was assessed using the
unpaired two-sample Student's t-test.
Human protein atlas
The HPA (https://www.proteinatlas.org/) is an open access
program that maps all human proteins in cells and tissues (15). The Pathology Atlas of the HPA depicts
the association between specific protein expression levels and the
OS time of the patients with LUAD (16). Here the HPA was used to analyze the
relationship between IDs protein expression and survival time.
Kaplan-Meier plotter survival
analysis
To analyze the prognostic values of ID expression
levels in LUAD samples, the Kaplan-Meier plotter (www.kmplot.com) was used. This demonstrated the
association between the OS time of patients with LUAD with high- or
low-ID mRNA expression based on median expression as cut-off value.
The Log-rank was used to compare the difference between curves and
is presented on the webpage (17).
Statistical analysis
All data are presented as the mean ± standard
deviation (SD). All statistical analyses were performed using
one-way ANOVA or unpaired two sample t test (SPSS 22.0; IBM Corp.).
The survival curves were calculated using the Kaplan-Meier method
and statistically compared using a log-rank test. Differences were
considered statistically significant when the P-values were
<0.05, unless otherwise specified.
Results
mRNA expression levels of ID family
members in human lung cancer tissues
ID proteins (ID1, ID2, ID3 and ID4) exhibited varied
expression patterns in tumors and normal tissues, as well as in
other human malignancies. The difference in expression levels was
analyzed using the online tumor database Oncomine, which had a
total of 445, 457, 420 and 442 analyses describing ID1, ID2, ID3
and ID4 expression, respectively. Regarding lung cancer, 8 studies
reported that ID1 was significantly downregulated compared with
normal tissues. Of these, 6 studies revealed that ID1 was
significantly downregulated in LUAD compared with the normal
