Introduction
According to the 2017 WHO classification of tumors
of hematopoietic and lymphoid tissues, T cell acute lymphoblastic
leukaemia (T-ALL) accounts for ~25% of the cases of adult ALL and
~15% of childhood ALL cases globally (1). Clinically, T-ALL typically presents
with a high leukocyte count and frequently with a large mediastinal
or gastrointestinal mass (1).
Lymphadenopathy and hepatosplenomegaly are common (2). T-ALL compared with B cell acute
lymphoblastic leukaemia often manifests with relative sparing of
normal bone marrow hematopoiesis and is associated with a higher
risk of induction failure, early relapse and isolated central
nervous system relapse (1,2). Although, the use of combination
chemotherapy regimens, including vincristine, daunorubicin, and
prednisolone has gradually improved the clinical outcome of T-ALL
over the last few decades, majority of patients with T-ALL
eventually relapse (3,4). Allogenetic hematopoietic stem cell
transplantation may be the only potential curative therapy
(5) and is also essential to
identify novel agents suitable for elderly and weak patients with
T-ALL.
Linalool, a natural small molecular compound is
isolated from various oils, such as camphor leaf oil, galoin oil
and rosewood oil (6). Linalool is a
colorless liquid that can be mixed with ethanol and ether and is
insoluble in water and glycerol (7).
Currently, linalool is mainly used as an antimicrobial and
antiviral agent, perfume, deodorant, anticaries agent and
insecticide (8,9). In recent years, it has been proven that
linalool can inhibit the malignant proliferation of numerous human
malignant solid tumors, including hepatocellular carcinoma, breast
cancer, small cell carcinoma, and malignant melanoma (10–13).
Linalool has also been demonstrated to have antiproliferative
activity against some hematological diseases, including chronic
myelogenous leukaemia, acute promyelocytic leukaemia and Burkitt's
lymphoma (14–19). However, the effect of linalool on
T-ALL remains unclear.
Linalool is inexpensive and may be developed as a
novel therapeutic agent for tumors (20). In the present study, in order to
reveal the role of linalool on T-ALL the effects of linalool on
T-ALL cell lines as well as peripheral blood mononuclear cells
(PBMCs) from healthy donors were investigated.
Materials and methods
Agents and antibodies
Linalool (with purity ≥95% and molecular
weight=154.25 Da) was purchased from Sigma-Aldrich; Merck KGaA
(cat. no. 78-70-6) and its formula is provided in Fig. 1A. Linalool was dissolved in dimethyl
sulfoxide (DMSO) and used at the required concentration (30 µM
linalool was the most frequently used as it approached 50%
inhibition at 48 h). The final concentration of DMSO was <0.1%
in the RPMI-1640 culture medium (cat. no. 670089; Invitrogen;
Thermo Fisher Scientific Inc.). Mouse antibodies against p38
(1:1,000; cat. no. sc-398305), p-p38 (1:1,000; cat. no. sc-166182),
JNK (1:1,000; cat. no. sc-7345), p-JNK (1:1,000; cat. no. sc-6254),
β-actin (1:1,000; cat. no. sc-47778), poly (ADP-ribose) polymerase
1PARP-1 (1:1,000; cat. no. sc-8007), cleaved PARP-1 (1:1,000; cat.
no. sc-56196), were purchased from Santa Cruz Biotechnology Inc.
PD98059 (cat. no. 9900), SP600125 (cat. no. 8177), SB203580 (cat.
no. 5633) and rabbit primary antibodies against ERK1/2 (1:1,000;
cat. no. 4376), p-ERK1/2 (1:1,000; cat. no. 4370), Growth Arrest
And DNA Damage Inducible α (GADD45A) (1:1,000; cat. no. 4632S),
caspase-3 (1:1,000; cat. no. 9662), cleaved caspase-3 (1:1,000;
cat. no. 9664) and secondary antibodies (1:1,000; cat. nos. 7074
and 7076) were purchased from Cell Signaling Technology Inc.
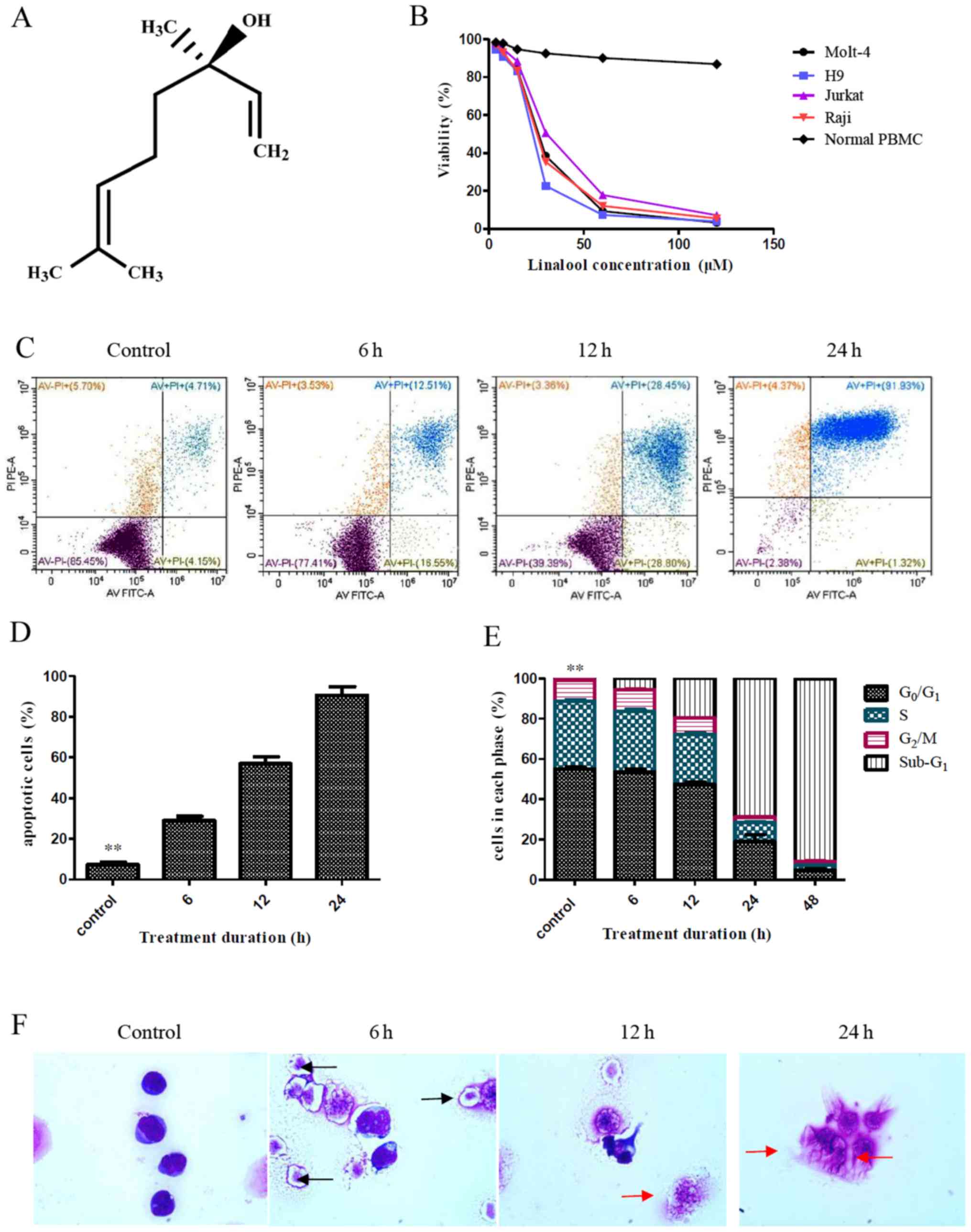 | Figure 1.Effects of linalool treatment on
T-ALL cells. (A) Molecular formula of linalool. (B) Linalool
inhibited the growth of Jurkat, H9, Molt-4 and Raji cells in a
dose-dependent manner, but it had almost no effects on the
viability of normal PBMCs. (C and D) Jurkat cells were treated with
30 µM linalool for 6, 12 or 24 h, respectively. The percentage of
apoptotic cells was significantly increased in a time-dependent
manner compared with the control group (treatment with DMSO only),
**P<0.01. (E) Treatment of Jurkat cells with 30 µM linalool
resulted in a significant decrease of G0/G1,
G2, and S phase cells with a concomitant increase of
sub-G1 phase cells compared with the control group
(treatment with DMSO only), **P<0.01. (F) Jurkat cells were
killed by 30 µM linalool in a time-dependent manner, apoptotic
bodies (black arrows) were present after 6 h treatment and
apoptotic cellular debris (red arrows) were increased after 12 h
treatment, and plenty of apoptotic cellular debris was found for 24
h (Wright-Giemsa staining; magnification ×1000). All experiments
were repeated at least three times. PI, propidium iodide; T-ALL, T
