Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common malignant tumor globally and >700,000 people die from
this disease every year (1).
Patients with HCC do not present with obvious symptoms until
advanced stages of the disease, making treatment difficult and
ineffective (2). Hence, it is vital
to identify some potential biomarkers for the diagnosis or
prognosis of HCC.
The extracellular environment of solid tumors is
acidic, with a pH between 6.5 and 6.9, whereas normal tissues is
alkaline, with a pH between 7.2 and 7.5 (3). Tissue acidosis can result in tumor
progression (4). Therefore,
interventions targeting the acidic microenvironment of tumors may
provide new therapeutic opportunities. However, the mechanism of
action behind the generation of the acidic microenvironment in
tumorigenesis is still in the initial stage, with further research
urgently needed.
In order to maintain the stability of the pH, tumor
cells transport acidic substances to the outside or transport
extracellular basic substances into the cells through a series of
transport proteins, such as the H+-ATPase and the
Na+/H+ exchanger 1 (5). The transmembrane protein (TMEM) family
contains proteins that span the entire width of the lipid bilayer
and are permanently immobilized (6).
A number of TMEMs act as channels, allowing specific substances to
be transported between the intracellular and extracellular
environment (7). However, the
functions of TMEMs are unclear, and investigations into their
functions are urgently required.
TMEMs are present in various cell types and are
involved in a number of important physiological processes. For
example, TMEM16A acts as a calcium-activated chloride channel
(8,9), whereas TMEM132A may be involved in
brain development during the embryonic stages (10). Furthermore, studies have confirmed
that TMEMs may serve an important role in tumor growth and
development (11,12). For instance, TMEM88 binds to
disheveled proteins and promotes the invasion and metastasis of
non-small cell lung cancer by activating the p38/glycogen synthase
kinase 3β/Snail pathway (12).
Recently, Yang et al demonstrated that one
type of CI− channels was activated by an acidic
extracellular pH, and it participated in the proton-activated
CI− (PAC) currents; this channel protein was confirmed
to be TMEM206, also termed PAC or pacc1 (13). However, little is known about
TMEM206. Based on the results of the aforementioned study, it may
be speculated that TMEM206 also serve a fundamental role in
tumorigenesis and progression. To improve the understanding of
TMEM206, the present study aimed to examine the expression
profiling and prognostic values of TMEM206 in human cancers using
multiple public databases, and potential co-expression genes that
may be associated with the dysregulation of TMEM206 in
hepatocellular carcinoma (HCC) were explored, which may be
beneficial for the further study of TMEM206.
Materials and methods
Expression profiling of TMEM206 in
human cancers
Gene expression profiling interactive analysis
(GEPIA, http://gepia.cancer-pku.cn) and
Oncomine (http://www.oncomine.org) databases were
used to explore the potential features of TMEM206 in human tumors
and normal tissues. GEPIA software is a web tool that analyzes the
RNA expression data from The Cancer Genome Atlas (TCGA) and
Genotype-Tissue Expression (GTEx) projects (https://www.cancer.gov/) (14). The Oncomine database is a web server
used for bioinformatics services containing 715 datasets that
provides a large-scale, high quality and consistent analytical
method for gene expression profile analysis (15).
Pan-cancer survival analysis
The Kaplan-Meier plotter (http://kmplot.com//analysis) (16) was used to assess the prognostic roles
of TMEM206 in 10,461 cancer samples. The forest plot was
constructed using the R programming language (17). The Human Protein Atlas (HPA) dataset
(https://www.proteinatlas.org/) (18,19) is
based on TCGA and was used to analyze the association between
TMEM206 RNA and protein expression and overall survival rate of
patients with HCC. Additionally, the UCSC Xena browser (http://xena.ucsc.edu) (20) was used to evaluate the association
between TMEM206 mRNA expression levels and the prognosis of
patients with HCC. Due to the crossing of survival curves, Simon's
two-stage test was used rather than the log-rank test (21,22).
Potential transcription regulatory
mechanisms of TMEM206
The cBioPortal for Cancer Genomics (http://www.cbioportal.org) was used as a web resource
to analyze and visualize the genomics data. The cBioPortal was also
used to obtain the potential co-expression genes of TMEM206 in HCC
(|Spearman's correlation coefficient |>0.4) (23). Additionally, the potential
co-expression genes of TMEM206 were extracted from Gene Expression
Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) datasets GSE36376
and GSE76427, separately, which have previously been used to
analyze gene expression for tumor and adjacent non-tumor tissues in
HCC (24,25). To identify the genes co-expressed
with TMEM206, a Spearman's correlation test was used by the R
platform (|Spearman's correlation coefficient|>0.4) (26). Finally, using online Venn software
(http://bioinformatics.psb.ugent.be/webtools/Venn/),
the intersection of these gene lists was determined as the
co-expression genes of TMEM206 in HCC.
The miRWalk (27,28),
miRDB (29) and TargetScan (30) databases were used to identify
potential interacting microRNAs (miRNAs/miRs) (TargetScan,
context++ score percentile >80; miRDB, score >80 and miRWalk:
Score >0.8). The shared miRNAs among the three sets of
prediction results were regarded as potential miRNAs that target
TMEM206 mRNA.
