Introduction
In the Global Cancer Statistical Analysis of 2018,
the number of breast cancer cases was 2,088,849, which was only
lower than the number of lung cancer cases, however, due to the
relatively good prognosis the number of deaths from breast cancer
was much lower compared with those from lung cancer (1). The disease status of the axillary lymph
nodes is the most important prognostic factor for patients with
early-stage breast cancer (2). The
most common predictors of node metastasis include lymphovascular
invasion, age, tumor size and tumor grade; additionally, the
predictive value is influenced by casting-type calcifications on
mammography, receptor states, tumor location and the method of
detection (3–8). However, no combination of these
predictors of axillary lymph node status can currently replace
histopathology and the surgical resection of lymph nodes (9). Moreover, the histopathological
diagnosis of removed lymph nodes via axillary lymph node dissection
is thought to be the most effective method to assess the disease
(10). Unfortunately, the anatomical
disruption caused by axillary lymph node dissection can result in
side effects, such as nerve injury, lymphedema and other
complications (11).
It has been demonstrated that breast cancer usually
spreads to one or a few lymph nodes, known as the sentinel lymph
nodes (SLNs), before spreading to other axillary nodes (12). Therefore, the use of SLN
identification and sampling procedures, referred to as SLN
biopsies, can be a reliable treatment strategy in patients with
early-stage breast cancer, as it reduces the need for axillary
lymph node dissection and avoids the associated morbidity (13–17).
Based on the aforementioned principles, SLN metastasis detection
may be a key method for assessing the spread of breast cancer to
the axillary lymph nodes. However, the underlying molecular
mechanism of SLN metastasis in breast cancer remains unknown.
Long non-coding RNAs (lncRNAs) are non-protein
coding transcripts of >200 nucleotides in length. Previous
studies have reported that lncRNAs participate in various
biological processes involved in tumorigenesis, metastasis and
proliferation (18–21). Therefore, lncRNAs may be considered
as diagnostic biomarkers for numerous types of cancer, such as
gastric, bladder, colorectum and prostate cancer (22–25). For
example, lncRNA-BANCR has been associated with lymph node
metastasis in colorectal cancer (26). The function of lncRNAs in breast
cancer has also been studied. For instance, lncRNA-SNHG15 regulates
microRNA (miRNA/miR)-211-3p and promotes cell proliferation,
migration and invasion of breast cancer (27). Additionally, lncRNA-MAPT-AS1 inhibits
cell proliferation and migration by regulating MAPT expression in
breast cancer (28). Therefore, the
aforementioned studies have indicated that lncRNAs may be important
for regulating breast cancer processes and may be potential
biomarkers of the disease. However, the role served by lncRNAs in
SLN metastasis of breast cancer is yet to be elucidated.
To screen for markers that can be used to identify
whether SLN has metastasized in breast cancer, in the present
study, the SLNs of patients with breast cancer were collected and
RNA sequencing (RNA-seq) was used to identify the key lncRNAs
involved in SLN metastasis. Furthermore, reverse
transcription-quantitative PCR (RT-qPCR) was conducted to analyze
the expression levels of lncRNAs among specimens with or without
SLN metastasis.
Materials and methods
Patient review and specimen
collection
The database of the Peking University Shenzhen
Hospital (Shenzhen, China) was reviewed between January 2018 and
December 2018 in breast cancer, and patients in the early stages (I
and II) of breast cancer were included in the present study. The
patients were >18 years old, had not received surgical
contraindication, chemotherapy or endocrine therapy, and had no
other malignancy or immune diseases. Samples were obtained from 46
patients with an age range of 26–72 years. The mean age of 26
patients with SLN metastasis was 47±2.726 years and that of 20
patients without SLN metastasis was 47±3.674 years. All the
patients were female. Patients were studied prospectively and data
were collected with regards to age; metastasis-relevant parameters
including disease history, tumor position, lymph node metastasis by
sentinel lymph node biopsy (SLNB) and pattern of the axillary lymph
nodes; and TNM stage according to the guidelines from the American
Joint Committee on Cancer (29).
Mammary areola injection was performed for the ultrasound contrast
to search for SLNs. Subsequently, punch biopsy was conducted in
order to collect the specimens. The patients underwent breast
cancer resection and intraoperative SLN biopsy within 48 h of
puncture. The present study was approved by the Institute Research
Medical Ethics Committee of the Peking University Shenzhen
Hospital, and informed consent was provided orally and in writing
by all patients.
