Introduction
Oral squamous cell carcinoma (OSCC) is the eighth
most common tumor worldwide, and accounts for 2.1% of deaths caused
by all cancers (1,2). Oral cancer consists of a group of
neoplasms that affect regions of the oral cavity, pharyngeal
regions and salivary glands (3).
Although great advances have been made in the treatment of OSCC,
including surgery, chemotherapy, radiotherapy and hormonotherapy,
the 5-year survival rate of OSCC remains less than 50%, mainly due
to cancer metastasis to lymph nodes (4). Therefore, there is an urgently need to
explore the molecular mechanisms underlying OSCC to improve
therapeutic efficacy and the survival rate of OSCC patients.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNAs more than 200 nucleotides in length. Increasing
evidence suggests that lncRNAs are notable molecular markers
involved in the regulation of gene expression and cancer
progression (5,6). Dysfunction of lncRNAs are closely
related to the incidence of human diseases, including tumors,
degenerative neurological diseases and other major diseases that
seriously endanger human health (7,8). Thus,
currently lncRNAs are a ‘hot spot’ of tumor research. lncRNA
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was
first discovered in non-small cell lung cancer (NSCLC), and is
located on chromosome 11q13.1 which is highly conserved (9). MALAT1 was reported to be upregulate in
colorectal cancer (CRC) and to promote cell proliferation,
metastasis and invasion (10).
However, the molecular mechanisms of MALAT1 in mediating the
progression of OSCC have not been well investigated.
MicroRNAs (miRNAs/miRs) are a large family of short
non-coding RNAs, consisting of 18–25 nucleotides, and have been
found to negatively regulate target genes via binding to the 3′-UTR
(3′-untranslated region) (11).
miR-101 has been reported to be downregulated in OSCC cells and to
act as a tumor suppressor in OSCC development (12). It is worth mentioning that miR-101
was suggested to be a potential target of MALAT by Starbase.
However, the exact role of miR-101 in MALAT-mediated OSCC
progression needs further investigation.
As previously elucidated, miRNAs are involved in
tumor development by targeting their mRNAs. Enhancer of zeste 2
polycomb repressive complex 2 subunit (EZH2) has been verified as a
direct target of miR-101 in various types of cancers, including
CRC, laryngeal squamous cell carcinoma and retinoblastoma (13–15).
However, whether EZH2 serves as the target of miR-101 in the
modulation of OSCC progression has not yet been clarified.
Materials and methods
Tissue specimens
Twenty paired OSCC tissues and adjacent normal
tissues were obtained from The Affiliated Yantai Yuhuangding
Hospital of Qingdao University (Yantai, Shandong, China) between
March 2015 and April 2018. All patients (age range, 35–75 years,
mean 61.3 years; 16 male and 4 female) were not treated with
radiotherapy or chemotherapy prior to surgery. Liquid nitrogen was
used to flash-freeze all the tissue specimens, and then the
collected tissues were stored at −80°C for further use. All
patients signed written informed consent. This study was approved
by the Ethics Committee of the Affiliated Yantai Yuhuangding
Hospital of Qingdao University (no. 20140506).
Cell culture
OSCC cell lines (HSC3, SCC9, SCC15 and SCC25) and
primary normal human oral keratinocyte (NHOK) cells were purchased
from the BeNa Culture Collection (BNCC, Shanghai, China). RPMI-1640
medium containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 µg/ml streptomycin were used to
culture OSCC cells and then cells were incubated at 37°C with 5%
CO2 atmosphere.
Cell transfection
Small interference RNA (siRNA), scrambled
oligonucleotide (NC) vector, MALAT1 siRNA, MALAT1 vector, miR-101
mimic, miR-101 inhibitor, negative control (NC)-mimic,
NC-inhibitor, EZH2 plasmid and NC-plasmid were designed and
synthesized by GenePharma Co. They were transfected into SCC9 cells
by Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Forty-eight hours after transfection, the efficiency was validated
by quantitative polymerase chain reaction (qPCR). miRNA sequences
were as follows: miR-101 mimic sense, 5′-UACAGUACUGUGAUAACUGAA−3′
and antisense, 5′-CAGUUAUCACAGUACUGUAUU−3′; control-mimic sense,
5′-UUCUCCGAACGUGUCACGUTT−3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT−3′; miR-101 inhibitor,
5′-UUCAGUUAUCACAGUAUGUA-3′; and inhibitor-control,
5′-CAGUACUUUUGUGUAGUACAA−3′.
CCK-8 assay
Cell Counting Kit-8 (CCK-8, Beyotime Institute of
Biotechnology) was applied for testing SCC9 cell viability. SCC9
cells were seeded onto 96-well plates at 1×103
cells/well with RPMI-1640 medium for 24, 48, 72 and 96 h in an
incubator under normal conditions (37°C, 5% CO2). Then,
CCK-8 solution (10 µl) was added for incubation for another 2 h at
37°C. Absorbance of each well was detected using a microplate
reader (Bio-Rad Laboratories, Inc.) at 450 nm.
Transwell invasion assay
SCC9 cells were inoculated in a prepared Transwell
chamber (8-µm pore size; Corning Inc.) with Matrigel (BD
Biosciences), and the cell density of each group was adjusted with
serum-free RPMI-1640 culture medium. Briefly, 5×104 SCC9
cells were added to the upper chamber and RPMI-1640 culture medium
containing 20% FBS was added to the lower chamber. The cells on the
upper chamber were removed with a cotton swab. The cells that
migrated to the lower chamber were fixed with 4% paraformaldehyde
and stained with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA)
for 15 min at room temperature. The number of transmembrane cells
from 5 random different fields was selected under a microscope
(magnification, ×100; SZ61; Olympus).
Dual luciferase activity assay
Through Starbase online software (http://starbase.sysu.edu.cn/index.php),
we searched for the potential target miRNAs of MALAT1. The MIMAT
number of miR-101-3p is MIMAT0000099 (miRBase). Next, through the
TargetScan online software (http://www.targetscan.org/vert_71/), we searched for
the potential target genes of miR-101. MALAT1 and EZH2 sequences
containing °the wild-type (WT) or mutated (MuT)-type miR-101
binding sites were cloned into the pGL3 luciferase reporter vector
(pGL3-empty; Promega Corp.), respectively. SCC9 cells were
co-transfected with miR-101 mimic along with MALAT1-WT/-MuT
reporter or EZH2-WT/-MuT reporter using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), respectively. After a
48-h transfection, luciferase activity was examined using the Dual
Luciferase Reporter Assay system (Promega Corp.). The values were
normalized to those obtained for pre-miR negative control
transfection.