Functional annotation of a
co-expression gene network of TMEM206
Kyoto Encyclopedia of Genes and Genomes (KEGG)
(31) and gene ontology (GO)
(32,33) are commonly used for the functional
annotations of genes. MetaScape (https://metascape.org/gp/index.html#/main/step1) is an
integrated analytics platform that integrates multiple annotation
datasets. The functional enrichment analysis was performed using
these platforms to analyze the function of TMEM206 co-expression
genes (34).
Results
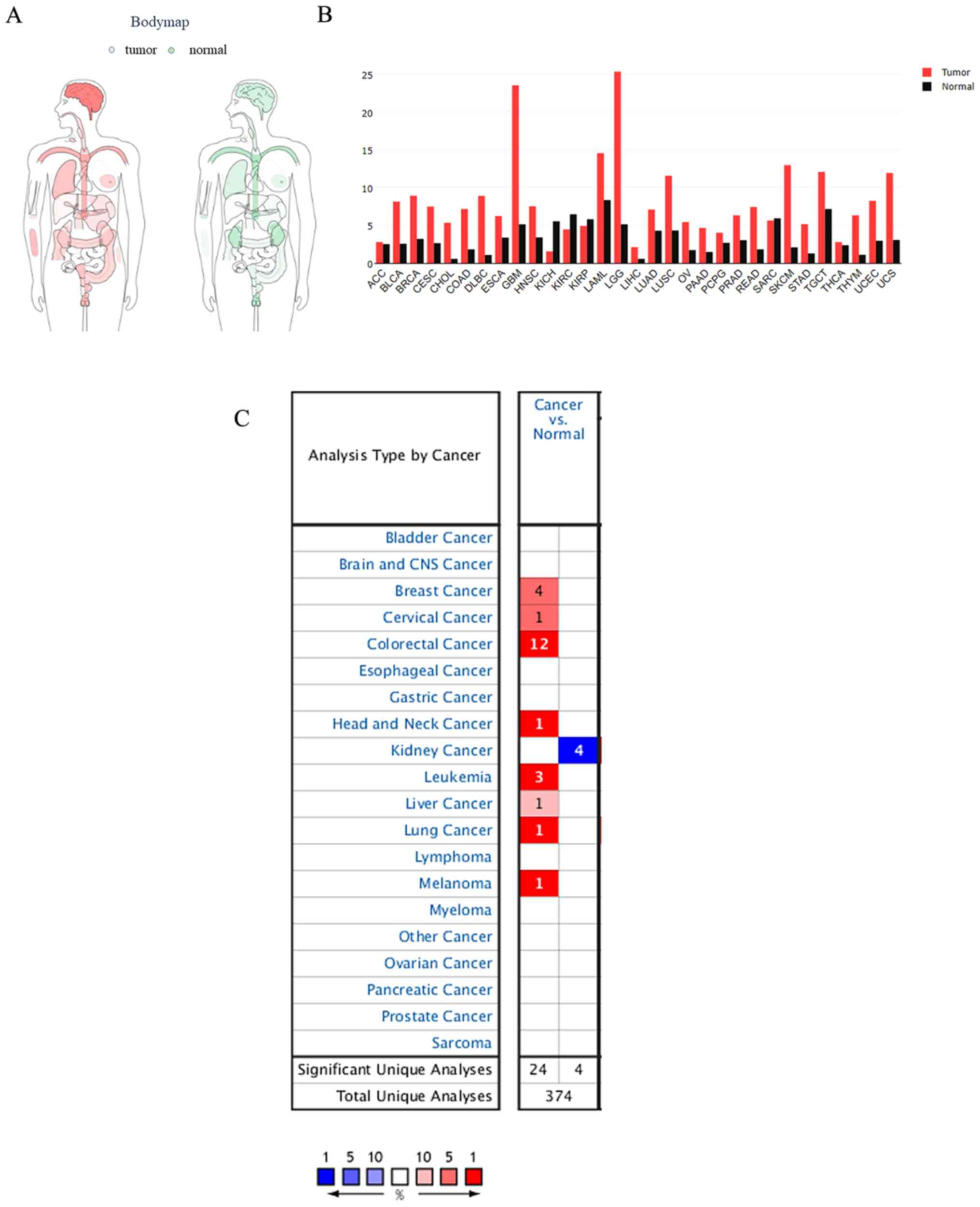
Expression of TMEM206 in cancer
The distribution of TMEM206 expression in various
tissues of the human body based on TCGA and GTEx projects was
obtained from the GEPIA website (Fig.
1A). TMEM206 was observed to be abundant in human tissues,
especially in the brain and blood (Fig.
1B). Subsequently, the differential expression of TMEM206 in
human tumor types and normal control tissues was investigated using
the Oncomine database; the results demonstrated that 30 out of 150
tumors exhibited TMEM206 upregulation, and eight exhibited
downregulation compared with that in normal tissues (Fig. 1C). Although, TMEM206 was abundant in
normal and malignant tissues, significant increases in TMEM206
expression level were observed in colorectal, breast and liver
cancer and lymphoma, and decreased levels were observed in kidney
cancer compared with those in the corresponding normal tissues.