RNA-seq
Total RNA from the tissues was purified using an
RNeasy Mini kit (Qiagen GmbH). RNA integrity was evaluated based on
the RNA integrity number (RIN) value using an Agilent Bioanalyzer
2100 (Agilent Technologies, Inc.). RNA clean-up was performed using
an RNA Clean XP kit (Beckman Coulter, Inc.) and the DNA residue was
removed with an RNase-free DNase Set (Qiagen GmbH). The quality and
concentration of the RNA were determined using NanoDrop 2000
(Thermo Fisher Scientific, Inc.). The ribosomal RNA was removed
using a NEBNext rRNA Depletion kit (New England BioLabs, Inc.).
Subsequently, 1 µg total RNA was used for library preparation using
a VAHTSTM mRNA-seq v2 library Prep kit (Vazyme Biotech Co., Ltd.),
according to the manufacturer's protocol. Briefly, the RNA was
fragmented and the double strand cDNA was synthesized.
Subsequently, end-polishing was performed and the cDNA fragments
were ligated with adapters. The ligated cDNA was amplified using 15
cycles of PCR for 10 sec at 98°C, 30 sec at 60°C and 30 s at 72°C
using a PCR master mixture (Illumina, Inc.) and subjected to
universal PCR amplification using DNA polymerase I (New England
BioLabs, Inc.) in order to obtain a library sufficient for
sequencing. The Agilent Bioanalyzer 2100 was used to evaluate the
quality of the library and an Illumina Hiseq 4000 (Illumina, Inc.)
was used for RNA-seq. SOAP (http://soap.genomics.org.cn/) was used to calculate
lncRNAs and mRNAs expression. Differentially expressed lncRNAs and
mRNAs were screened using R software version 3.1 (30) according to the following criteria:
False discovery rate (FDR) ≤0.001 and fold-change >2.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis
The Database for Annotation, Visualization and
Integrated Discovery (https://david.ncifcrf.gov/) was used to annotate the
potential functions in various signaling pathways of the
differentially expressed mRNAs. Subsequently, the functional
annotation of parental genes was predicted via GO functional
annotation (http://geneontology.org/page/go-enrichment-analyses).
KEGG pathway annotation (http://www.genome.jp/kegg/pathway.html) was also used
to identify the relevant pathways of the differentially expressed
mRNAs.
RT-qPCR
Six lncRNAs with lengths of 500–3,000 bp were
selected for assessment based on information from the lncRNASNP2
database (http://bioinfo.life.hust.edu.cn/lncRNASNP/) and their
predicted association with breast cancer. lncRNA expression levels
were verified by RT-qPCR. RNA was extracted according to the above
RNA-seq method. Gene expression was analyzed via RT-qPCR. RNA was
reverse-transcribed into cDNA using the cDNA Synthesis Kit system
(Promega Corporation) according to the manufacturer's protocol at
42°C for 15 min and then 95°C for 3 min. The qPCR reaction was
performed using GoTaq qPCR Master mix (Promega Corporation), and
qPCR amplification was performed using an ABI 7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Thermocycling
conditions for qPCR were as follows: 40 cycles of 20 sec at 95°C
followed by 30 sed at 60°C and 30 sec at 72°C. The relative mRNA
expression levels were calculated by 2−ΔΔCq method
(31). Primers for qPCR are
presented in Table I. GAPDH was used
as an internal control.
 | Table I.Primers used for quantitative
PCR. |
Table I.
Primers used for quantitative
PCR.
| Gene | Sequence, 5′→
3′ | Product length,
bp |
|---|
| lnc-ACAN-2:1 | F:
CAAAGGGGAGCCAAGGTAGG | 149 |
|
| R:
GGGTGAGCGTTCAGATTCCA |
|
| lnc-ZPBP2-4:1 | F:
CCTAGACGGCAGCTTAGGAC | 100 |
|
| R:
TTGTGGCAGTGTAAACCCCT |
|
| lnc-GATA3-16:1 | F:
CGAGGAGGCAGTGTGACAAA | 178 |
|
| R:
CTCTAGGAAGTGGAGGCACC |
|
| lnc-ACOX3-5:1 | F:
TTCATCATCTCGTGGGACGC | 96 |
|
| R:
GTGTCCAGCCTATTGGGACC |
|
|
lnc-ANGPTL1-3:3 | F:
AGTTGGGGACGCTAGAATGC | 109 |
|
| R:
TGTTGCCTATCCTCGCTGTT |
|
| lnc-GJA10-12:1 | F:
TCCAAGCTGTCCTGTACGAAG | 99 |
|
| R:
GCTGCTGATGCAAGCTGAAA |
|
| GAPDH | F:
GTCTCCTCTGACTTCAACAGCG | 235 |
|
| R:
ACCACCCTGTTGCTGTAGCCAA |
|
Statistical analysis
The data from three experimental repeats was shown
as mean ± standard deviation. Data comparisons were performed using
unpaired Student's t-tests and χ2 tests. Receiver
operating characteristic (ROC) curve was drawn to analyze the
specificity and sensitivity of lncRNA as a disease diagnosis.