Real-time-polymerase chain reaction
(qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was applied for extracting total RNA from OSCC
tissues and cells. Complementary DNA (cDNA) was synthesized using
the PrimeScript RT reagent kit (Takara). Then, RNA was reverse
transcribed using reverse transcription kit (Takara). RT-PCR was
performed using the SYBR-Green RT-PCR kit (Bio-Rad Laboratories,
Inc.). The expression levels were analyzed using the
2−ΔΔCq method (16).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were used
as the internal reference for MALAT1 and miR-101, respectively. The
primers are shown in Table I.
 | Table I.Primer sequences for real-time
fluorescence quantification PCR. |
Table I.
Primer sequences for real-time
fluorescence quantification PCR.
| Gene | Primer
sequences |
|---|
| GAPDH | Forward primer:
5′-GCACCGTCAAGGCTGAGAAC-3′ |
|
| Reverse primer:
5′-ATGGTGGTGAAGACGCCAGT-3′ |
| U6 | Forward primer:
5′-CTCGCTTCGGCAGCACA-3′ |
|
| Reverse primer:
5′-AACGCTTCACGAATTTGCGT-3′ |
| lncRNA MALAT1 | Forward primer:
5′-GGGTGTTTACGTAGACCAGAACC-3′ |
|
| Reverse primer:
5′-CTTCCAAAAGCCTTCTGCCTTAG-3′ |
| miR-101-3p | Forward primer:
5′-CGGCGGATCTTTGATAGACT-3′ |
|
| Reverse primer:
5′-GGAGGTGCTGGATGGAGTTA-3′ |
| EZH2 | Forward primer:
5′-TTCATGCAACACCCAACACT-3′ |
|
| Reverse primer:
5′-GAGAGCAGCAGCAAACTCCT-3′ |
Western blot analysis
Total proteins were extracted using RIPA lysis
buffer (25 mmol/l Tris-HCl + 150 mmol/l NaCl + 1% NP-40 + 1% sodium
deoxycholate + 0.1% SDS, pH 7.6; Beyotime Institute of
Biotechnology) supplemented with 1% protease inhibitors (100X;
Roche Diagnostics) and phenylmethanesulfonyl fluoride (PMSF, 100
mmol/l; Sigma-Aldrich; Merck KGaA). The protein concentration was
detected by BCA kit (Beyotime Institute of Biotechnology). Equal
amounts (20 µg) of extracted proteins were separated by 10%
SDS-PAGE and then transferred to NC membranes. Then, the NC
membranes were blocked with 5% non-fat milk for 1 h at room
temperature, and subsequently cultured with the primary antibodies,
rabbit monoclonal anti-EZH2 (dilution 1:1,000, ab191080, Abcam),
rabbit monoclonal anti-caspase-3 (dilution 1:1,000, ab197202,
Abcam) and rabbit monoclonal anti-caspase-8 (dilution 1:1,000,
ab25901, Abcam). The membrane was washed with TBST, and incubated
with horseradish peroxidase conjugated goat anti-rabbit secondary
antibody (ab6721, Abcam) at room temperature for 1 h. Finally, the
enhanced chemiluminescence (ECL) was applied for detecting the
immune complexes. GAPDH was used as the internal control. The bands
were subjected to quantification using the ImageJ imaging
processing program (National Institutes of Health, Bethesda, MD,
USA).
Statistical analysis
All data are expressed as mean ± standard deviation
(SD). Each experiment was repeated at least three times. The
Student's t-test, one-way analysis of variance (ANOVA) and Pearson
product moment correlation coefficient were used for statistical
analysis in GraphPad Prism 6.0 software (GraphPad Software, Inc.).
Kaplan-Meier curves and log-rank test were performed to analyze the
survival rate of OSCC patients with different levels of MALAT1.
Spearman's correlation analysis was implemented to validate the
correlation between the levels of MALAT1 and miR-101 in OSCC
tissues. P<0.05 was considered statistically significant.
Results
MALAT1 is upregulated in OSCC tissues
and cell lines
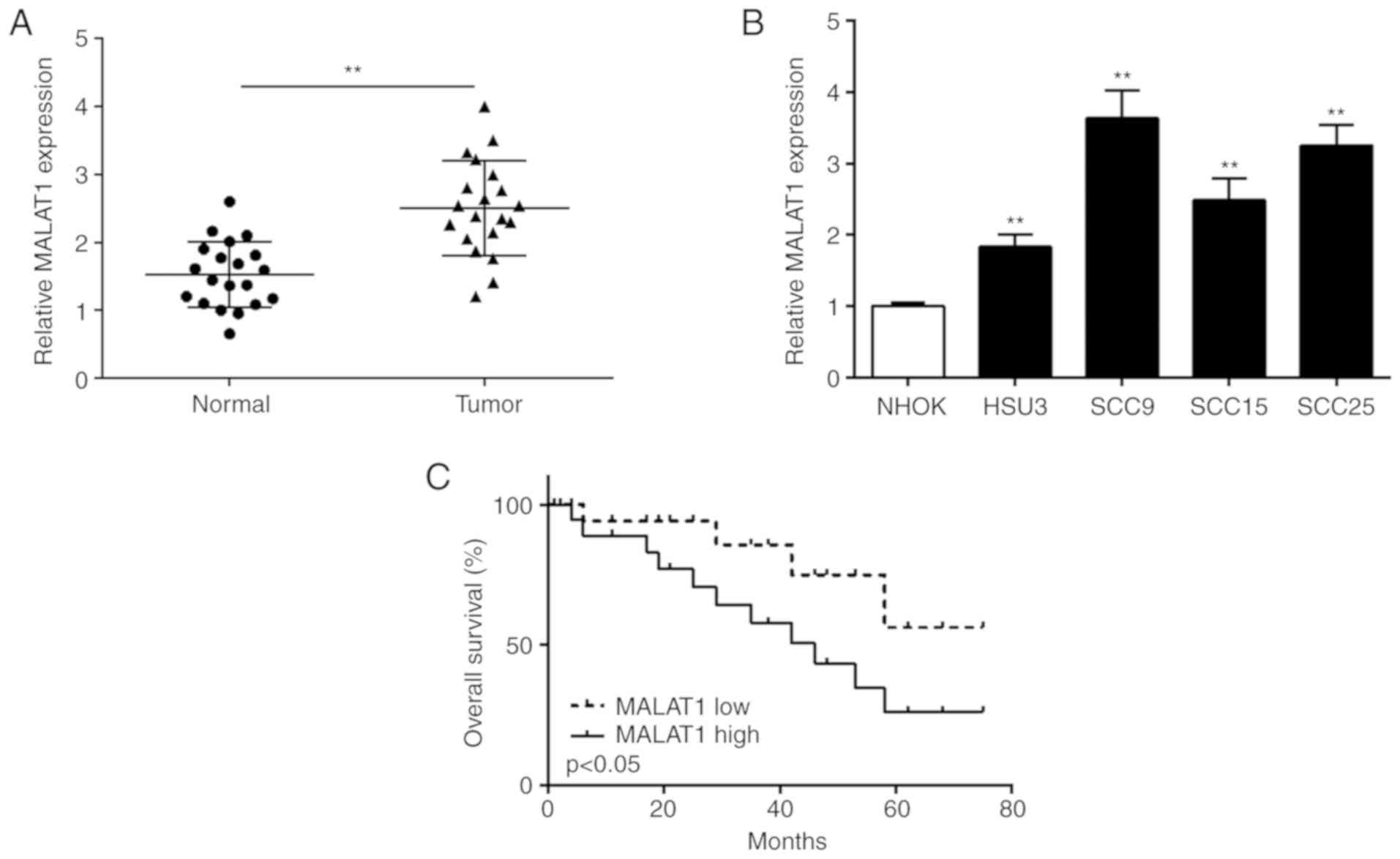
qPCR was carried out to detect MALAT1 expression in
OSCC tissues and cell lines. As shown in Fig. 1A, the expression of MALAT1 was
significantly increased in the OSCC tissues compared to that noted
in the normal tissues. Then, MALAT1 expression was detected in OSCC
cell lines using qPCR. As shown in Fig.