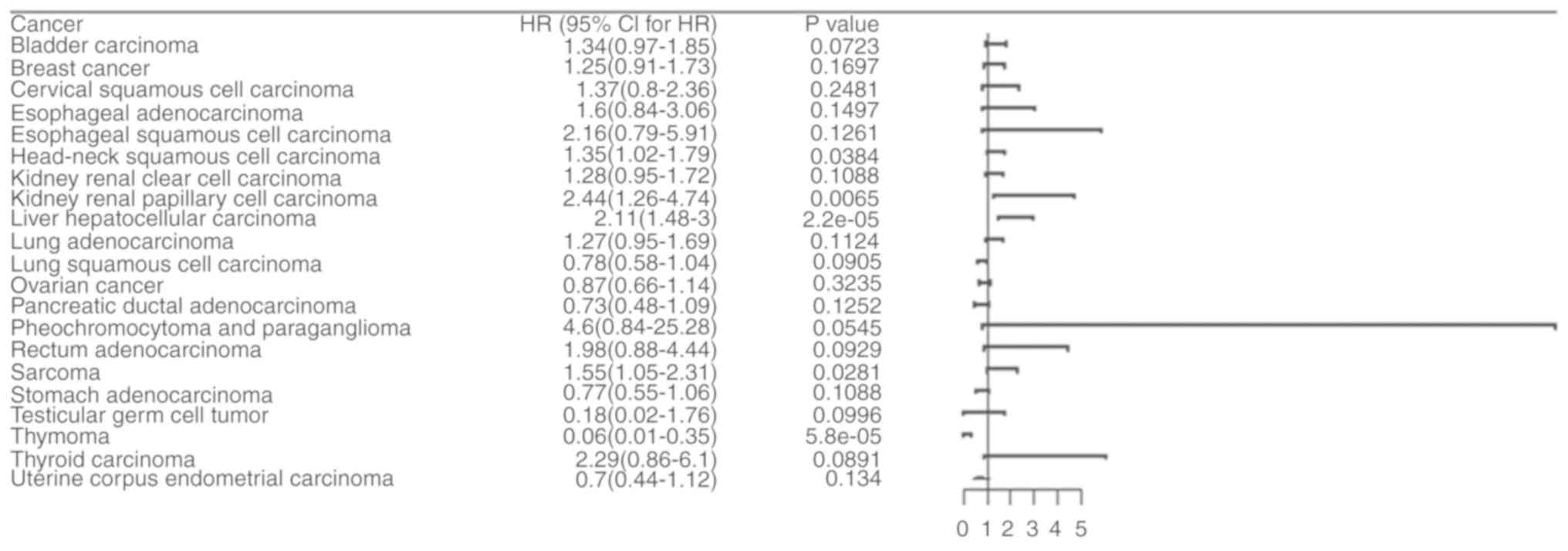
Pan-cancer survival analysis of
TMEM206 mRNA expression suggests a prognostic role of TMEM206 in
HCC
The Kaplan-Meier plotter datasets were used for
pan-cancer survival analysis. The results demonstrated that TMEM206
exhibited notable prognostic significance in 5 of the 21 analyzed
types of tumors (Table SI).
Heterogeneity among various types of cancer was observed (Fig. 2). A higher expression level of
TMEM206 was associated with a poor prognosis of head and neck
squamous cell carcinoma, renal papillary cell carcinoma, HCC and
sarcoma, while a lower expression level of TMEM206 was associated
with a poor prognosis of thymoma (Fig.
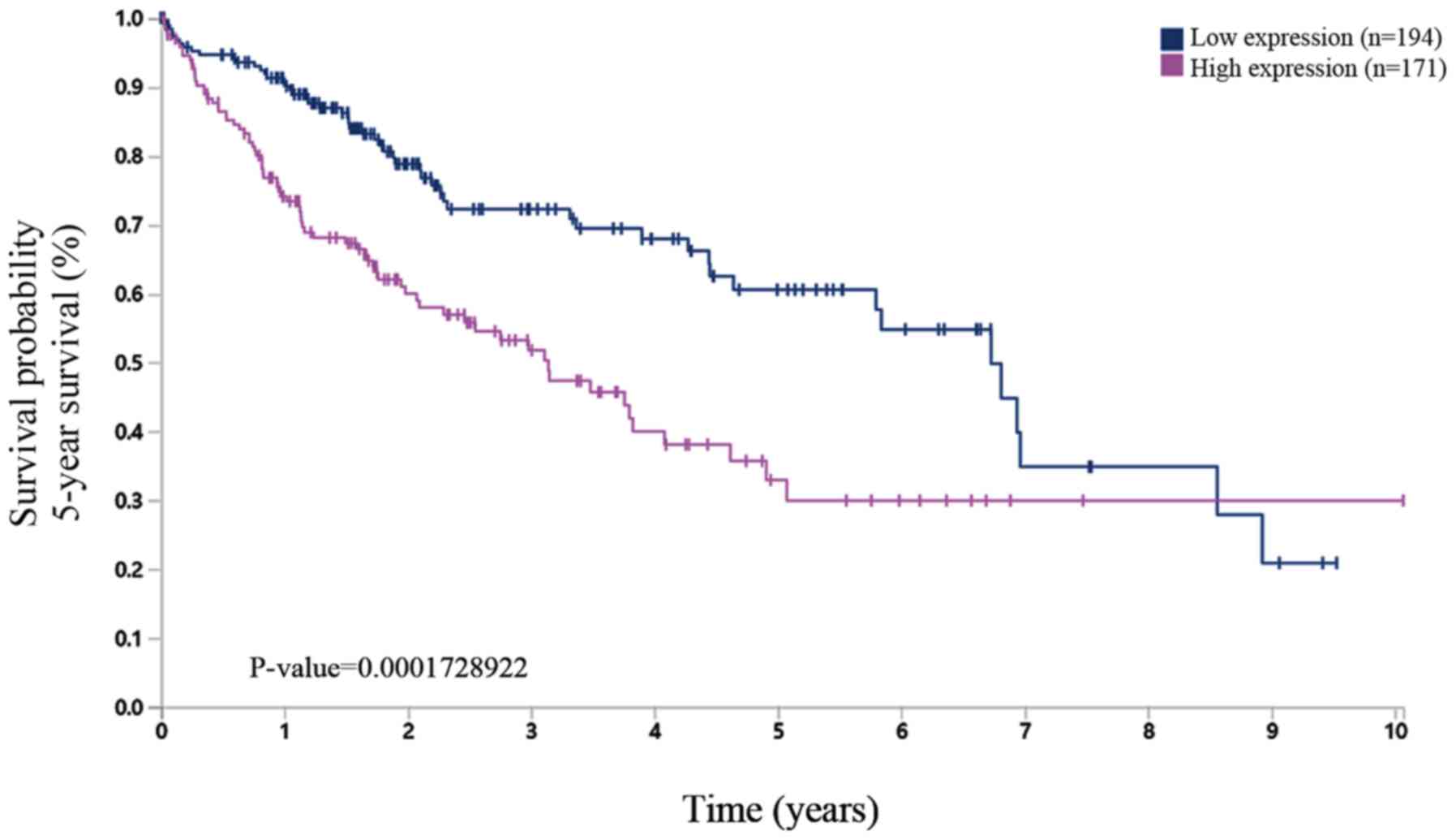
2). Subsequently using the HPA dataset, which is based on
experimental data, it was found that high protein expression level