cell acute lymphoblastic leukaemia; PBMCs, peripheral blood
mononuclear cells. |
Cell lines and cell culture
Human T-ALL cell lines (Jurkat, H9, Molt-4 and Raji)
were obtained from Zhejiang University (Hangzhou, China). The cells
were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher
Scientific Inc.) supplemented with 10% fetal calf serum (FCS;
Invitrogen; Thermo Fisher Scientific Inc.), 2 mmol/l glutamine, 0.1
mg/ml streptomycin, and 100 U/ml penicillin and incubated at 37°C
in a 5% CO2 humidified incubator. All experiments using
these cell lines were performed within 6 months of receipt or
thawing after −80°C cryopreservation, the cells were passaged every
2 days with fresh medium. In all experiments, cells were used in
logarithmic growth phase. As a normal cell line was not available,
PBMCs were chosen for comparison in the present study. PBMCs were
collected from healthy donors after informed written consent was
obtained (n=7, 4 males and 3 females; age range, 25.5–34.2 years,
with the median age of 27.4 years). All were from the Second
Affiliated Hospital, School of Medicine, Zhejiang University
(Hangzhou, China), and 2 ml of blood was obtained each donor. PBMCs
were isolated with Ficoll-Hypaque gradients by centrifugation from
fresh blood (19). This study was
approved by the Ethics Committee of The Second Affiliated Hospital,
School of Medicine, Zhejiang University (Hangzhou, China) (approval
number, IR2020001187) according to the guidelines of the
Declaration of Helsinki.
Cell viability assay
Cell viability was measured using the Cell Counting
Kit-8 (CCK-8) reagent (Donjindo Molecular Technologies, Inc.)
according to the manufacturer's protocol, the OD value was used for
measuring absorbance (21). Briefly,
one experiment was performed to investigate the effect of linalool
on T-ALL viability, human T-ALL cell lines (Jurkat, H9, Molt-4, and
Raji cells) and normal PBMCs were cultured in 96-well plates
(2×105 cells/ml), and then linalool (3.75, 7.50, 15.00,
30.00, 60.00 and 120.00 µM, respectively) was added to the cells.
Another experiment was performed to investigate the effect of
linalool and ERK1/2-selective inhibitor PD98059, JNK inhibitor
SP600125 or P38 inhibitor SB203580 on T-ALL viability, Jurkat cells
were cultured in 96-well plates (2×105 cells/ml), and 10
µmol/l ERK1/2-selective inhibitor PD98059, 20 µmol/l JNK inhibitor
SP600125 or 10 µmol/l P38 inhibitor SB203580 was added to the cell
suspensions for 1 h. Subsequently, 30 µM linalool was added to the
cell suspensions for 9 h. The cells were then harvested and
measured by the CCK-8 assay. The control group consisted of cells
treated with DMSO only.
Cell cycle analysis
Flow cytometry (FCM) was performed to determine the
effect of linalool on the cell cycle following a standard protocol
(21,22). Briefly, Jurkat cells
(2×105 cells/ml) were treated with 30 µM linalool for 6,
12, 24 or 48 h at 37°C. The cells were washed and fixed in 70%
ice-cold ethanol overnight at 4°C. Subsequently, the cells were
harvested and incubated in PBS with 100 mg/ml propidium iodide
(PI), 0.1% Triton X-100 for permeabilization and 100 µg/ml RNase A
for 30 min at 4°C (Beyotime Institute of Biotechnology). The
PI-stained cells were subjected to cell cycle profiling analysis
using a flow cytometer (FACSCalibur; BD Biosciences). The cell
cycle distribution was analysed using ModFit LT software version
3.1 (Verity Software House, Inc.).
Apoptosis assay
Apoptosis was investigated using an Annexin
V-fluorescein isothiocyanate (FITC) apoptosis kit (cat. no. K101;
BioVision, Inc.) according to the manufacturer's instructions
(21,22). Briefly, in one experiment, Jurkat
cells were treated with 30 µM linalool for 6, 12 or 24 h at 37°C,
respectively. In another experiment, Jurkat cells were pre-treated
with 10 µmol/l PD98059, 20 µmol/l SP600125, or 10 µmol/l SB203580
for 1 h and then 30 µM linalool was added to the cells suspensions
for 9 h at 37°C. The cells were harvested and washed with ice-cold
PBS 3 times and adjusted to 1×106 cells/ml with binding
buffer. Subsequently, the cells were incubated with 5 µl
FITC-labelled Annexin V and 5 µl PI at room temperature in the dark
for 15 min. Finally, the stained cells were analysed using a flow
cytometer (FACSCalibur, BD Biosciences). The data were analysed
using FlowJo software version 10.0.7 (Tree Star, Inc.). The control
group was DMSO only.
Morphological analysis
Jurkat cells (2×105 cells/ml) were
cultured with 30 µM linalool for various time periods (0, 6, 12 or
24 h). Cytospin slides (4–8 µm) were prepared, cells were fixed
with 100% methanol at room temperature for 1 min, and stained with
Wright-Giemsa staining (Baso Diagnostics Inc.) for 10 min at room
temperature. Cell morphology was observed with a light microscope
(Olympus Corporation, magnification ×1,000).