tissues. Regarding ID2, 5 studies revealed that it was
significantly downregulated in lung cancer and, of these, 4 studies
described the significant downregulation of ID2 in LUAD tissues.
ID3 was revealed to be significantly downregulated in 4 studies of
LUAD tissues. Moreover, ID4 was demonstrated to be downregulated in
9 studies on lung cancer, 7 of which were specifically describing
LUAD tissues compared with normal tissues (Fig. 1).
Downregulation of ID mRNA levels
according to UCLAN database
To further explore the expression of ID proteins in
LUAD, data was analyzed using the UCLAN database and it was
observed that the entire ID family, comprising ID1, ID2, ID3 and
ID4, were all significantly downregulated in LUAD compared with
that in normal tissues (Fig. 2).
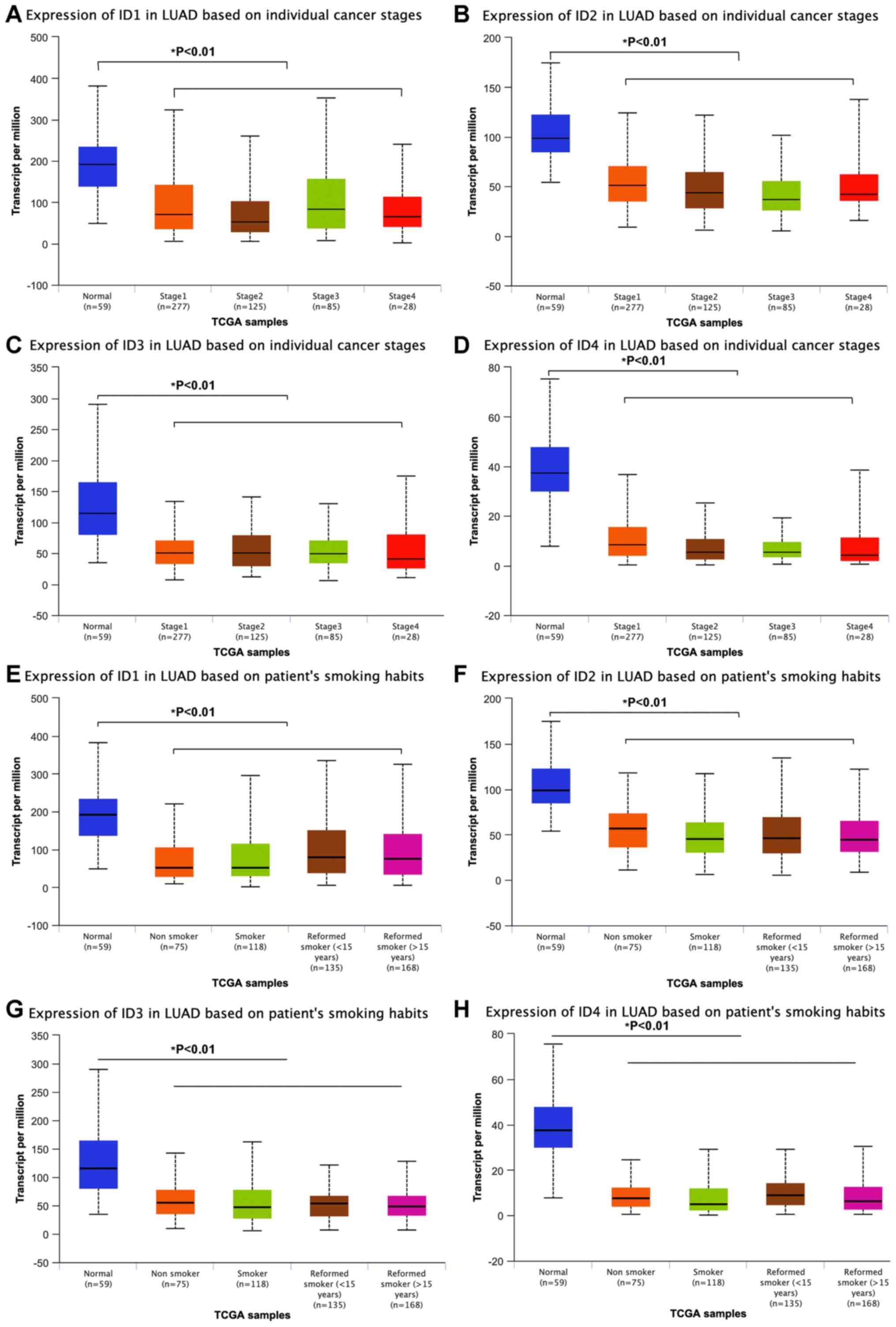
Downregulation of the ID family
according to TCGA database of different clinical features
ID family expression was further analyzed between
different stages of LUAD and it was observed that all ID proteins
were downregulated between stage I and IV in LUAD compared with
those in the normal tissues (Fig.
3A-D). Moreover, smoking status did not significantly influence
expression levels, as IDs expressions were significantly
downregulated in LUAD compared with that in the normal tissues,
irrespective of the smoking history (Fig. 3E-H). The current data indicate that
IDs serve an important role in the tumorigenesis rather than in
progression of LUAD, and that their expression is not significantly
associated with tobacco usage.
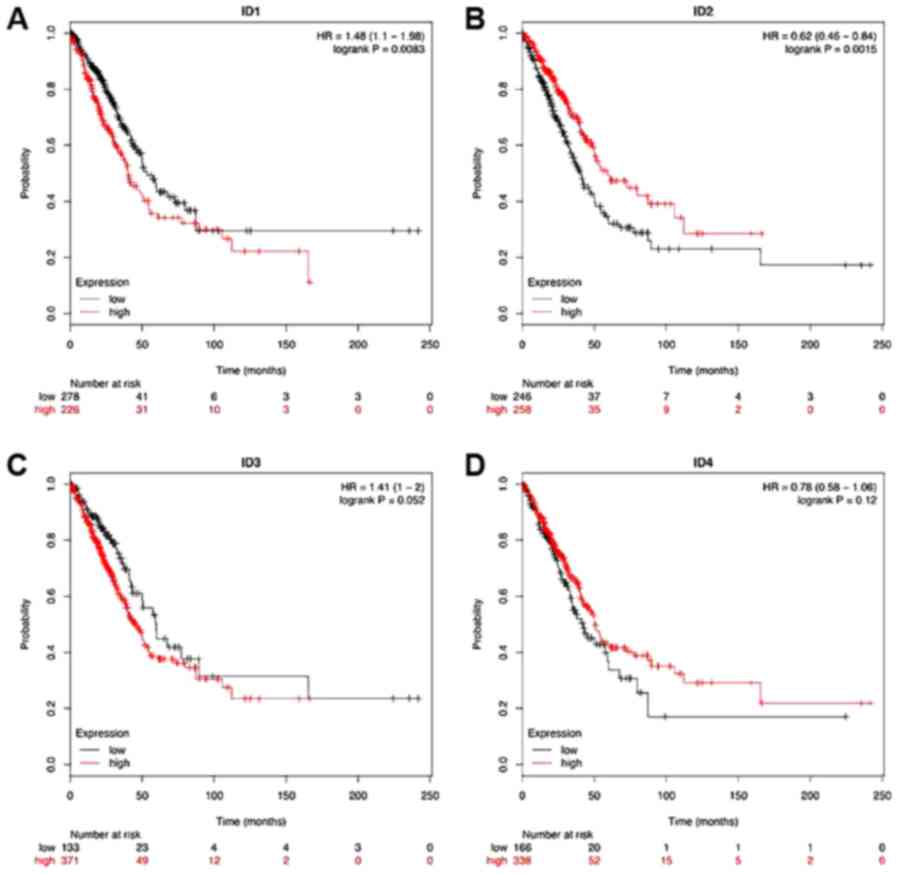
Dysregulation of ID1 and ID2 mRNA
expression, and its association with OS time
Subsequently, data from the KM plotter was analyzed
to identify whether members of this family are associated with the
survival time of patients with LUAD. It was observed that higher
expression of ID1 was associated with poor survival in LUAD