of TMEM206 was related to a poor survival rate of HCC (Fig. 3). Therefore, the present study
focused on TMEM206 in HCC for the subsequent investigation.
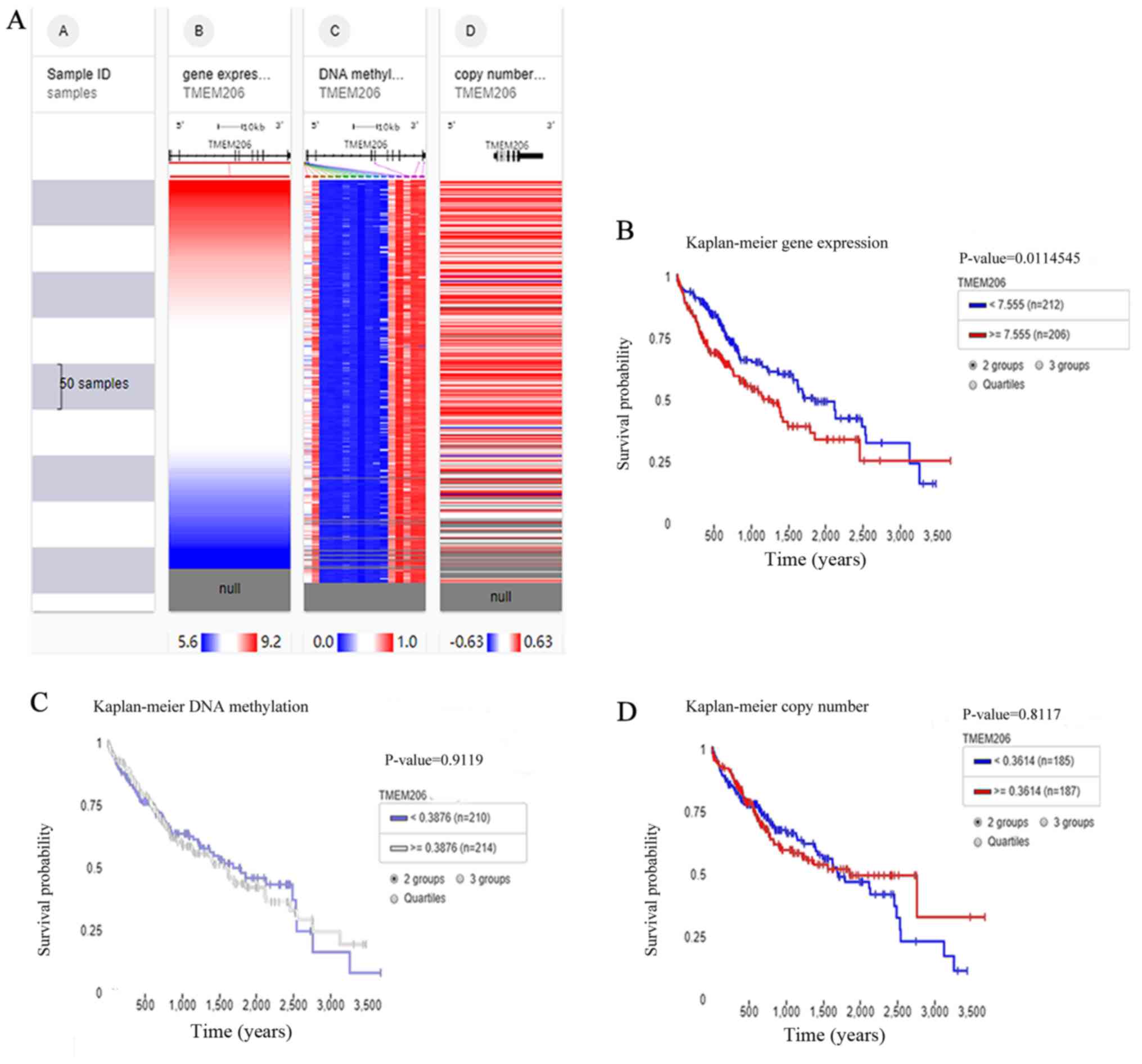
The UCSC Xena browser was used to evaluate the
association between the DNA variants and upregulation of TMEM206 in
HCC. Expression was colored red to blue for high to low expression
(Fig. 4A). Survival analysis using
the Simon's two-stage test (low expression, n=212 and high
expression, n=206, respectively) demonstrated that high expression
of TMEM206 mRNA was an unfavorable prognostic marker for patients
with HCC (Fig. 4B). However, the
methylation status and copy number variant of TMEM206 DNA were not
associated with survival in HCC (Fig. 4C
and D). As there was no significant difference, the Simon's