RNA extraction, cDNA library
construction and Illumina sequencing
Jurkat cells 2×105 cells/ml) were treated
with 30 µM linalool for 9 h and then total RNA from Jurkat cells
before and after treatment with linalool was isolated using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific Inc.) for 5 min at
4°C according to the manufacturer's instructions. RNA integrity was
evaluated using an Agilent Bioanalyser 2100 (Agilent Technologies,
Inc.). Paired-end libraries were constructed with the TruSeq RNA
Sample Preparation kit (Illumina Inc.; cat. no. RS-122-2001)
following the TruSeq RNA Sample Preparation Guide. Briefly,
polyadenylated RNA was isolated and fragmented into ~200 bp
fragments. The first-strand cDNA was synthesized using random
hexamer primers, and the second strand cDNA was synthesized using
DNA Polymerase I and RNase H, followed by an end repair process
with the addition of a single ‘A’ base and ligation of the
adapters. Library quality control was performed with 2100
Bioanalyzer (Agilent Technologies Inc.). The DNA library
concentration was diluted to 10 nM with hybridization buffer
(Illumina Inc.) for library normalization. DNA sequencing was
performed on the Illumina HiSeq 2500 platform. Synthesis sequencing
was performed with the HiSeq Rapid SBS Kit v2 (cat. no. 402-4023;
Illumina Inc.), generating single-fragment reads (2×150 bp; PE)
with two fragment end-to-end assemblies. The DNA library was loaded
into the flow cell and placed in a cBot device (Illumina, Inc.).
The cycle sequencing procedure was performed with the HiSeq Rapid
SBS Kit v2 according to the manufacturer's instructions. In brief,
the libraries were bound to immobilized oligos on flow cells, DNA
was synthesized through polymerase activity from the free 3 end and
cyclic bridge amplification, denaturation, linearization, 3 end
blocking, reverse strand denaturation and re-amplification
procedures for clustering were performed. The FASTQ file obtained
from the present study was analyzed on the web-based Galaxy
platform (https://usegalaxy.org/).
Real-time quantitative PCR
(RT-qPCR)
Briefly, RNA was extracted from Jurkat cells with
TRIzol reagent (Thermo Fisher Scientific Inc.) according to the
manufacturer's protocol. cDNA was obtained by a High Capacity cDNA
Archive kit (Applied Biosystems; Thermo Fisher Scientific Inc.).
The reverse transcription reactions were as follows: 10 min at
25°C, 120 min at 37°C and 5 min at 85°C. The samples were then
placed on ice. The PCR solution was a master mix that included SYBR
Green Mastermix (Beijing Solarbio Science & Technology Co.,
Ltd.), forward primer, reverse primer and 10 ng template cDNA. The
final concentration of primers in the PCR solution was 0.45 µmol/l.
GADD45A and heat shock protein 27 (HSP27) sequences were amplified
using the following gene-specific primers. GADD45A, forward,
5′-GAGAGCAGAAGACCGAAAGGA-3′ and reverse,
5′-CACAACACCACGTTATCGGG-3′; HSP27, forward,
5′-ACGGTCAAGACCAAGGATGG-3′ and reverse, 5′-AGCGTGTATTTCCGCGTGA-3′;
GAPDH, forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′) was used as an internal control to
normalize the PCR results. The reaction was performed on a 7500
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific
Inc.). The amplification conditions for the assay were set as
follows: Initial denaturation at 95°C for 10 min, then 40 cycles of
denaturation at 95°C for 10 sec and annealing/extension at 60°C for
60 sec. The PCR results were analysed using the comparative
2−ΔΔCq method (23) using
the AB Prism software (Applied Biosystems; Thermo Fisher Scientific
Inc.).
Gene Ontology (GO) analysis and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis
Clean reads of each developmental stage sample were
mapped to transcriptome assembly results by Bowtie with a maximum
mismatch set to 2 (22) and <2
mismatches were allowed in the alignment. The number of mapped
clean reads for each UniGene was calculated and normalized to
fragments per kb per million reads, which is a widely used method
to calculate the levels of gene expression (24). GO analysis (25) and KEGG pathway (26) enrichment analysis of mRNA were
performed with the R language version 3.4.4 cluster Profiler
(27), org.Hs.eg.db genome-wide
annotation (https://bioconductor.org/packages/org.Hs.eg.db/), the
topGO package (https://bioconductor.org/packages/topGO/), the
pathview R package (https://bioconductor.org/packages/pathview/) and the
enrichplot package (https://github.com/GuangchuangYu/enrichplot). The
ggplot2 package (http://ggplot2.tidyverse.org) was used to create the
graphics (28). Differentially
expressed genes were identified with an absolute fold-change ≥2 as
the cut off value (29). Heatmap was
performed with the R language version 3.4.4.
Western blotting
Jurkat cells (2×105 cells/ml) were
treated with 30 µM linalool for 6 h, 12 h or 24 h at 37°C, and then
the cells were harvested and washed with ice-cold PBS 3 times.
Proteins were extracted using lysis buffer (Beyotime Institute of
Biotechnology) that contained protease inhibitors (Sigma-Aldrich;
Merck KGaA) and a phosphatase Inhibitor Cocktail Set II (cat. no.
524625; Sigma-Aldrich; Merck KGaA; added each solution at 1:100
(v/v) dilution to cell lysates) was used in the lysis buffer for
detection of phosphorylated proteins. Protein concentration was
analysed with a bicinchoninic acid (BCA) protein assay kit
(Beyotime Institute of Biotechnology). Total protein (40 µg/lane)
was separated using 10–12% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis gels and transferred to a polyvinyl difluoride
membrane (Bio-Rad Laboratories Inc.). The membrane was subsequently
blocked with TBST (0.05% Tween-20) containing 5% skimmed milk for 1
h at 4°C and probed overnight with the corresponding primary
antibodies (ERK, p-ERK, GADD45A, cleaved PARP, caspase-3, cleaved
caspase-3, JNK, p-JNK, P38, p-P38, and β-actin) at 4°C. The
membrane was washed and incubated with secondary antibodies (goat
anti-rabbit IgG, conjugated to horse-radish peroxidase) for 1 h at
room temperature and then reacted with SuperSignal West Pico
chemiluminescent substrate (Pierce; Thermo Fisher Scientific Inc.)
for visualization (24). β-actin was
used as the loading control. ImageJ software version 1.8.0 was used
for densitometry analysis (National Institutes of Health).
Statistical analysis
Data are presented as the mean ± SD of at least 3
biological replicates. Multiple group comparisons were performed
using ANOVA followed by the post hoc Tukey's test and comparisons
between two groups were performed using unpaired Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference. Statistical analysis was conducted using GraphPad Prism
version 5.0 software (GraphPad Software, Inc.).
Results
Linalool inhibits the growth of T-ALL
cells
A CCK-8 assay was used to analyse cell viability, it
demonstrated that linalool inhibited the growth of Jurkat, H9,
Molt-4 and Raji cells in a dose-dependent manner (Fig. 1B). The 50% inhibition
(IC50) at 48 h was calculated using GraphPad Prism
version 5.0 software and it demonstrated that the IC50
at 48 h of Jurkat, H9, Molt-4 and Raji cells was 31.35, 22.16,
25.80 and 25.19 µM, respectively (data not shown). In contrast,
treatment with 30 µM linalool had almost no effects on the
viability of normal PBMCs, and there was a very slight decrease in
the percentage of viable cells (86.84±2.04%) after treatment with
120 µM linalool (Fig. 1B).
Linalool induces apoptosis of T-ALL
cells
Jurkat cells were treated with 30 µM linalool for 6,
12 or 24 h, respectively. The percentage of apoptotic Jurkat cells
increased in a time-dependent manner from 28.92±3.70% at 6 h to
56.92±5.92% at 12 h to 90.50±7.38% at 24 h (Fig. 1C and D). Significant differences were
observed compared to the control group which had a percentage of
apoptotic Jurkat cells of 7.34±1.89% (all P<0.01; Fig. 1C and D). Wright-Giemsa staining
demonstrated that Jurkat cells were killed after treatment with 30
µM linalool in a time-dependent manner as apoptotic bodies (black
arrows) were present after treatment with linalool for 6 h and
apoptotic cellular debris (red arrows) was increased after
treatment with linalool for 12 h and plenty of apoptotic cellular
debris was found for 24 h (Fig.