1B, MALAT1 expression was significantly increased in all OSCC
cell lines (HSC3, SCC9, SCC15 and SCC25) in comparison with that in
normal NHOK cells. OSCC patients were divided into two subgroups
(low/high MALAT1 level) using the median level of MALAT1 as a
cut-off value. Kaplan-Meier survival analysis displayed that the
overall survival was shorter in OSCC patients with high expression
of MALAT1 compared to that with low expression of MALAT1 (Fig. 1C). We subsequently chose SCC9 for
further research because it exhibited the highest MALAT1 expression
in the 6 cell lines.
MALAT1 promotes OSCC cell
proliferation and invasion
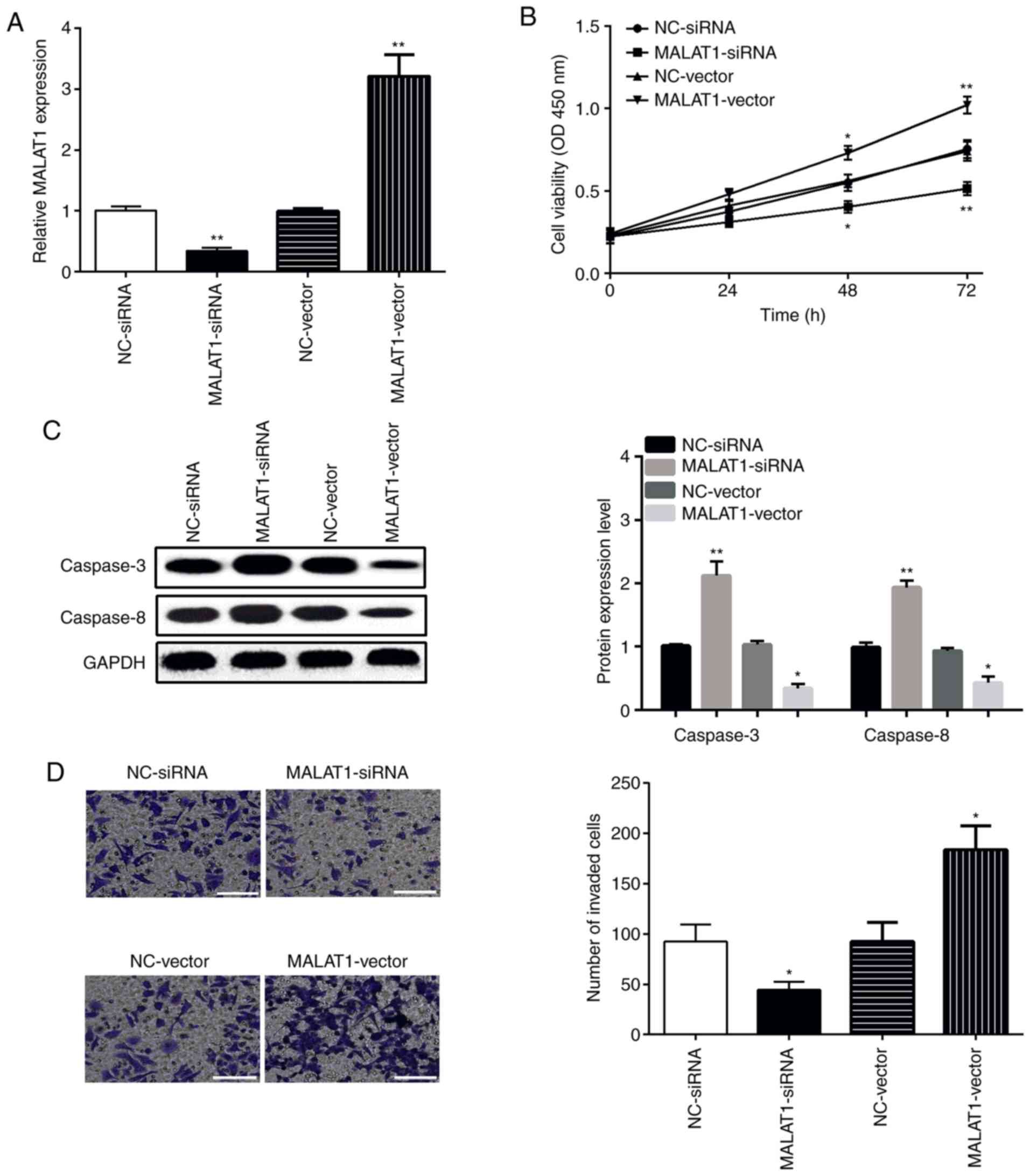
To clarify the role of MALAT1 in proliferation and
invasion, MALAT1-vector or MALAT1-siRNA was transfected into SCC9
cells. Expression of MALAT1 was determined by qPCR. The findings
showed that MALAT1 was downregulated by MALAT1-siRNA, while
upregulated by MALAT1-vector when compared with the relevant NC
control (Fig. 2A). The viability and
invasive ability of SCC9 cells were measured by CCK-8 and Transwell
assays, respectively. As shown in Fig.