two-stage test was not used inspite of the crossover of survival
curves.
Functional annotation of the TMEM206
co-expression gene network
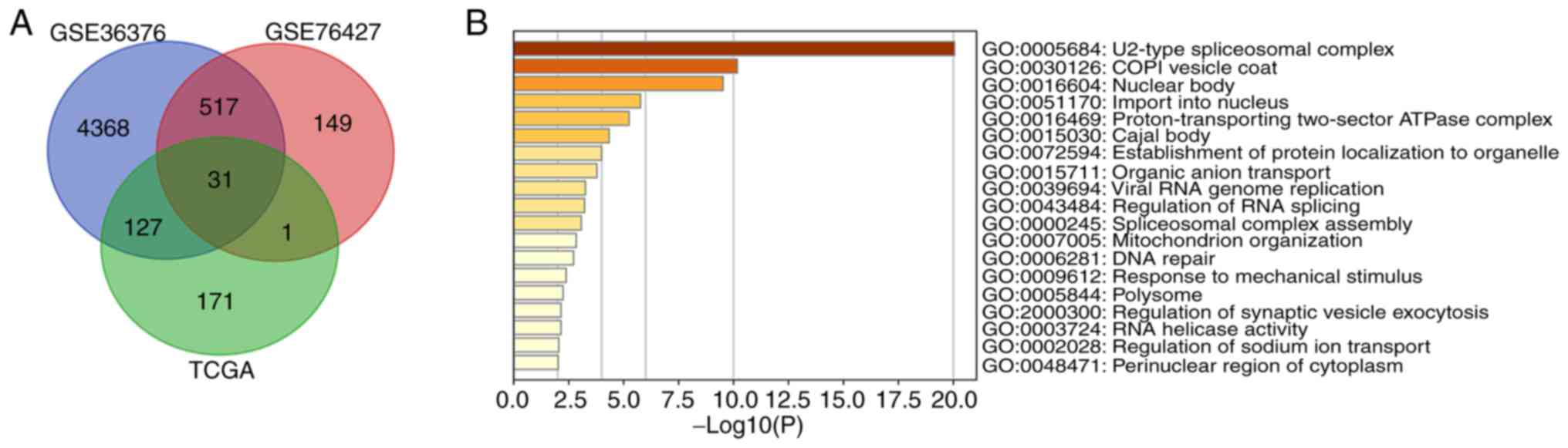
Co-expression gene analysis was used to further
investigate the possible functions of TMEM206 in HCC. Data were
acquired from cBioPortal and the GEO datasets GSE36376 and GSE76427
(|Spearman's correlation coefficient |>0.4|). The intersections
of the three gene lists in the Venn diagram were considered to be
potential co-expression genes for TMEM206 (Fig. 5A; Table
SII). MetaScape was used to analyze functional gene enrichment;
the most significantly enriched GO term was the ‘U2-type
spliceosomal complex’, which suggested that may regulate the
expression of TMEM206 and the co-expression genes associated with
it (Fig. 5B).