1F).
Linalool reduces T-ALL cell
accumulation at the G0/G1 phase
Following treatment of Jurkat cells with 30 µM
linalool, G0/G1, S and G2/M phase
cells were significantly decreased in a time-dependent manner.
After 48 h of treatment with linalool, the percentage of
G0/G1 phase cells in the control group (DMSO
treatment only) decreased from 55.02±1.54 to 4.89±0.91%, the
percentage of S phase cells decreased from 33.68±1.13 to 2.37±0.12%
and the percentage of G2/M phase cells decreased from
10.51±0.57 to 2.06±0.03% (all P<0.01; Fig. 1E). In contrast, the percentage of
sub-G1 phase cells in the control group increased from
0.73±0.09 to 90.01±3.17% following 48 h of treatment with linalool
(P<0.01; Fig. 1E).
Linalool inhibits T-ALL cell survival
with involvement of the MAPK signaling pathway
RNA sequencing demonstrated that 3,512 genes were
significantly differentially expressed before and after treatment
with 30 µM linalool for 9 h. Enrichment of mRNAs in the regulatory
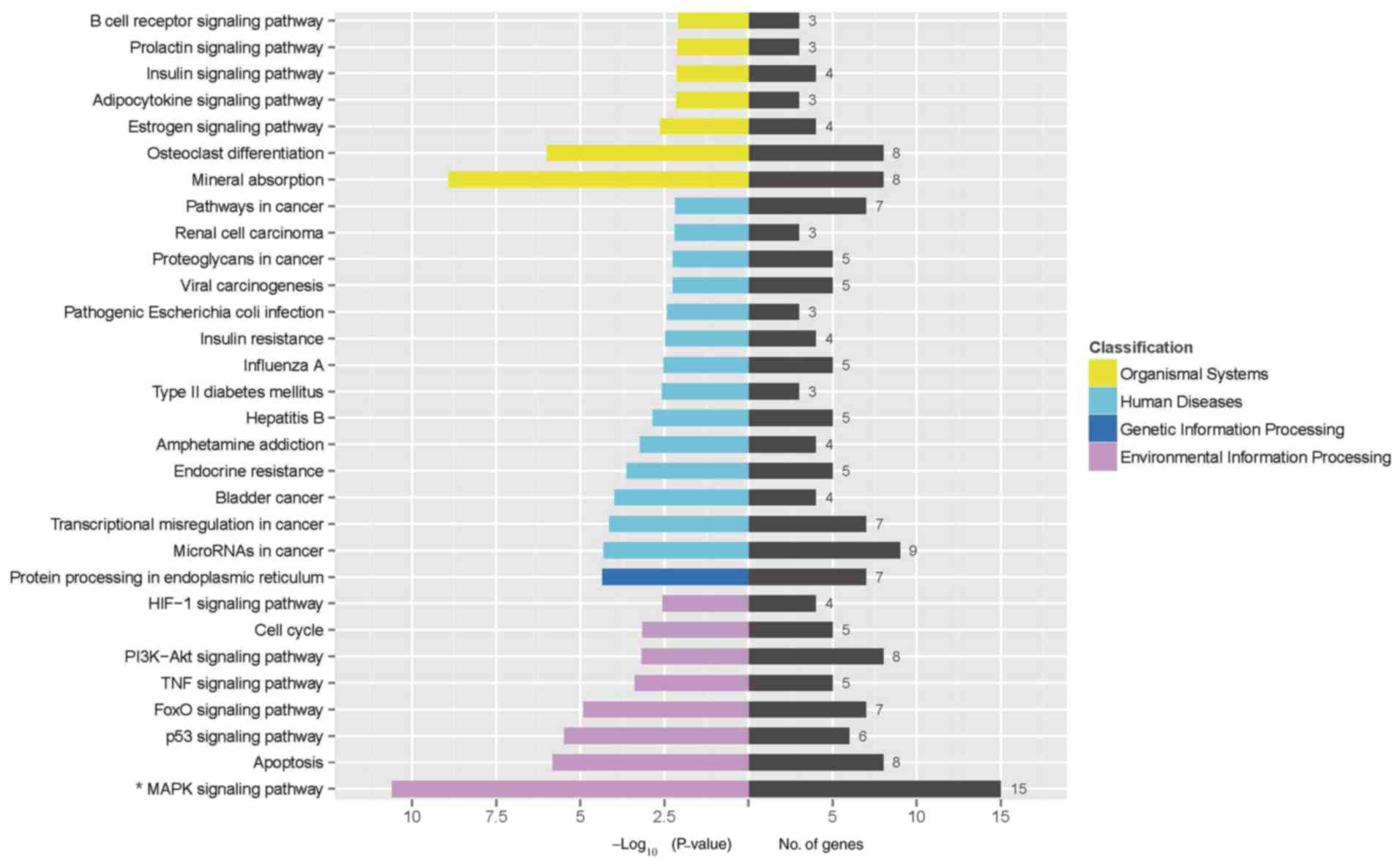
network was assessed by analysis of KEGG pathways and it was found
that the MAPK signaling pathway (the -log10 value was
10.62) was significantly involved in the effect of linalool
treatment on Jurkat cells (Fig. 2).
Subsequently, a GO analysis was conducted to determine the
functional roles of these differentially expressed genes. The
differently expressed genes enriched in GO terms were associated
with ‘protein binding’, ‘regulation of biological process’ and
‘response to stimulus’ (the -log10 value were 36.2,
38.6, and 38.5, respectively) (Fig.