2B, MALAT1 overexpression promoted SCC9 cell viability, while
MALAT1 knockdown inhibited the viability of SCC9 cells. The results
of western blot analysis displayed that the levels of caspase-3 and
caspase-8 were decreased by the MALAT1-vector, while increased by
MALAT1-siRNA (Fig. 2C).
Additionally, the findings of the Transwell assay displayed that
the invasiveness of the SCC9 cells in the MALAT1-vector group was
significantly increased, whereas this ability was decreased in the
MALAT1-siRNA group (Fig. 2D). The
above-mentioned data indicate that overexpression of MALAT1
promotes the proliferation and invasion of OSCC cells.
MALAT1 directly interacts with miR-101
in OSCC cells
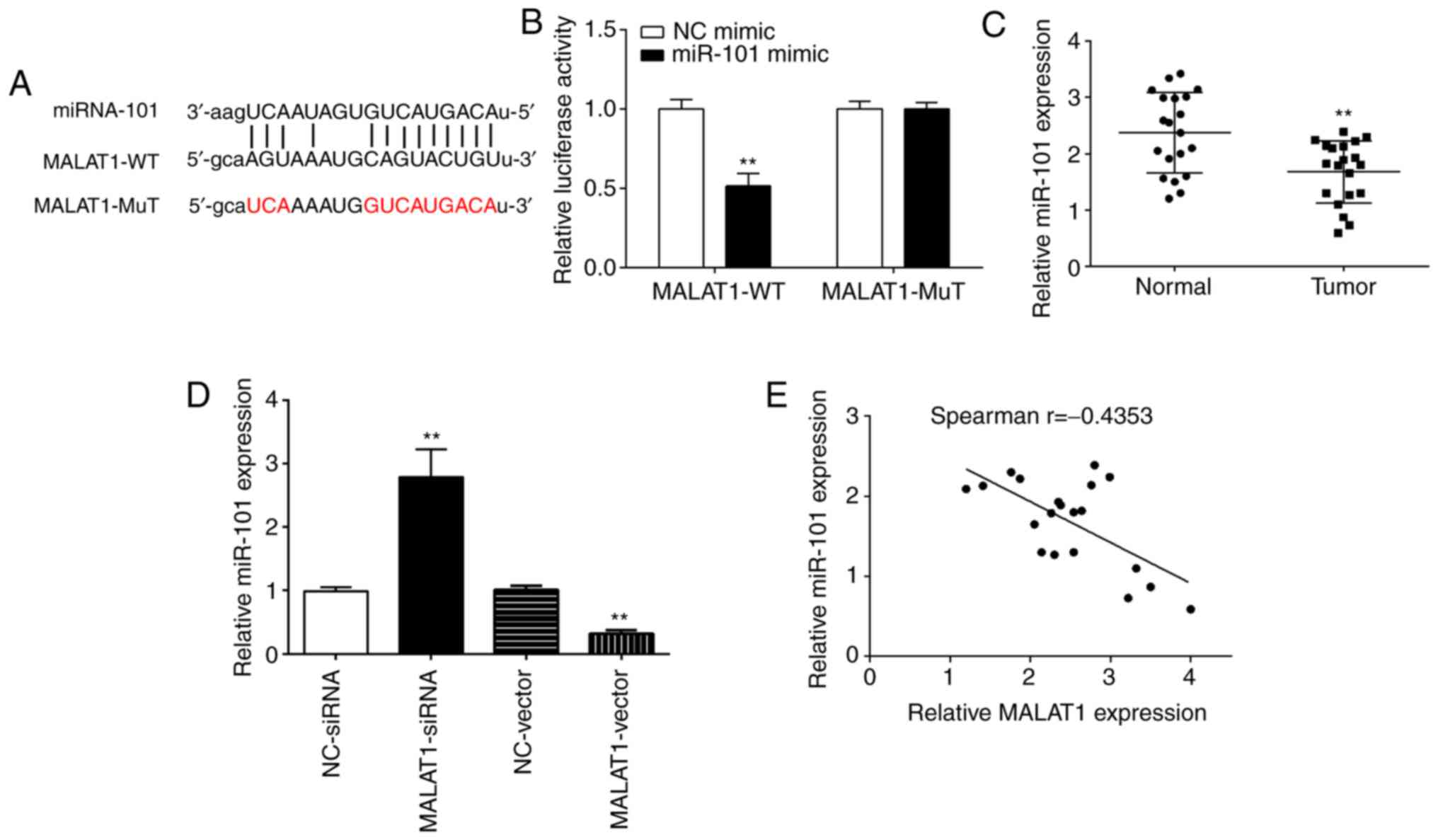
The bioinformatics analysis Starbase online software
(http://starbase.sysu.edu.cn/index.php) was first
performed to detect the potential miRNAs regulated by MALAT1. It
was identified that miR-101 was the most probable target miRNA of
MALAT1; miR-101 shared complementary binding sites with the 3′-UTR
of MALAT1 (Fig. 3A). Dual-luciferase
assay was then performed to confirm the bioinformatical prediction.
As shown in Fig. 3B, miR-101 mimic
significantly downregulated the luciferase activity of MALAT1
3′-UTR-WT in SCC9 cells. However, there was no significantly change
in the luciferase activity of MALAT1 3′UTR-MuT. Furthermore, the
result of the qPCR analysis revealed that miR-101 was downregulated
in the OSCC tissues when compared with the normal tissues (Fig. 3C). Notably, overexpression of MALAT1
inhibited miR-101 expression, while silencing of MALAT1 expression
enhanced miR-101 expression in the SCC9 cells (Fig. 3D). More strikingly, a negative
correlation was noted between MALAT1 expression and miR-101 in the
OSCC tissues (Fig. 3E). Taken
together, these results suggest that MALAT1 directly targets
miR-101 in regulating OSCC progression.