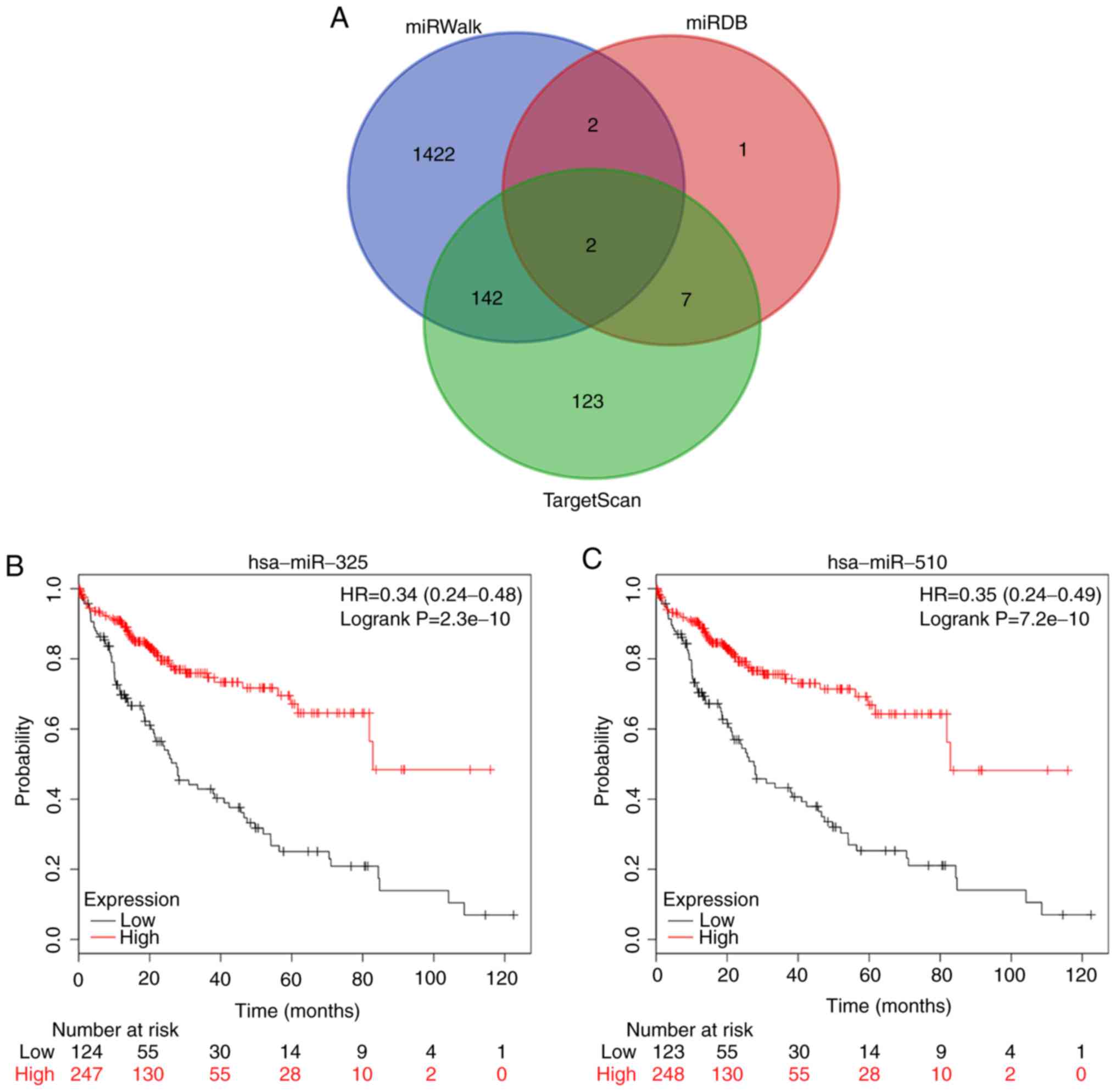
In addition, the miRWalk, miRDB and TargetScan
databases were used to identify the potential miRNAs involved in
the regulation of TMEM206. With strict screening criteria, two
microRNAs, hsa-miR-325 and hsa-miR-510-5p (also termed has-miR-510)
that may bind to TMEM206 mRNA were identified (Fig. 6A). Kaplan-Meier survival analysis
demonstrated that hsa-miR-325 and hsa-miR-510-5p were associated
with the prognosis of patients with HCC. High expression levels of
hsa-miR-325 and hsa-miR-510-5p were favorable for overall survival
(P=2.3×10−10 and P=7.2×10−10, respectively;
Fig. 6B and C), suggesting that
hsa-miR-325 and hsa-miR-510-5p expression levels were negatively
associated with TMEM206 in HCC.
Discussion
Numerous studies have demonstrated that TMEM
expression is up- or downregulated in tumor tissues compared with
that in adjacent healthy or benign tissues (35,36). For
example, TMEM97 acts as prognostic biomarker for non-small cell
lung cancer (35). Furthermore, a
previous study has reported that TMEM proteins are involved in
tumorigenesis and cancer development, such as TMEM45A, which
participates in the proliferation and invasion of ovarian cancer
cells (37). Therefore, an improved
understanding of the TMEM family may help identify their functions
in various types of cancer and improve therapeutic strategies.
TMEM206 is a newly identified transmembrane protein,
Yang et al (13) performed
unbiased RNA interference screening and demonstrated that TMEM206
was essential for the widely observed PAC currents
(ICl,H), which are involved in acid-sensing ion
channels. Little is known about the role of TMEM206 in tumors. Zhao
et al (38) reported that
TMEM206 mRNA and protein expression levels were higher in
colorectal cancer (CRC) tissues compared with adjacent non-tumor
tissues, overexpression of TMEM206 promoted the proliferation and
invasion of CRC cells and positively regulated the levels of
phospho-AKT and its downstream signaling pathway components,
suggesting that TMEM206 promoted the development and progression of
CRC by enhancing the interactions between the AKT and ERK signaling
pathways.