S1). A distinct set of upregulated genes associated with the
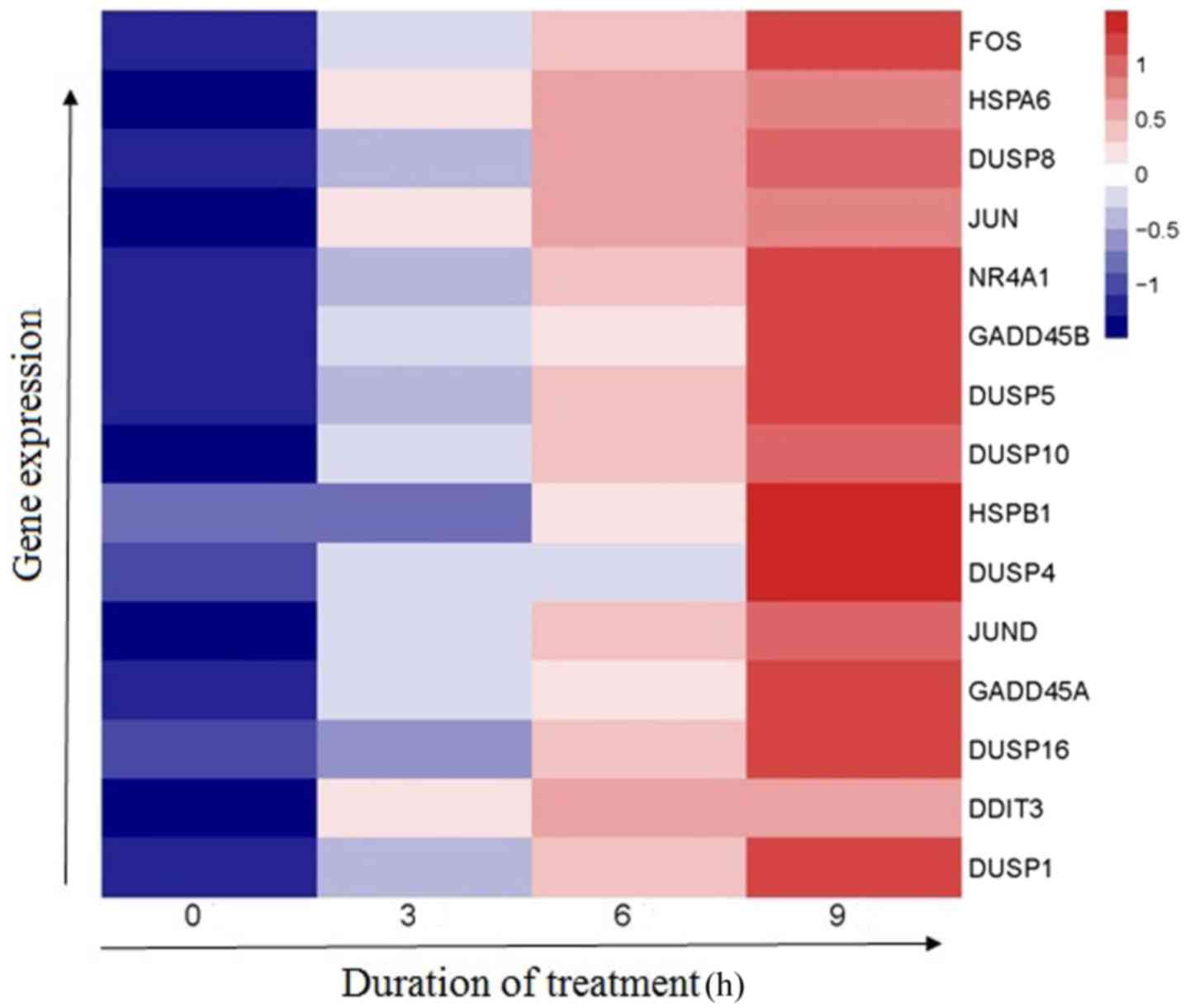
MAPK signaling pathway was identified using heatmap (Fig. 3).
To verify the RNA sequencing results, the mRNA
levels of GADD45A and HSPB1 (HSP27) which are components of the
MAPK signaling pathway (30) were
assessed by RT-qPCR in Jurkat cells. Consistent with the sequencing
results, GADD45A mRNA levels (1.02±0.02; 1.56±0.28; 2.63±0.41 and
4.87±0.70) and HSP27 mRNA levels (1.00±0.01, 2.04±0.51, 3.65±0.74,
and 4.48±0.82) were significantly upregulated in a time-dependent
manner (0, 6, 12 or 24 h) following treatment with 30 µM linalool
(P<0.05; Fig. 4A and B).
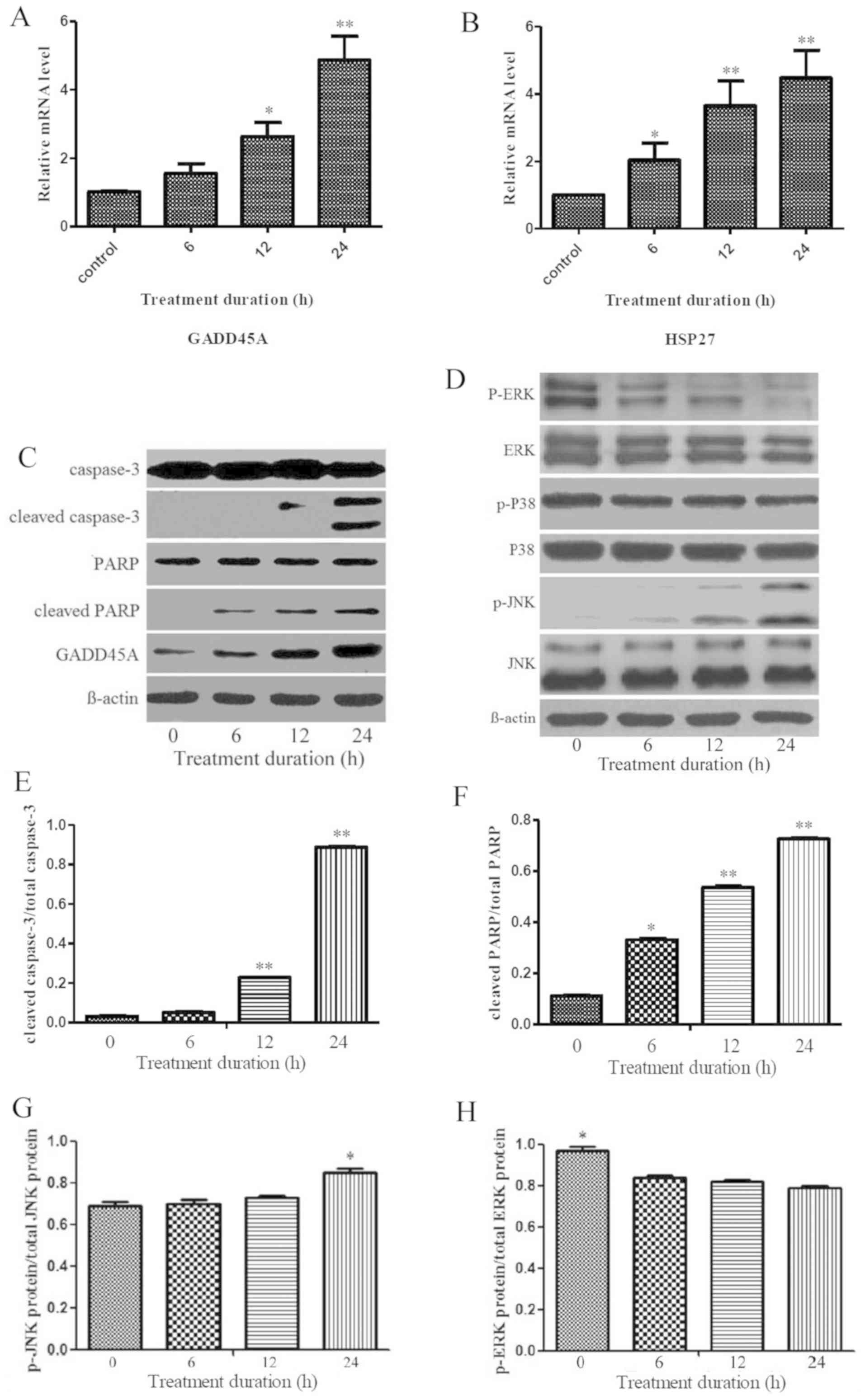 | Figure 4.Linalool inhibited T-ALL cells
survival by p-ERK suppression and p-JNK activation. (A and B)
RT-qPCR results demonstrating that GADD45A and HSP27 mRNA levels
were significantly upregulated in a time-dependent manner after
treatment with 30 µM linalool, *P<0.05, **P<0.01 compared
with control group. (C and D) Western blotting results following 30
µM linalool treatments of Jurkat cells. p-JNK, GADD45A, cleaved
caspase-3 and cleaved PARP protein expression were upregulated in a
time-dependent manner, while p-ERK protein expression was
downregulated. In addition, ERK, p38, p-P38 and JNK protein
expression showed no obvious differences compared to the 0 h group.