miR-101 is associated with
MALAT1-mediated OSCC cell proliferation and invasion
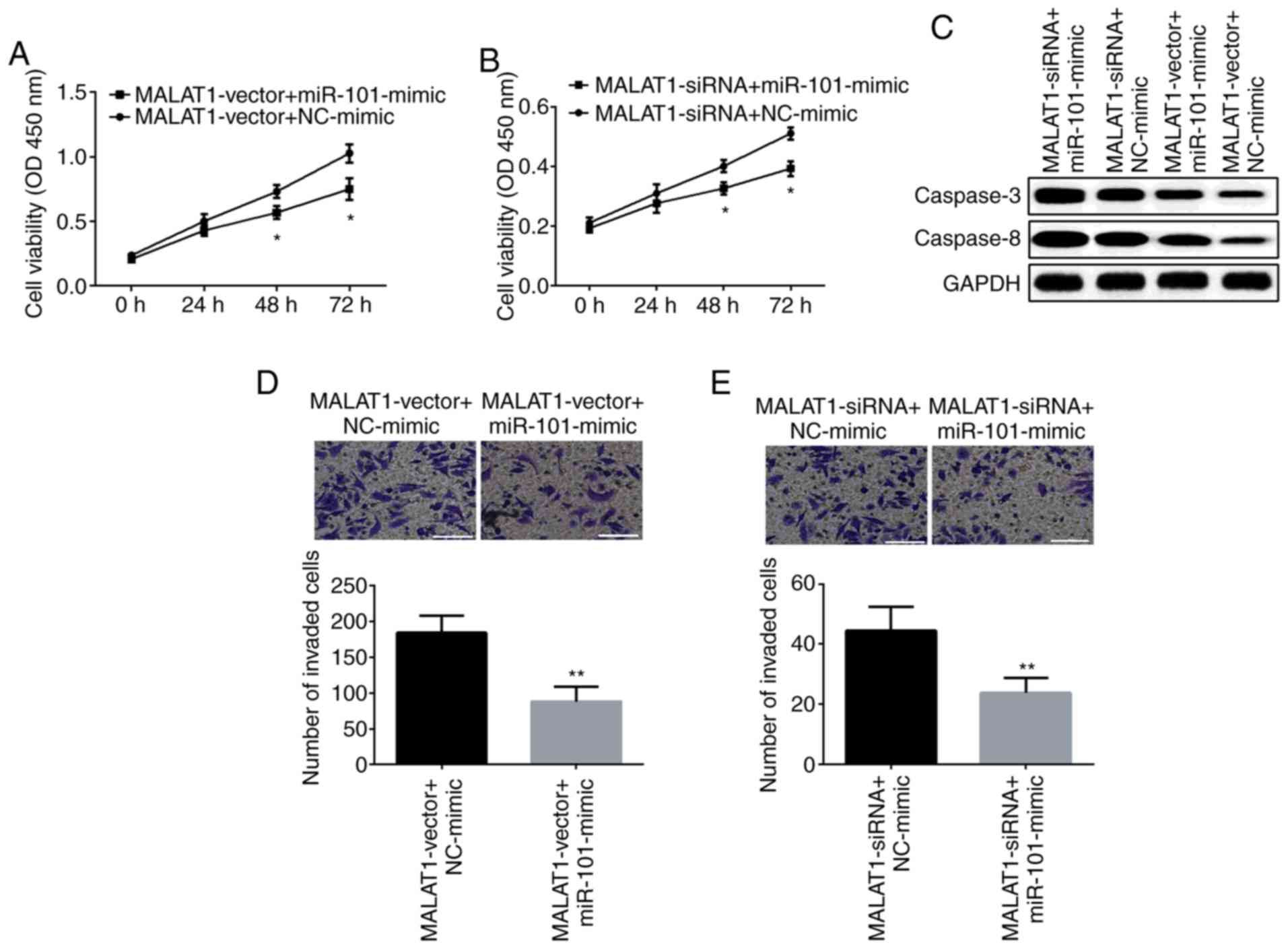
Next, we explored the biological effect of miR-101
as regulated by lncRNA MALAT1 on OSCC cell proliferation and
invasion. SCC9 cells were treated with the MALAT1-vector or
MALAT1-siRNA, simultaneously with the miR-101 mimic. The results of
the CCK-8 assay demonstrated that the viability of SCC9 cells was
increased by the MALAT1-vector yet significantly reduced by the
miR-101 mimic (Fig. 4A). However,
the viability of SCC9 cells was inhibited by MALAT1-siRNA which was
further suppressed by miR-101 mimic (Fig. 4B). Moreover, the miR-101 mimic was
able to reverse the inhibitory effect of the MALAT1-vector on
expression of caspase-3 and caspase-8, while the miR-101 mimic
attenuated the MALAT1-siRNA-induced facilitating effect on SCC9
cell apoptosis (Fig. 4C). A similar
trend for the Transwell assay is showed in Fig. 4D and E, overexpression of miR-101
inhibited the promotive role of the MALAT1-vector in SCC9 cell
invasion, otherwise, miR-101 mimic was able to reverse the
suppression of MALAT1-siRNA in regards to cell invasion.
Collectively, these findings indicate that MALAT1 facilitates cell
proliferation and invasion via suppressing miR-101 in OSCC