A previous study has reported that the TMEM family
is widely expressed in mammalian cells (39), which is in accordance with the
present study. In the present study, the results of the expression
profiling analysis demonstrated that TMEM206 expression was
significantly increased in colorectal cancer, breast cancer,
lymphoma and liver cancer, and reduced in the kidney cancer
datasets. In terms of a prognostic role for TMEM206, the present
study demonstrated that TMEM206 was a significant prognostic factor
for certain types of tumor and served either protective or
unfavorable roles depending on the tumor type. These contrary
results make the role of TMEM206 dubious.
Further analysis of data from patients with HCC
demonstrated that the upregulation of TMEM206 was an unfavorable
prognostic factor for HCC. TMEM206 was associated with the overall
survival probability and 5-year survival rate of patients with
liver cancer in The Kaplan Meier plotter and HPA. In addition, in
the present study, the UCSC Xena browser was used to evaluate the
association between the DNA variants and upregulation of TMEM206 in
HCC. However, no significant associations were observed between the
copy number variant or the extent of methylation of TMEM206 and the
prognosis of patients with HCC. GEO and cBioPortal were used to
determine genes co-expressed with TMEM206 and 31 genes were
discovered. Using MetaScape, the signalling pathways that
co-expression genes of TMEM206 may participate in were identified
and the term U2-type spliceosomal complex was most significant.
Finally, the present study identified two potential miRNAs,
hsa-miR-325 and hsa-miR-510-5p, that may target TMEM206.
Hsa-miR-325 and hsa-miR-510-5p participate in tumor
development (40–43). Hsa-miR-325 has been identified as a
potential biomarker in human bladder cancer and low expression of
hsa-miR-325 was associated with poor overall survival in patients
with bladder cancer (40). In
addition, downregulation of hsa-miR-325 has been demonstrated to
promote the progression of non-small cell lung cancer and HCC
(41,42). Chen et al (43) found that hsa-miR-510-5p acted as a
tumor suppressor in renal cell carcinoma by reducing cell
proliferation, migration and inducing apoptosis.
In conclusion, the results of the present study
demonstrated that upregulation of TMEM206 is a potential prognostic
indicator for HCC. However, studies into the role of TMEM206 are
still at the early stage, and further research is required to
clarify its role in HCC and other types of tumors.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81860276).
Availability of data and materials
The datasets used/and or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ, SYL, XY, YQW and YXC contributed toward data
analysis, drafting and revising the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zeng Z, Dong J, Li Y, Dong Z, Liu Z, Huang
J, Wang Y, Zhen Y and Lu Y: The expression level and diagnostic
value of microRNA-22 in HCC patients. Artif Cells Nanomed
Biotechnol. 48:683–686. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heimbach JK, Kulik LM, Finn RS, Sirlin CB,
Abecassis MM, Roberts LR, Zhu AX, Murad MH and Marrero JA: AASLD
guidelines for the treatment of hepatocellular carcinoma.
Hepatology. 67:358–380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Griffiths JR: Are cancer cells acidic? Br
J Cancer. 64:425–427. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alfarouk KO, Ahmed SB, Ahmed A, Elliott
RL, Ibrahim ME, Ali HS, Wales CC, Nourwali I, Aljarbou AN, Bashir
AH, et al: The interplay of dysregulated pH and electrolyte
imbalance in cancer. Cancers (Basel). 7:8982020. View Article : Google Scholar
|
|
5
|
White SH: Biophysical dissection of
membrane proteins. Nature. 459:344–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jung SJ, Jung Y and Kim H: Proper
insertion and topogenesis of membrane proteins in the ER depend on
sec63. Biochim Biophys Acta Gen Subj. 1863:1371–1380. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marx S, Dal Maso T, Chen JW, Bury M,
Wouters J, Michiels C and Le Calvé B: Transmembrane (TMEM) protein
family members: Poorly characterized even if essential for the
metastatic process. Semin Cancer Biol. 60:96–106. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hartzell HC, Yu K, Xiao Q, Chien LT and Qu
Z: Anoctamin/TMEM16 family members are Ca2+-activated Cl-channels.
J Physiol. 587:2127–2139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang YD, Cho H, Koo JY, Tak MH, Cho Y,
Shim WS, Park SP, Lee J, Lee B, Kim BM, et al: TMEM16A confers
receptor-activated calcium-dependent chloride conductance. Nature.
455:1210–1215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oh-hashi K, Imai K, Koga H, Hirata Y and
Kiuchi K: Knockdown of transmembrane protein 132A by RNA
interference facilitates serum starvation-induced cell death in
neuro2a cells. Mol Cell Biochem. 342:117–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y, Song K, Zhang Y, Xu H, Zhang X,
Wang L, Fan C, Jiang G and Wang E: TMEM17 promotes malignant
progression of breast cancer via AKT/GSK3β signaling. Cancer Manag
Res. 10:2419–2428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Yu X, Jiang G, Miao Y, Wang L,
Zhang Y, Liu Y, Fan C, Lin X, Dong Q, et al: Cytosolic TMEM88
promotes invasion and metastasis in lung cancer cells by binding
DVLS. Cancer Res. 75:4527–4537. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang J, Chen J, Del Carmen Vitery M,
Osei-Owusu J, Chu J, Yu H, Sun S and Qiu Z: PAC, an evolutionarily
conserved membrane protein, is a proton-activated chloride channel.