(E and F) The relative ratios of cleaved PARP/total PARP and
cleaved caspase-3/total caspase-3 were significantly upregulated in
a time-dependent manner after treatment with 30 µM linalool
compared with 0 h group (*P<0.05). (G) The ratio of p-JNK
protein/total JNK protein demonstrated an upward trend in a
time-dependent manner and the corresponding ratio at 24 h was
obviously higher than that at 0, 6 and 12 h. (H) The ratio of p-ERK
protein/total ERK protein demonstrated a downward trend in a
time-dependent manner, the corresponding ratio at 0 h was
significantly higher compared with that at 6, 12, and 24 h
(P<0.05). The experiments were repeated at least three times.
T-ALL, T cell acute lymphoblastic leukaemia; GADD45A, Growth Arrest
And DNA Damage Inducible α; PARP, Poly (ADP-ribose) polymerase; p,
phosphorylated; RT-q, reverse-transcription quantitative; HSP 27,
heat shock protein 27. |
Linalool inhibits T-ALL cell survival
with the involvement of p-ERK suppression and p-JNK activation
Jurkat cells were treated with 30 µM linalool at 0,
6, 12 or 24 h. Western blot analysis demonstrated that p-JNK and
GADD45A protein expression were upregulated in a time-dependent
manner, while p-ERK protein expression was downregulated in a
time-dependent manner (Fig. 4C and
D). ERK, p38, p-p38 and JNK protein expression showed no
obvious differences following treatment with linalool (Fig. 4C and D). The relative ratio of
cleaved caspase-3/total caspase-3 and cleaved PARP/total PARP was
significantly upregulated in a time-dependent manner after
treatment with 30 µM linalool (Fig. 4E
and F). In addition, the ratio of p-JNK protein/total JNK
protein (0.69±0.02, 0.70±0.02, 0.73±0.01 and 0.85±0.02)
demonstrated an upward trend in a time-dependent manner following
treatment with 30 µM linalool with 24 h treatment demonstrating
significant difference compared to the other time points tested
(P<0.05; Fig. 4G). Conversely,
the ratio of p-ERK protein/total ERK protein (0.97±0.02, 0.84±0.01,
0.82±0.01 and 0.79±0.01) demonstrated a downward trend in a
time-dependent manner where the ratio at 0 h was significantly
higher compared with 6, 12 and 24 h (P<0.05; Fig. 4H) indicating a suppression of p-ERK
protein. For the ratio of p-p38 protein/total p38 protein, no
significant difference was found after treatment with 30 µM
linalool at 0 h (0.97±0.02), 6 h (0.97±0.01), 12 h (0.95±0.01) or
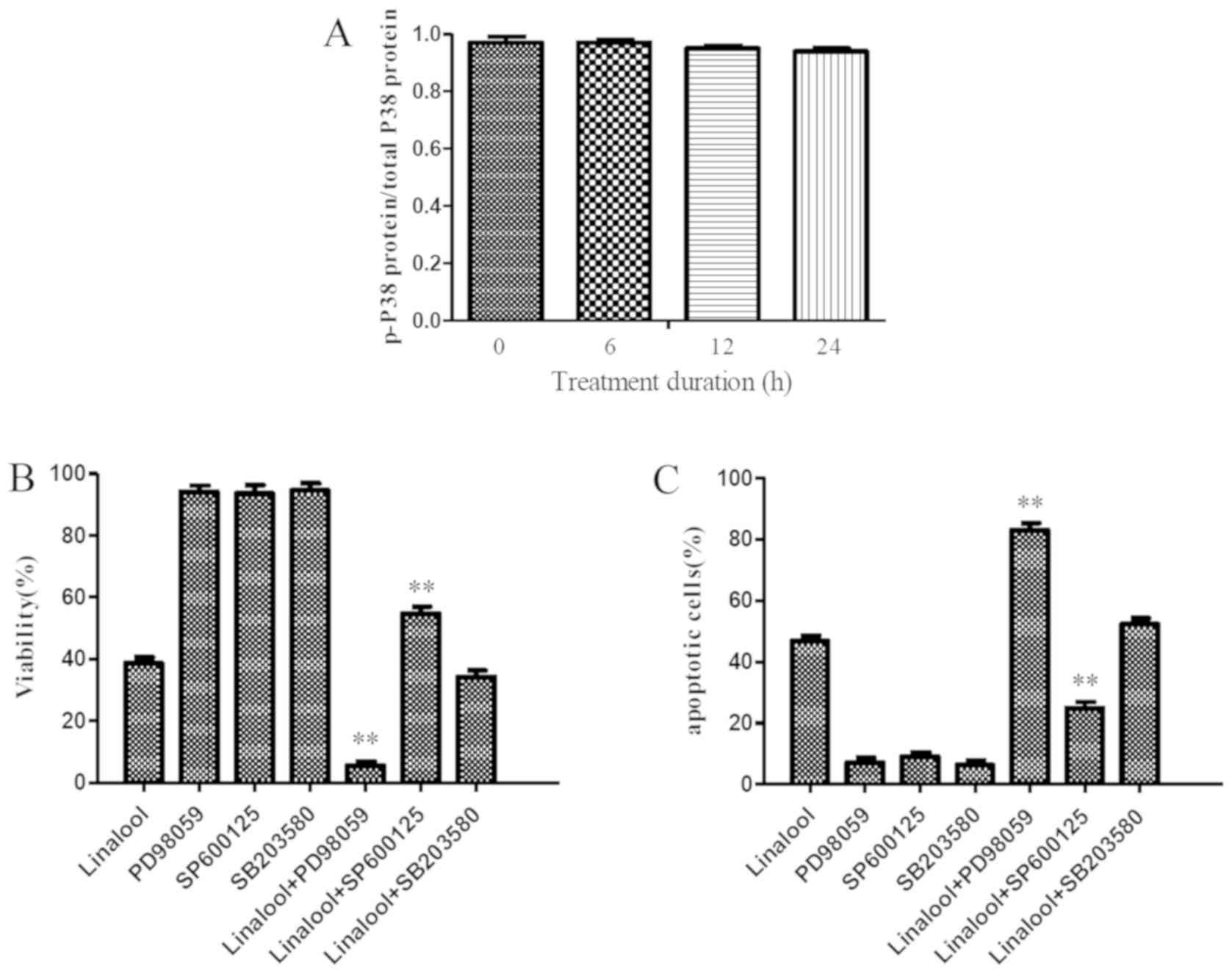
24 h (0.94±0.01) (Fig. 5A).
To further determine the role of ERK, JNK and p38 in
linalool-induced Jurkat cell inhibition and apoptosis, specific
inhibitors were used for 1 h before treatment with 30 µM linalool
for 9 h. With the addition of 10 µmol/l ERK inhibitor PD98059, cell
viability was significantly reduced compared with the use of 30 µM
linalool alone (5.57±2.12 vs. 38.70±3.24; P<0.001; Fig. 5B) and the number of apoptotic cells
was significantly increased compared with the use of 30 µM linalool
alone (83.0±4.03 vs. 46.72±3.15; P<0.001; Fig. 5C). In addition, the addition of 20
µmol/l JNK inhibitor SP600125 compared with the use of 30 µM
linalool alone, significantly reversed the linalool-mediated
inhibition of Jurkat cell viability (55.33±3.35 vs. 38.70±3.24;
P<0.01; Fig. 5B) and apoptosis
(24.9±3.47 vs. 46.72±3.15; P<0.01; Fig. 5C). However, the addition of 10 µmol/l
p38 inhibitor SB203580 demonstrated no significant effect on cell
viability and apoptosis compared to the use of 30 µM linalool alone
(P>0.05; Fig. 5B and C). The
aforementioned results demonstrated that the JNK inhibitor SP600125
significantly reversed the linalool-mediated growth inhibition and
apoptosis, the ERK1/2-selective inhibitor PD98059 enhanced the
linalool-induced growth inhibition and apoptosis and the inhibition
of p38 activity with SB203580 resulted in a very modest increase in
growth inhibition and apoptosis.