(P<0.05 vs. P=0.0083; Fig. 4A).
However, higher ID2 mRNA expression was significantly associated
with a longer OS time in LUAD (P<0.05; Fig. 4B). Notably, regarding ID3 and ID4,
there was no significant difference in OS time between the high-
and low-expression group, with the median expression used as the
cut-off value (both P>0.05; Fig. 4C
and D).
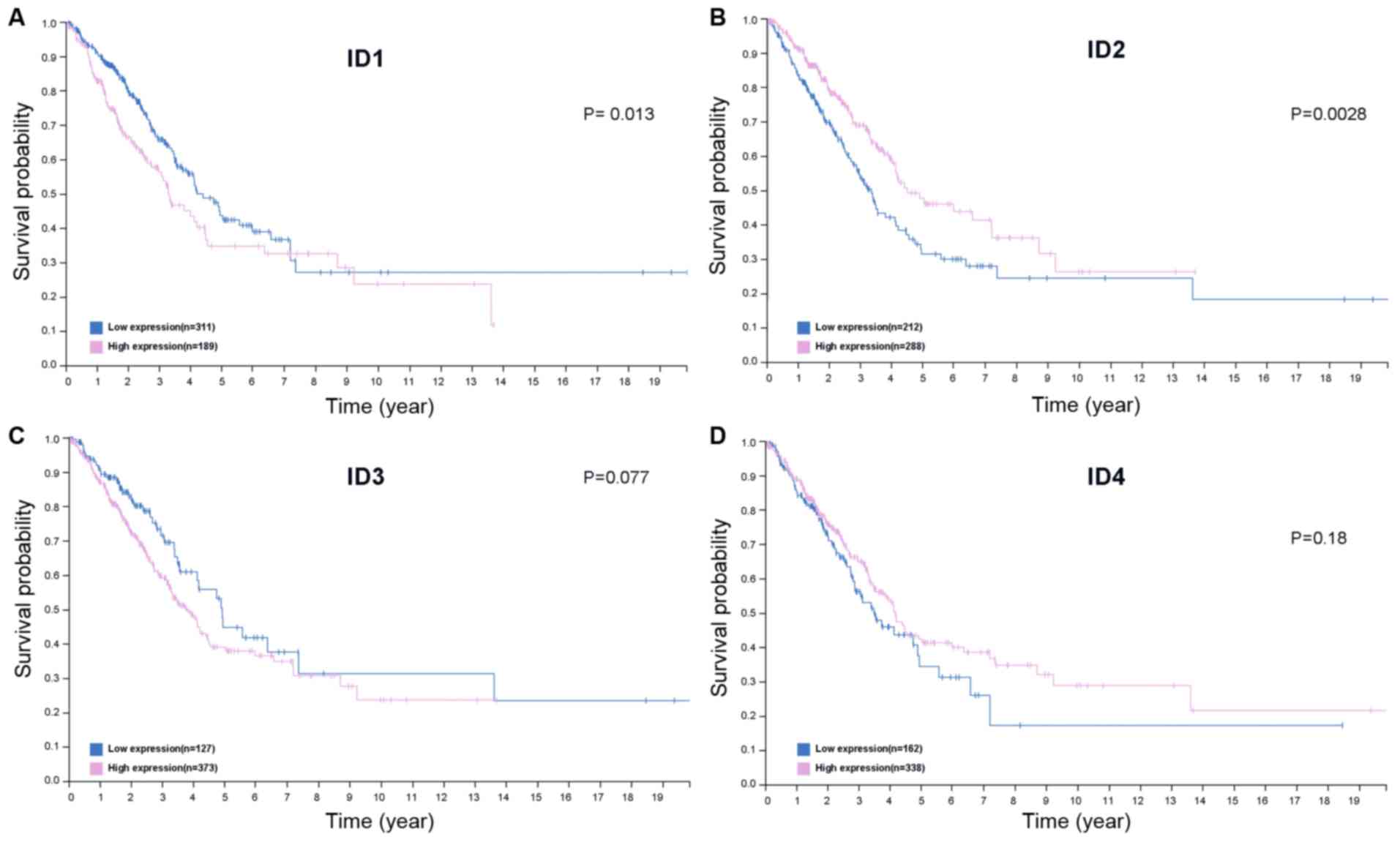
Upregulation of ID1 and ID2 protein
expression is associated with OS time in patients with LUAD
Data from the HPA database were analyzed to discover
whether there was an association between level of protein
expression and OS time. It was observed that increased ID2 protein
expression was positively associated with improved survival time
while high ID1 protein was associated with poor survival time in
patients with LUAD (Fig. 5A and B;
P<0.05). However, the protein expression levels of ID3 and ID4
did not have any significant influence on the OS time of the
patients with LUAD (P=0.077, Fig.
5C; P=0.18, Fig. 5D).
Discussion
LUAD treatment modality has been revolutionized by
the implementation of targeted therapy against genes such as EGFR
and ALK, which has improved clinical outcomes significantly
(18,19). Other targeted treatments include
those against more uncommon mutations such as BRAF V600E and ROS1
(20,21). Immunotherapy can to be used alone or
in combination with chemotherapy to treat LUAD as the first-line
therapeutic option, depending on the expression level of PD-L1
(22–24). This paradigm shift in targeted
treatment options is based on the underlying mechanisms of LUAD
progression, such as the EGFR signaling pathway and ALK
translocations.
IDs are a family of transcription regulators
containing a highly conserved HLH domain. These proteins influence
a variety of tumor processes, including tumorigenesis, progression,
tumor differentiation, cell cycle progression and metastasis
(8). Moreover, IDs are the