Science. 364:395–399. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Menyhárt O, Nagy Á and Győrffy B:
Determining consistent prognostic biomarkers of overall survival
and vascular invasion in hepatocellular carcinoma. R Soc Open Sci.
5:1810062018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Uhlen M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thul PJ, Åkesson L, Wiking M, Mahdessian
D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science.
356:63402017. View Article : Google Scholar
|
|
20
|
Goldman M, Craft B, Hastie M, Repečka K,
Kamath A, McDade F, Rogers D, Brooks AN, Zhu J and Haussler D: The
UCSC Xena platform for public and private cancer genomics data
visualization and interpretation. bioRxiv 326470. 2019.
|
|
21
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10:e01167742015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qiu P and Sheng J: A two-stage procedure
for comparing hazard rate functions. J Royal Statistical Soc.
70:191–208. 2008.
|
|
23
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lim HY, Sohn I, Deng S, Lee J, Jung SH,
Mao M, Xu J, Wang K, Shi S, Joh JW, et al: Prediction of
disease-free survival in hepatocellular carcinoma by gene
expression profiling. Ann Surg Oncol. 20:3747–3753. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grinchuk OV, Yenamandra SP, Iyer R, Singh
M, Lee HK, Lim KH, Chow PK and Kuznetsov VA: Tumor-adjacent tissue
co-expression profile analysis reveals pro-oncogenic ribosomal gene
signature for prognosis of resectable hepatocellular carcinoma. Mol
Oncol. 12:89–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang S, Jing H, Huang Z, Huang T, Lin S,
Liao M and Zhou J: Identification of key candidate genes in
neuropathic pain by integrated bioinformatic analysis. J Cell
Biochem. 121:1635–1648. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sticht C, De La Torre C, Parveen A and
Gretz N: MiRWalk: An online resource for prediction of microRNA
binding sites. PLos One. 13:e02062392018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du A, Zhao S, Wan L, Liu T, Peng Z, Zhou
Z, Liao Z and Fang H: MicroRNA expression profile of human
periodontal ligament cells under the influence of Porphyromonas
gingivalis LPS. J Cell Mol Med. 20:1329–1338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu W and Wang X: Prediction of functional
microRNA targets by integrative modeling of microRNA binding and
target expression data. Genome Biol. 20:182019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
ELife. 4:e050052015. View Article : Google Scholar
|
|
31
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
The Gene Ontology Resource, . 20 years and
still Going strong. Nucleic Acids Res. 47:D330–D338. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ding H, Gui X, Lin X, Chen R, Ma T, Sheng
Y, Cai H and Fen Y: The prognostic effect of MAC30 expression on
patients with non-small cell lung cancer receiving adjuvant
chemotherapy. Technol Cancer Res Treat. 16:645–653. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang H, Zou L, Ma K, Yu J, Wu H, Wei M and
Xiao Q: Cell-Specific mechanisms of TMEM16A
Ca2+-activated chloride channel in cancer. Mol Cancer.
16:1522017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo J, Chen L, Luo N, Yang W, Qu X and
Cheng Z: Inhibition of TMEM45A suppresses proliferation, induces
cell cycle arrest and reduces cell invasion in human ovarian cancer
cells. Oncol Rep. 33:3124–3130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao J, Zhu D, Zhang X, Zhang Y, Zhou J
and Dong M: TMEM206 promotes the malignancy of colorectal cancer
cells by interacting with AKT and extracellular signal-regulated
kinase signaling pathways. J Cell Physiol. 234:10888–10898. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ullrich F, Blin S, Lazarow K, Daubitz T,
von Kries JP and Jentsch TJ: Identification of TMEM206 proteins as
pore of PAORAC/ASOR acid-sensitive chloride channels. ELife.
8:e491872019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lin T, Zhou S, Gao H, Li Y and Sun L:
MicroRNA-325 is a potential biomarker and tumor regulator in human
bladder cancer. Technol Cancer Res Treat. 17:15330338187905362018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Z, Han Y, Sun G, Liu X, Jia X and Yu
X: MicroRNA-325-3p inhibits cell proliferation and induces
apoptosis in hepatitis B virus-related hepatocellular carcinoma by
down-regulation of aquaporin 5. Cell Mol Biol Lett. 24:132019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yao S, Zhao T and Jin H: Expression of
MicroRNA-325-3p and its potential functions by targeting HMGB1 in
non-small cell lung cancer. Biomed Pharmacother. 70:72–79. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen D, Li Y, Yu Z, Li Y, Su Z, Ni L, Yang
S, Gui Y and Lai Y: Downregulated microRNA-510-5p acts as a tumor
suppressor in renal cell carcinoma. Mol Med Rep. 12:3061–3066.
2015. View Article : Google Scholar : PubMed/NCBI
|