Discussion
Although, the effects of treatments for T-ALL have
dramatically improved over the years, the prognosis of T-ALL is
still poor (1,2). There is a pressing need for the
development of more effective and safer treatments (2–4).
Linalool can inhibit the growth of a variety of tumor cells,
including hepatocellular carcinoma, breast cancer, small cell
carcinoma and malignant melanoma (9–15). In
the present study, linalool inhibited the viability of 4 human
T-ALL cell lines in a dose-dependent manner. The IC50 at
48 h of Jurkat cells was higher compared with that of H9, Molt-4 or
Raji cells, hence Jurkat cells were chosen in the present study for
the subsequent analysis. In addition, as a normal cell line was not
available PBMCs were chosen for comparison in the present study.
The viability of PBMCs from normal individuals was unaffected when
the cells were treated with 30 µM linalool. Annexin V-FITC/PI
analysis conducted in the present study revealed that linalool
inhibits the viability of Jurkat cells by inducing the apoptosis of
leukemia cells, which was also verified by morphological
analysis.
Cell cycle control is a major event in cellular
division and the disruption of the normal cell cycle serves an
important role in the development of tumors (31). A large number of antitumor natural
compounds, such as baicalin and homoharringtonine, have
demonstrated to induce cell death and apoptosis in close
association with cell cycle arrest at the G1 phase
(31,32). Notably, the present study
demonstrated that Jurkat cells treated with linalool had reduced
cell accumulation at the G0/G1 phase, which
contains numerous non-proliferating leukaemia cells (33). The findings of the present study
indicate that linalool may affect proliferating and
non-proliferating T-ALL cells. The latter are highly related to
traditional chemotherapeutic drug resistance and tumor recurrence
(3,4). The effect of linalool on
non-proliferating T-ALL cells should be investigated in future
studies.
The development of next-generation sequencing
technology has provided new tools for exploring the differential
gene expression of tumors before and after treatment (34). In the present study, the HiSeq X
Series sequencing platform was used to perform RNA sequencing
analysis on Jurkat cells before and after treatment with linalool.
The present study identified 3,512 genes were statistically
differentially expressed before and after treatment with 30 µM
linalool for 9 h. In the present study, KEGG pathway analysis and
Gene Ontology enrichment analysis were conducted. The KEGG pathways
analysis demonstrated that the MAPK signaling pathway is
significantly involved in the effect of linalool treatment on
Jurkat cells and the GO enrichment analysis demonstrated that the
differently expressed genes enriched in GO terms were associated
with ‘protein binding’, ‘regulation of biological process’ and
‘response to stimulus’.
The MAPK signaling pathways are known to function as
crucial components of cell proliferation and apoptosis in tumor
cells and have been identified as chemotherapeutic targets for
sensitizing tumor cells to apoptosis (35). In the present study, RT-qPCR was
performed to confirm the RNA sequencing results in Jurkat cells and
the effect of linalool treatment on the mRNA levels of GADD45A and
HSP27 from the MAPK signaling pathway was assessed. Consistent with
the RNA sequencing results, exposure to linalool resulted in a
significant increase in the mRNA levels of GADD45A and HSP27. The
findings of the present study revealed that linalool inhibits T-ALL
cells survival through the involvement of the MAPK signaling
pathway.
MAPKs in mammals include JNK, p38 MAPK, and ERK
(36). Western blotting conducted in
the present study demonstrated that the p-JNK protein expression
level significantly increased following linalool treatment and
inhibition of JNK activity by the JNK inhibitor SP600125
significantly reversed the linalool-mediated growth inhibition and
apoptosis in Jurkat cells. In addition, linalool treatment exerted
a significant inhibitory effect on p-ERK1/2 protein expression and
the ERK1/2-selective inhibitor PD98059 enhanced the
linalool-induced growth inhibition and apoptosis in Jurkat cells.
However, p-p38 protein expression and p38 mRNA levels were not
obviously different following linalool treatment and the inhibition
of p38 activity with SB203580 resulted in a very modest increase in
growth inhibition and apoptosis. Taken together, the findings of
the present study indicated that the JNK and ERK pathways, but not
the p38 pathway are involved in linalool-induced apoptosis of
Jurkat cells.
In conclusion, the present study revealed that
linalool can preferentially inhibit the viability of T-ALL cells
without any significant toxicity in PBMCs from normal individuals.
In addition, it was demonstrated that linalool inhibits T-ALL cell
survival with involvement of MAPK signaling pathways, JNK
activation and ERK inhibition, which serve functional roles in
apoptosis induction. Linalool may be developed as a novel anti
T-ALL leukaemia agent. Future animal experiments should be
conducted for the comparison of the effect and safety of linalool
with other traditional chemotherapy drugs.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by a grant from the National
Natural Science Foundation of China (grant no. 81400107).
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
XZ conceived and designed the study. XG, BW and XL
performed the experiments. LY prepared the figures and analysed the
data. XG wrote the manuscript. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
The Second Affiliated Hospital, School of medicine, Zhejiang
University (Hangzhou, China; approval number:, IR2020001187)
according to the guidelines of the Declaration of Helsinki. A
written consent form was obtained from all the participating
healthy donors.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Swerdlow SH, Campo E, Pileri SA, Harris
NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz
AD and Jaffe ES: The 2016 revision of the world health organization
classification of lymphoid neoplasms. Blood. 127:2375–2390. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gascoyne RD, Campo E, Jaffe ES, Chan WC,
Chan JK, Rosenwald A, Stein A and Swerdlow SH: Diffuse large B-cell
lymphoma, not otherwise specified. World health organization
classification of haematopoietic and lymphoid tissues of tumors.
4th. IARC Press; Lyon, France: pp. 291–297. 2016
|
|
3
|
Kato K, Uike N, Wake A, Yoshimitsu M,
Tobai T, Sawayama Y, Takatsuka Y, Fukuda T, Uchida N, Eto T, et al:
The outcome and characteristics of patients with relapsed adult T
cell leukemia/lymphoma after allogeneic hematopoietic stem cell
transplantation. Hematol Oncol. 37:54–61. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vadillo E, Dorantes-Acosta E, Pelayo R and
Schnoor M: T cell acute lymphoblastic leukemia (T-ALL): New
insights into the cellularorigins and infiltration mechanisms
common and unique among hematologic malignancies. Blood Rev.