cells.
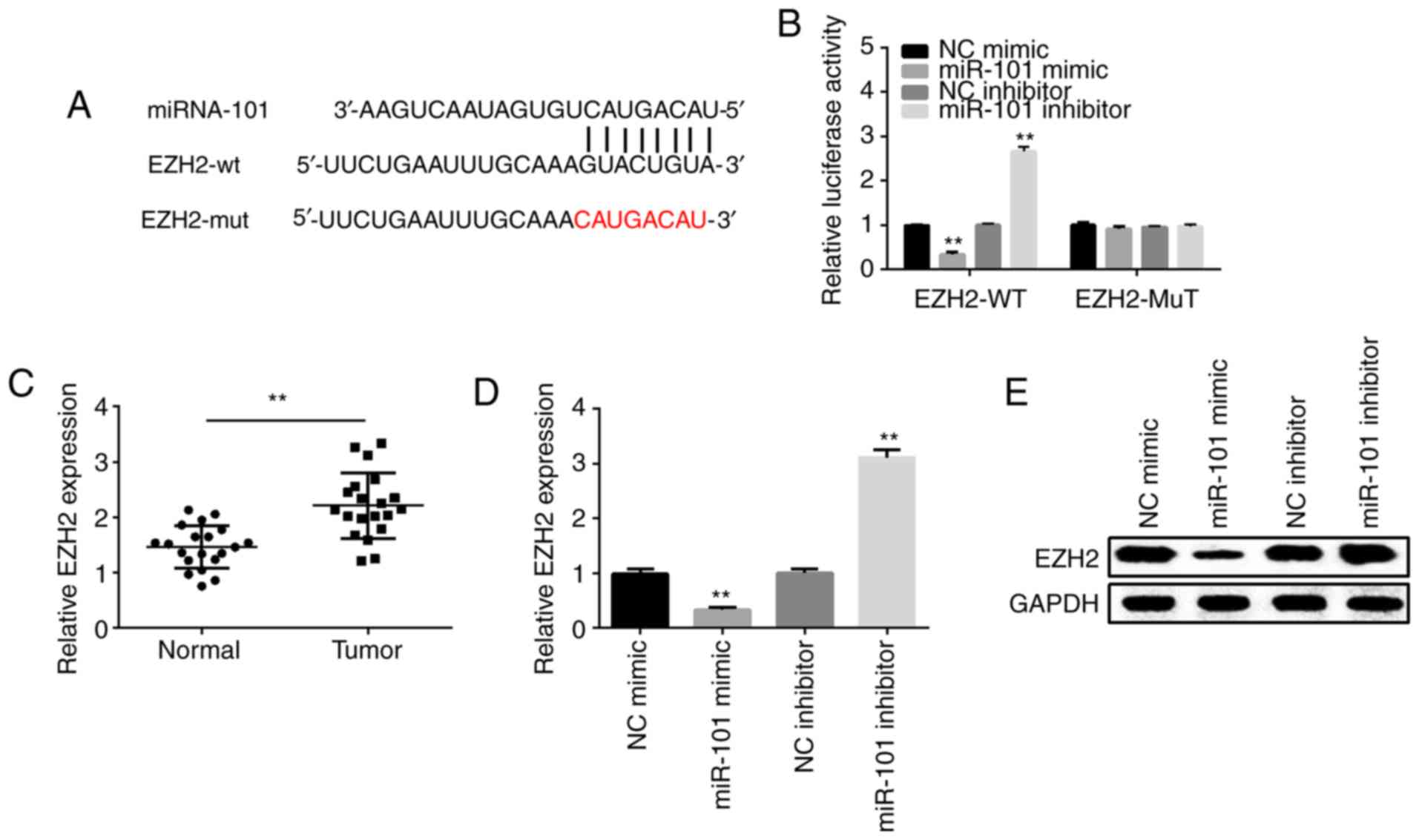
EZH2 is a target gene of miR-101 in
OSCC cells
TargetScan online software (http://www.targetscan.org/vert_71/) was used to detect
the potential target of miR-101. It was found that EZH2 contains a
putative binding site for miR-101 as shown in Fig. 5A. The results of dual-luciferase
reporter assay indicated that the miR-101 mimic significantly
reduced the luciferase activity of EZH2 3′-UTR-WT in the SCC9
cells. However, no significant change was noted in the EZH2
3′-UTR-MuT in SCC9 cells (Fig. 5B).
Moreover, the result of qPCR indicated that EZH2 expression was
significantly higher in OSCC tissues in comparison with that in the
normal tissues (Fig. 5C).
Overexpression of miR-101 suppressed EZH2 expression, while
silencing of miR-101 expression enhanced EZH2 expression both at
the mRNA and protein levels (Fig. 5D and
E). These findings suggest that EZH2 is a direct target of
miR-101.
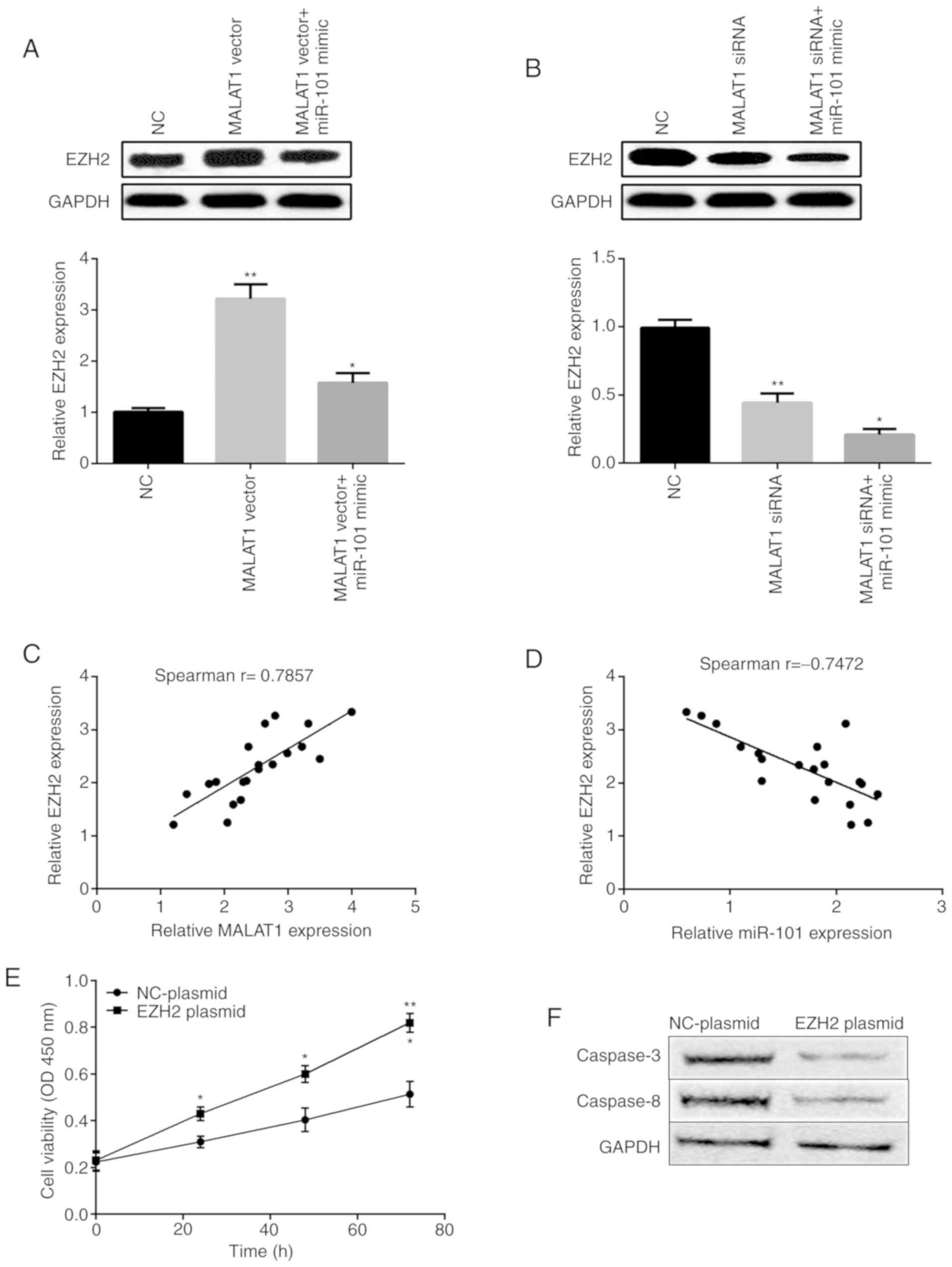
EZH2 acts as a downstream gene of
MALAT1 in OSCC cells
Then, we investigated whether EZH2 is regulated by
MALAT1 through miR-101. As shown in Fig.
6A, EZH2 mRNA and protein expression was significantly enhanced
by MALAT1 overexpression and then inhibited by miR-101 mimic.