downstream targets of a number of oncogenic pathways, making them
potential targets for cancer therapeutics (25). However, from the known literature,
the different ID members have contradictory roles in distinct types
of cancer, and the role of IDs in LUAD is yet to be elucidated. In
the present study, the mRNA expression of the ID family members was
identified using the Oncomine database. It was revealed that IDs
were differentially expressed between different types of human
malignancies. In lung cancer, all ID family members were
significantly downregulated. Notably, IDs were decreased mostly
when observing the histology of the LUAD compared with that in the
normal tissues. To confirm the aforementioned results, data from
UCLAN (a large accessible database containing data from TCGA and
MET500) were investigated. It was revealed that all IDs were
significantly downregulated in LUAD tissues compared with those in
normal tissues, irrespective of stage or smoking history,
indicating their role in tumorigenesis and tumor progression.
Castañon et al (26) reported
that ID1 is upregulated in NSCLC tissues and that this is
associated with shorter OS time. ID family members may influence
cell fate determination, differentiation and cell proliferation by
binding to bHLH. However, each individual ID has diverse role in
these processes. It was found that ID1, ID3 and ID4 were
co-expressed in LUAD. Moreover, in the study by Castañon et
al (26), the expression levels
of ID1 and ID3 were demonstrated to be associated with each other,
and were also associated with a poor clinical outcome in patients
with locally advanced NSCLC treated with definitive
chemoradiotherapy. In the present study, it was revealed that all
ID family members were downregulated in LUAD, irrespective of stage
or smoking habits. However, to understand the distinct mechanisms
underlying the roles of the different IDs in LUAD progression,
further research is needed.