32:36–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thol F and Ganser A: Treatment of relapsed
acute myeloid leukemia. Curr Treat Options Oncol. 21:662020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vázquez-Sánchez D, Galvão JA, Mazine MR,
Gloria EM and Oetterer M: Control of staphylococcus aureus biofilms
by the application of single and combined treatments based in plant
essential oils. Int J Food Microbiol. 286:128–138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Burdock GA and Carabin IG: Safety
assessment of coriander (Coriandrum sativum L.) essential
oil as a food ingredient. Food Chem Toxicol. 47:22–34. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Elliott JF, Ramzy A, Nilsson U, Moffat W
and Suzuki K: Severe intractable eyelid dermatitis probably caused
by exposure to hydroperoxides of linalool in a heavily fragranced
shampoo. Contact Dermatitis. 76:114–115. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ramzi A, Ahmadi H, Sadiktsis I and Nilsson
U: A two-dimensional non-comprehensive reversed/normal phase
high-performance liquid chromatography/tandem mass spectrometry
system for determination of limonene and linalool hydroperoxides. J
Chromatogr A. 1566:102–110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jabir MS, Taha AA, Sahib UI, Taqi ZJ,
Al-Shammari AM and Salman AS: Novel of nano delivery system for
linalool loaded on gold nanoparticles conjugated with CALNN peptide
for application in drug uptake and induction of cell death on
breast cancer cell line. Mater Sci Eng C Mater Biol Appl.
94:949–964. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Elansary HO, Abdelgaleil SA, Mahmoud EA,
Yessoufou K, Elhindi K and El-Hendawy S: Effective antioxidant,
antimicrobial and anticancer activities of essential oils of
horticultural aromatic crops in northern Egypt. BMC Complement
Altern Med. 18:2142018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sakurai K, Tomiyama K, Yaguchi Y and
Asakawa Y: Characteristic odor of the Japanese liverwort
(Leptolejeunea elliptica). J Oleo Sci. 69:767–770. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kubatka P, Kello M, Kajo K, Samec M, Jasek
K, Vybohova D, Uramova S, Liskova A, Sadlonova V, Koklesova L, et
al: Chemopreventive and therapeutic efficacy of cinnamomum
zeylanicum L. Bark in experimental breast carcinoma: Mechanistic in
vivo and in vitro analyses. Molecules. 25:13992020. View Article : Google Scholar
|
|
14
|
Chang MY, Shieh DE, Chen CC, Yeh CS and
Dong HP: Linalool induces cell cycle arrest and apoptosis in
leukemia cells and cervical cancer cells through CDKIs. Int J Mol
Sci. 16:28169–28179. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Miyashita M and Sadzuka Y: Effect of
linalool as a component of humulus lupulus on doxorubicin-induced
antitumor activity. Food Chem Toxicol. 53:174–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He X, Zhu Y, Lin YC, Li M, Du J, Dong H,
Sun J, Zhu L, Wang H, Ding Z, et al: PRMT1-mediated FLT3 arginine
methylation promotes maintenance of FLT3-ITD+acute myeloid
leukemia. Blood. 134:548–560. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maeda H, Yamazaki M and Katagata Y:
Kuromoji (Lindera umbellata) essential oil-induced apoptosis
and differentiation in human leukemiaHL-60 cells. Exp Ther Med.
3:49–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saab AM, Tundis R, Loizzo MR, Lampronti I,
Borgatti M, Gambari R and Menichini F, Esseily F and Menichini F:
Antioxidant and antiproliferative activity of Laurus nobilis
L. (Lauraceae) leaves and seeds essential oils against K562 human
chronic myelogenous leukaemia cells. Nat Prod Res. 26:1741–1745.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gu Y, Ting Z, Qiu X, Zhang X, Gan X, Fang
Y, Xu X and Xu R: Linalool preferentially induces robust apoptosis
of a variety of leukemia cells via upregulating p53 and
cyclin-dependent kinase inhibitors. Toxicology. 268:19–24. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Souza-Junior FJ, Luz-Moraes D, Pereira FS,
Barros MA, Fernandes LM, Queiroz LY, Maia CF, Maia JG and
Fontes-Junior EA: Aniba canelilla (Kunth) mez (Lauraceae): A
review of ethnobotany, phytochemical, antioxidant,
anti-inflammatory, cardiovascular, and neurological properties.
Front Pharmacol. 11:6992020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li CJ: Flow cytometry analysis of cell
cycle and specific cell synchronization with butyrate. Methods Mol
Biol. 1524:149–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li JJ, Wang W, Wang XQ, He Y, Wang SS and
Yan YX: A novel strategy of identifying circRNA biomarkers in
cardiovascular disease by meta-analysis. J Cell Physiol.
234:21601–21612. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang YL, Fang LJ, Zhong LY, Jiang J, Dong
XY and Feng Z: Hub genes and key pathways of traumatic brain
injury: Bioinformatics analysis and in vivo validation. Neural
Regen Res. 15:2262–2269. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Y, Xia L, Guo Q, Zhu J, Deng Y and Wu
X: Identification of chemoresistance-associated key genes and
pathways in high-grade serous ovarian cancer by bioinformatics
analyses. Cancer Manag Res. 12:5213–5223. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao M, Zhang S, Luo C, He X, Wei S, Jiang
W, He F, Lin Z, Yan M and Dong W: Transcriptome analysis of starch
and sucrose metabolism across bulb development in Sagittaria
sagittifolia. Gene. 649:99–112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gu Y, Zhang J, Ma X, Kim BW, Wang H, Li J,
Pan Y, Xu Y, Ding L, Yang L, et al: Stabilization of the c-myc
protein by CAMKIIγ promotes T cell lymphoma. CancerCell. 2:115–128.
2017.
|
|
30
|
Yang L, Zhang C, Chen J, Zhang S, Pan G,
Xin Y, Lin L and You Z: Shenmai injection suppresses multidrug
resistance in MCF-7/ADR cells through the MAPK/NF-κB signalling
pathway. Pharm Biol. 58:276–285. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baell JB, Leaver DJ, Hermans SJ, Kelly GL,
Brennan MS, Downer NL, Nguyen N, Wichmann J, McRae HM, Yang Y, et
al: Inhibitors of histone acetyltransferases KAT6A/B induce
senescence and arrest tumor growth. Nature. 560:253–257. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao Y, Liu H, Wang H, Hu H, He H, Gu N,
Han X, Guo Q, Liu D, Cui S, et al: Baicalin inhibits breast cancer
development via inhibiting IĸB kinase activation in vitro and in
vivo. Int J Oncol. 53:2727–2736. 2018.PubMed/NCBI
|
|
33
|
Kong Y, Zhao S, Tian H and Hai Y: GAS2
promotes cell proliferation and invasion and suppresses apoptosis
in pediatric T-cell acute lymphoblastic leukemia and activates
wnt/β-catenin pathway. Onco Targets Ther. 13:1099–1108. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rheinbay E, Parasuraman P, Grimsby J, Tiao
G, Engreitz JM, Kim J, Lawrence MS, Taylor-Weiner A,
Rodriguez-Cuevas S, Rosenberg M, et al: Recurrent and functional
regulatory mutations in breast cancer. Nature. 547:55–60. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Athuluri-Divakar SK, Vasquez-Del Carpio R,
Dutta K, Baker SJ, Cosenza SC, Basu I, Gupta YK, Reddy MV, Ueno L,
Hart JR, et al: A small molecule RAS-mimetic disrupts RAS
association with effector proteins to block signaling. Cell.
165:643–655. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Johannessen CM, Johnson LA, Piccioni F,
Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo
JA, Root DE and Garraway LA: A melanocyte lineage program confers
resistance to MAP kinase pathway inhibition. Nature. 504:138–142.
2013. View Article : Google Scholar : PubMed/NCBI
|