However, silencing of MALAT1 expression inhibited EZH2 expression
while miR-101 mimic further suppressed EZH2 expression (Fig. 6B). In addition, the results of
Pearson's correlation coefficient demonstrated that EZH2 expression
was positively associated with MALAT1 expression (Fig. 6C), but negatively correlated with
miR-101 expression in OSCC tissues (Fig.
6D). As shown in Fig. 6E, EZH2
overexpression facilitated SCC9 cell viability. The results of the
western blot analysis displayed that the levels of caspase-3 and
caspase-8 were decreased by the EZH2 plasmid (Fig. 6F). This may provide an added
advantage to establish the oncogenic and tumor-suppressive roles of
MALAT1 and miR-101 via EZH2.
Discussion
Long non-coding RNAs (lncRNAs) are a class of
transcripts with more than 200 nucleotides that do not exhibit
protein-coding abilities (17).
Recent research has shown that lncRNAs play a pivotal role in the
development of many tumors, but the related mechanism remains to be
investigated (18). In this study,
we revealed that metastasis associated lung adenocarcinoma
transcript 1 (MALAT1) was upregulated in oral squamous cell
carcinoma (OSCC) tissues and cell lines (HSC3, SCC9, SCC15 and
SCC25).
MALAT1 is an important lncRNA in tumor progression.
Previous studies have shown that MALAT1 is upregulated in
colorectal cancer (CRC), pancreatic ductal adenocarcinoma and head
and neck squamous cell carcinoma (19–21).
Moreover, Xu et al displayed that MALAT1 overexpression
promoted the viability and migration of CRC (22). Pan et al found that
overexpression of MALAT1 enhanced the invasion and migration of
hepatocellular carcinoma (23).
Huang et al implicated the role of MALAT1 in facilitating
the angiogenesis of breast cancer (24). Zhu et al found that MALAT1
displayed facilitating effect on gastric cancer cell proliferation
and metastasis (25). Moreover, Lin
et al displayed that MALAT1 was upregulated in gallbladder
cancer and its high expression was associated with poor prognosis
(26). Stone et al showed
high expression of MALAT1 in breast cancer and MALAT1 was
associated with tumor development (27). Moreover, Zhou et al revealed
that MALAT1 was elevated in OSCC and promoted tumor growth and
metastasis (28). Our findings
revealed that MALAT1 enhanced OSCC cell proliferation and invasion,
which supporting previous observations that MALAT1 acts as an
oncogene in OSCC development (29).
The results of this study also displayed that the levels of
caspase-3 and caspase-8 were decreased by MALAT1 overexpression,
while increased by silencing of MALAT1. In the present study, we
chose to investigate caspase-3 and caspase-8 but not caspase-9,
which may be the limitation of the present study.
As the development of RNA-sequencing technologies,
many non-coding RNAs have been identified. lncRNAs possess
important functions in development, immunology and cancer. For
example, lncRNA CASC2 was found to be downregulated in OSCC and
markedly inhibited cell proliferation via targeting microRNA-21
(30). On the other hand, Zhang
et al showed that lncRNA PAPAS conferred a promotive effect
on OSCC development as an oncogene by modulating the TGF-β1
signaling pathway (31). In
addition, lncRNA LEF1-AS1 was found to act as an oncogene in OSCC
progression and silencing of LEF1-AS1 suppressed cell
proliferation, migration and tumor growth through the Hippo
signaling pathway (32).
Recently, a growing number of reports have suggested
that lncRNAs act as ‘sponges’ to bind specific miRNAs and regulate
their function. Previous research has established that MALAT1
promoted hepatocellular carcinoma cell migration and invasion by
targeting miR-142-3p (33). Chang
and Hu identified the biological function of MALAT1 in promoting
OSCC development via targeting the miR-125b/STAT3 axis (29). Here, we showed that MALAT1 is a
target of miR-101 by bioinformatics analysis and luciferase
reporter assays. Thus, our research is the first to report that
MALAT1 regulates OSCC proliferation and invasion by targeting
miR-101.
By using TargetScan, we predicted Enhancer of zeste
2 polycomb repressive complex 2 subunit (EZH2) as a direct target
of miR-101. Moreover, luciferase reporter assay further determined
EZH2 as a direct target of miR-101. EZH2 has been reported to be
involved in OSCC progression as the target of several miRNAs, such
as miR-200, miR-138 and miR-32 (34–36).
Here, we demonstrated that MALAT1 can regulate EZH2 expression by
modulating miR-101 expression. Overexpression of MALAT1
significantly promoted EZH2 expression, while treatment with the
miR-101 mimic attenuated EZH2 expression. Conversely, knockdown of
MALAT1 inhibited EZH2 expression, while transfection with the
miR-101 mimic further suppressed EZH2 expression.
In conclusion, the present study aimed to discover
the biological function and molecular mechanism of lncRNA MALAT1 in
OSCC development. The findings showed that MALAT1 was upregulated,
while miR-101 was downregulated in OSCC tissues and cell lines.
MALAT1 modulated OSCC progression by negatively regulating miR-101.
Moreover, EZH2 was determined as the target of miR-101 by
bioinformatics analysis. Taken together, MALAT1 regulates EZH2
expression by modulating miR-101, which may be considered as a
novel target in OSCC treatment.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LX first author contributed significantly to
analysis and manuscript preparation. WW as the co-first author
performed the data analyses and wrote the manuscript. HX as the
corresponding author contributed to the conception of the study.
JZ, SL and XY helped perform the analyses with constructive
discussions. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Affiliated Yantai Yuhuangding Hospital of Qingdao University.
Signed written informed consents were obtained from the patients
and/or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Choi S and Myers JN: Molecular
pathogenesis of oral squamous cell carcinoma: Implications for
therapy. J Dent Res. 87:14–32. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnson NW, Jayasekara P and Amarasinghe
AA: Squamous cell carcinoma and precursor lesions of the oral
cavity: Epidemiology and aetiology. Periodontol 2000. 57:19–37.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sinevici N and O'sullivan J: Oral cancer:
Deregulated molecular events and their use as biomarkers. Oral
Oncol. 61:12–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang S, Hui Y, Li X and Jia Q: Silencing
of lncRNA CCDC26 restrains the growth and migration of glioma cells
in vitro and in vivo via targeting miR-203. Oncol Res.
26:1143–1154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang JJ, Wang DD, Du CX and Wang Y: Long
noncoding RNA ANRIL promotes cervical cancer development by acting
as a sponge of miR-186. Oncol Res. 26:345–352. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao YL, Zhao ZS, Zhang MY, Han LJ, Dong YJ
and Xu B: Long noncoding RNA PVT1 facilitates cervical cancer
progression via negative regulating of miR-424. Oncol Res.