It has been reported that in methylated
glioblastomas, ID4 mRNA is significantly reduced and ID4
methylation is significantly associated with a favorable clinical
outcome (27). Other studies
reported that an increased expression level of ID4 along with a
concomitant loss of the BRCA proteins in triple-negative breast
cancer adversely affect the survival outcome of affected patients
(28). However, it was revealed that
higher ID2 mRNA expression predicted an improved survival outcome,
while the expression levels of both ID3 and ID4 mRNA did not exert
any significant effect on clinical outcome. Notably, ID1 was
downregulated in LUAD and the high mRNA expression group was
significantly associated with a poorer survival time in patients
with LUAD. Thus, contradictory results were obtained, which should
be validated on tumor samples. At the protein level, it was
revealed that high ID2 expression was associated with a longer OS
time, while high ID1 was associated with a shorter survival time.
Thus, the expression of ID1 and survival time is contradictory and
should be validated on tumor samples. Therefore, it was supposed
that amongst the ID family members, ID2 may serve a critical role
in the genesis and progression of LUAD, thereby having a
significant clinical outcome favoring survival of the patients with
LUAD.
Wen et al (12) revealed that ID2 upregulated notch3
expression via blocking the binding of E2A to an E-box motif in the
notch3 promoter; further decreased expression of ID2 was associated
with worse long-term metastasis-free survival in patients with
breast cancer (12). Moreover, Bae
et al (29) revealed that ID2
expression was significantly higher in the head and neck squamous
cell carcinoma (HNSCC) cells with stemness, compared with that in
differentiated HNSCC cells; furthermore, increased expression of
ID2 was closely associated with poorer post-treatment survival
rates in patients with HNSCC. By contrast, it was observed in the
current study that increased expression of ID2 was strongly
associated with better survival outcomes in patients with LUAD.
These differential findings indicate that different types of human
cancer have varied ID2 expression patterns.
Several potential limitations of the present study
should be noted; although it was demonstrated that IDs proteins
serve an important role in LUAD and may represent a therapeutic
target, all data were retrieved from public databases and not
validated using tumor and normal samples. In addition, Gene
Ontology and Kyoto Encyclopedia of Genes and Genomes pathway
analyses were not performed to identify genes associated with IDs
in LUAD, as the current study was focused on determining the role
of the ID family in the progression of LUAD. In the present study,
it was revealed that the mRNA expression of ID1 and ID2 was
associated with survival outcomes, and at the protein level, ID1
and ID2 was associated with OS time in patients with LUAD. However,
based on the contradictory results of ID1, it was proposed that ID2
may serve a critical role in the genesis and progression of LUAD.
To further elucidate the underlying molecular mechanism behind the
role of ID2 in LUAD, more in-depth studies are warranted to
investigate the exact functions of ID2 in tumorigenesis and the
development of LUAD.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available in the Oncomine (https://www.oncomine.org), UCLAN (http://ualcan.path.uab.edu/analysis.html), Kaplan
Meier-plotter (kmplot.com/analysis), and Human Protein Atlas
(https://www.proteinatlas.org).
Authors' contributions
XL, LM and ZZ participated in the design of the
study, data acquisition and analysis as well as drafting the
manuscript. LS and YQ were responsible for the data analysis. YZ
and YW participated in data acquisition, analysis, and
interpretation. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johnson DH, Schiller JH and Bunn PA Jr:
Recent clinical advances in lung cancer management. J Clin Oncol.
32:973–982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li P, Gao Q, Jiang X, Zhan Z, Yan Q, Li Z
and Huang C: Comparison of clinicopathological features and
prognosis between ALK rearrangements and EGFR mutations in
surgically resected early-stage lung adenocarcinoma. J Cancer.
10:61–71. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paesmans M: Prognostic and predictive
factors for lung cancer. Breathe. 9:112–121. 2012. View Article : Google Scholar
|
|
7
|
Ke J, Wu R, Chen Y and Abba ML: Inhibitor
of DNA binding proteins: Implications in human cancer progression
and metastasis. Am J Transl Res. 10:3887–3910. 2018.PubMed/NCBI
|
|
8
|
Sikder HA, Devlin MK, Dunlap S, Ryu B and
Alani RM: Id proteins in cell growth and tumorigenesis. Cancer
Cell. 3:525–530. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yokota Y: Id and development. Oncogene.