25:1391–1398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hua F, Li CH, Chen XG and Liu XP: Long
noncoding RNA CCAT2 knockdown suppresses tumorous progression by
sponging miR-424 in epithelial ovarian cancer. Oncol Res.
26:241–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
11
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu B, Lei D, Wang L, Yang X, Jia S, Yang
Z, Shan C, Yang X, Zhang C and Lu B: MiRNA-101 inhibits oral
squamous-cell carcinoma growth and metastasis by targeting zinc
finger E-box binding homeobox 1. Am J Cancer Res. 6:1396–1407.
2016.PubMed/NCBI
|
|
13
|
Jiang M, Xu B, Li X, Shang Y, Chu Y, Wang
W, Chen D, Wu N, Hu S, Zhang S, et al: O-GlcNAcylation promotes
colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2
regulatory feedback circuit. Oncogene. 38:301–316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen L, Jia J, Zang Y, Li J and Wan B:
MicroRNA-101 regulates autophagy, proliferation and apoptosis via
targeting EZH2 in laryngeal squamous cell carcinoma. Neoplasma.
66:507–515. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jin Q, He W, Chen L, Yang Y, Shi K and You
Z: MicroRNA-101-3p inhibits proliferation in retinoblastoma cells
by targeting EZH2 and HDAC9. Exp Ther Med. 16:1663–1670.
2018.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bertone P, Stolc V, Royce TE, Rozowsky JS,
Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et
al: Global identification of human transcribed sequences with
genome tiling arrays. Science. 306:2242–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: A new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang D, Yang Z, Long F, Luo L, Yang B, Zhu
R, Sang X and Cao G: Inhibition of MALAT1 reduces tumor growth and
metastasis and promotes drug sensitivity in colorectal cancer. Cell
Signal. 57:21–28. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yao Y, Chen X, Lu S, Zhou C, Xu G, Yan Z,
Yang J, Yu T, Chen W, Qian Y, et al: Circulating long noncoding
RNAs as biomarkers for predicting head and neck squamous cell
carcinoma. Cell Physiol Biochem. 50:1429–1440. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhuo M, Yuan C, Han T, Cui J, Jiao F and
Wang L: A novel feedback loop between high MALAT-1 and low
miR-200c-3p promotes cell migration and invasion in pancreatic
ductal adenocarcinoma and is predictive of poor prognosis. BMC
Cancer. 18:10322018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu Y, Zhang X, Hu X, Zhou W, Zhang P,
Zhang J, Yang S and Liu Y: The effects of lncRNA MALAT1 on
proliferation, invasion and migration in colorectal cancer through
regulating SOX9. Mol Med. 24:522018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pan Y, Tong S, Cui R, Fan J, Liu C, Lin Y,
Tang J, Xie H, Lin P, Zheng T and Yu X: Long non-coding MALAT1
functions as a competing endogenous RNA to regulate vimentin
expression by sponging miR-30a-5p in hepatocellular carcinoma. Cell
Physiol Biochem. 50:108–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang XJ, Xia Y, He GF, Zheng LL, Cai YP,
Yin Y and Wu Q: MALAT1 promotes angiogenesis of breast cancer.
Oncol Rep. 40:2683–2689. 2018.PubMed/NCBI
|
|
25
|
Zhu K, Ren Q and Zhao Y: lncRNA MALAT1
overexpression promotes proliferation, migration and invasion of
gastric cancer by activating the PI3K/AKT pathway. Oncol Lett.
17:5335–5342. 2019.PubMed/NCBI
|
|
26
|
Lin N, Yao Z, Xu M, Chen J, Lu Y, Yuan L,
Zhou S, Zou X and Xu R: Long noncoding RNA MALAT1 potentiates
growth and inhibits senescence by antagonizing ABI3BP in
gallbladder cancer cells. J Exp Clin Cancer Res. 38:2442019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stone JK, Kim JH, Vukadin L, Richard A,
Giannini HK, Lim SS, Tan M and Ahn EE: Hypoxia induces cancer
cell-specific chromatin interactions and increases MALAT1
expression in breast cancer cells. J Biol Chem. 294:11213–11224.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren
Y, Wu Y, Mei M, Zhang L and Wang X: Long Non-coding RNA MALAT1
promotes tumor growth and metastasis by inducing
epithelial-mesenchymal transition in oral squamous cell carcinoma.
Sci Rep. 5:159722015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang SM and Hu WW: Long non-coding RNA
MALAT1 promotes oral squamous cell carcinoma development via
microRNA-125b/STAT3 axis. J Cell Physiol. 233:3384–3396. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan L, Chen H, Bai Y, Wang Q and Chen L:
Long non-coding RNA CASC2 serves as a ceRNA of microRNA-21 to
promote PDCD4 expression in oral squamous cell carcinoma. Onco
Targets Ther. 12:3377–3385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang P, Liu Y, Li C, Zhang L, Liu Q and
Jiang T: LncRNA PAPAS promotes oral squamous cell carcinoma by
upregulating transforming growth factor-β1. J Cell Biochem.
120:16120–16127. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang C, Bao C, Zhang X, Lin X, Pan D and
Chen Y: Knockdown of lncRNA LEF1-AS1 inhibited the progression of
oral squamous cell carcinoma (OSCC) via Hippo signaling pathway.
Cancer Biol Ther. 20:1213–1222. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu Q, Xiang L, Chen Z, Liu X, Ou H, Zhou J
and Yang D: MALAT1 functions as a competing endogenous RNA to
regulate SMAD5 expression by acting as a sponge for miR-142-3p in
hepatocellular carcinoma. Cell Biosci. 9:392019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Guo W, Li Z, Wu Y, Jing C, Ren Y,
Zhao M, Kong L, Zhang C, Dong J, et al: Role of the EZH2/miR-200
axis in STAT3-mediated OSCC invasion. Int J Oncol. 52:1149–1164.
2018.PubMed/NCBI
|
|
35
|
Hong Y, He H, Sui W, Zhang J, Zhang S and
Yang D: Long non-coding RNA H1 promotes cell proliferation and
invasion by acting as a ceRNA of miR-138 and releasing EZH2 in oral
squamous cell carcinoma. Int J Oncol. 52:901–912. 2018.PubMed/NCBI
|
|
36
|
Zhang D, Ni Z, Xu X and Xiao J: miR-32
functions as a tumor suppressor and directly targets EZH2 in human
oral squamous cell carcinoma. Med Sci Monit. 20:2527–2535. 2014.
View Article : Google Scholar : PubMed/NCBI
|