20:8290–8298. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Antonângelo L, Tuma T, Fabro A, Acencio M,
Terra R, Parra E, Vargas F, Takagaki T and Capelozzi V: Id-1, Id-2,
and Id-3 co-expression correlates with prognosis in stage I and II
lung adenocarcinomapatients treated with surgery and adjuvant
chemotherapy. Exp Biol Med (Maywood). 241:1159–1168. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou XL, Zeng, Ye YH, Sun SM, Lu XF, Liang
WQ, Chen CF and Lin HY: Prognostic values of the inhibitor of
DNA-binding family members in breast cancer. Oncol Rep.
40:1897–1906. 2018.PubMed/NCBI
|
|
12
|
Wen XF, Chen M, Wu Y, Chen MN, Glogowska
A, Klonisch T and Zhang GJ: Inhibitor of DNA binding 2 inhibits
epithelial-mesenchymal transition via upregulation of Notch3 in
breast cancer. Transl Oncol. 11:1259–1270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin HY, Zeng, Liang YK, Wei XL and Chen
CF: GATA3 and TRPS1 are distinct biomarkers and prognostic factors
in breast cancer: Database mining for GATA family members in
malignancies. Oncotarget. 8:34750–34761. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thul PJ, Åkesson L, Wiking M, Mahdessian
D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science.
356:eaal33212017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nagy A, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep.
8:115152018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open- label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa
K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al:
First-line crizotinib versus chemotherapy in ALK-positive lung
cancer. N Engl J Med. 371:2167–2177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shaw AT and Solomon BJ: Crizotinib in
ROS1- rearranged non-small-cell lung cancer. N Engl J Med.
372:683–684. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Planchard D, Smit EF, Groen HJM, Mazieres
J, Besse B, Helland Å, Giannone V, D'Amelio AM Jr, Zhang P,
Mookerjee B and Johnson BE: Dabrafenib plus trametinib in patients
with previously untreated BRAFV600E-mutant metastatic
non-small-cell lung cancer: An open-label, phase 2 trial. Lancet
Oncol. 18:1307–1316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: Pembrolizumab versus Chemotherapy for PD-L1-Positive
Non-Small-Cell Lung Cancer. N Engl J Med. 375:1823–1833. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho
BC, Turna HZ, Castro G Jr, Srimuninnimit V, Laktionov KK,
Bondarenko I, et al: Pembrolizumab versus chemotherapy for
previously untreated, PD-L1-expressing, locally advanced or
metastatic non-small-cell lung cancer (KEYNOTE-042): A randomised,
open-label, controlled, phase 3 trial. Lancet. 393:1819–1830. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gandhi L, Rodríguez-Abreu D, Gadgeel S,
Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ,
Powell SF, et al: Pembrolizumab plus chemotherapy in metastatic
non-small-cell lung cancer. N Engl J Med. 378:2078–2092. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McAllister SD, Christian RT, Horowitz MP,
Garcia A and Desprez PY: Cannabidiol as a novel inhibitor of Id-1
gene expression in aggressive breast cancer cells. Mol Cancer Ther.
6:2921–2927. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Castañon E, Bosch-Barrera J, López I,
Collado V, Moreno M, López-Picazo JM, Arbea L, Lozano MD, Calvo A
and Gil-Bazo I: Id1 and Id3 co-expression correlates with clinical
outcome in stage III-N2 non-small cell lung cancer patients treated
with definitive chemoradiotherapy. J Transl Med. 11:132013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Martini M, Cenci T, D'Alessandris GQ,
Cesarini V, Cocomazzi A, Ricci-Vitiani L, De Maria R, Pallini R and
Larocca LM: Epigenetic silencing of Id4 identifies a glioblastoma
subgroup with a better prognosis as a consequence of an inhibition
of angiogenesis. Cancer. 119:1004–1012. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thike AA, Tan PH, Ikeda M and Iqbal J:
Increased ID4 expression, accompanied by mutant p53 accumulation
and loss of BRCA1/2 proteins in triple-negative breast cancer,
adversely affects survival. Histopathology. 68:702–712. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bae WJ, Koo BS, Lee SH, Kim JM, Rho YS,
Lim JY, Moon JH, Cho JH and Lim YC: Inhibitor of DNA binding 2 is a
novel therapeutic target for stemness of head and neck squamouscell
carcinoma. Br J Cancer. 117:1810–1818. 2017. View Article : Google Scholar : PubMed/NCBI
|