P<0.05 was considered to indicate a statistically significant
difference. SPSS version 19.0 (IBM Corp.) and GraphPad Prism 8
(GraphPad Software, Inc.) were used to conduct the statistical
analyses.
Results
Characteristics of the patients
involved in the present study
The inclusion criteria for the present study were
patients diagnosed with breast cancer and those suitable for
axillary lymph node dissection. The reagent for contrast-enhanced
ultrasound was subcutaneously injected near the mammary areola to
search for the position of the SLN biopsy. The patients were
categorized into SLN(+) or SLN(−) metastasis groups and their
characteristics are summarized in Table
II. Analysis of the data identified that the positive rate of
SLN metastasis was associated with the pattern of the axillary
lymph nodes (P=0.0001), but was not associated with age, disease
history, tumor position, molecular subtyping of breast cancer or
TNM stage (all P>0.05; Table
II).
 | Table II.Clinicopathological parameters of
patients with (n=26) and without (n=20) SLN metastasis. |
Table II.
Clinicopathological parameters of
patients with (n=26) and without (n=20) SLN metastasis.
| Parameter | SLN(+) | SLN(+) NA | SLN(−) | SLN(−) NA | P-value |
|---|
| Cases | 26 | – | 20 | – |
|
| Mean age ± SD,
years | 47.27±2.726 | – | 47.60±3.674 | – | 0.9414 |
| Disease history,
year |
|
|
|
|
|
| ≤1 | 19 | – | 15 | – | 0.8211 |
|
>1 | 7 | – | 5 | – |
|
| Tumor position |
|
|
|
|
|
| Left
breast | 17 | – | 13 | – | 0.9783 |
| Right
breast | 9 | – | 7 | – |
|
| Lymph node
metastasis by SLN biopsy |
|
|
|
|
|
| + | 26 | – | 1 | – | <0.0001 |
| − | 0 | – | 19 | – |
|
| Pattern of the
axillary lymph nodes |
|
|
|
|
|
|
Non-suspicious | 1 | – | 12 | – |
|
|
Suspicious | 25 | – | 8 | – |
|
| Human epidermal
growth factor receptor 2 |
|
|
|
|
|
| + | 3 | 9 | 1 | 1 | 0.5162 |
| − | 14 |
| 18 |
|
|
| Triple negative
breast cancer |
|
|
|
|
|
| + | 0 | 9 | 3 | 1 | 0.2310 |
| − | 17 |
| 16 |
|
|
| Luminal A |
|
|
|
|
|
| + | 0 | 9 | 2 | 1 | 0.4873 |
| − | 17 |
| 17 |
|
|
| Luminal B |
|
|
|
|
|
| + | 14 | 9 | 13 | 1 | 0.5631 |
| − | 3 |
| 6 |
|
|
| Tumor size, cm |
|
|
|
|
|
|
<2 | 1 | – | 5 | – | 0.0949 |
| ≥2 | 25 | – | 15 | – |
|
| N stage |
|
|
|
|
|
| N0 | 7 | – | 11 | – | 0.0531 |
| N1 | 19 | – | 9 | – |
|
| M stage |
|
|
|
|
|
| M0 | 25 | – | 20 | – | 1.0000 |
| M1 | 1 | – | 0 | – |
|
Analysis of differentially expressed
lncRNAs and mRNAs associated with SLN(+) and SLN(−) metastasis
The differentially expressed lncRNAs and mRNAs in
patients with SLN(+) metastasis and SLN(−) metastasis are presented
in Figs. 1 and 2 as volcano plots, scatter plots and
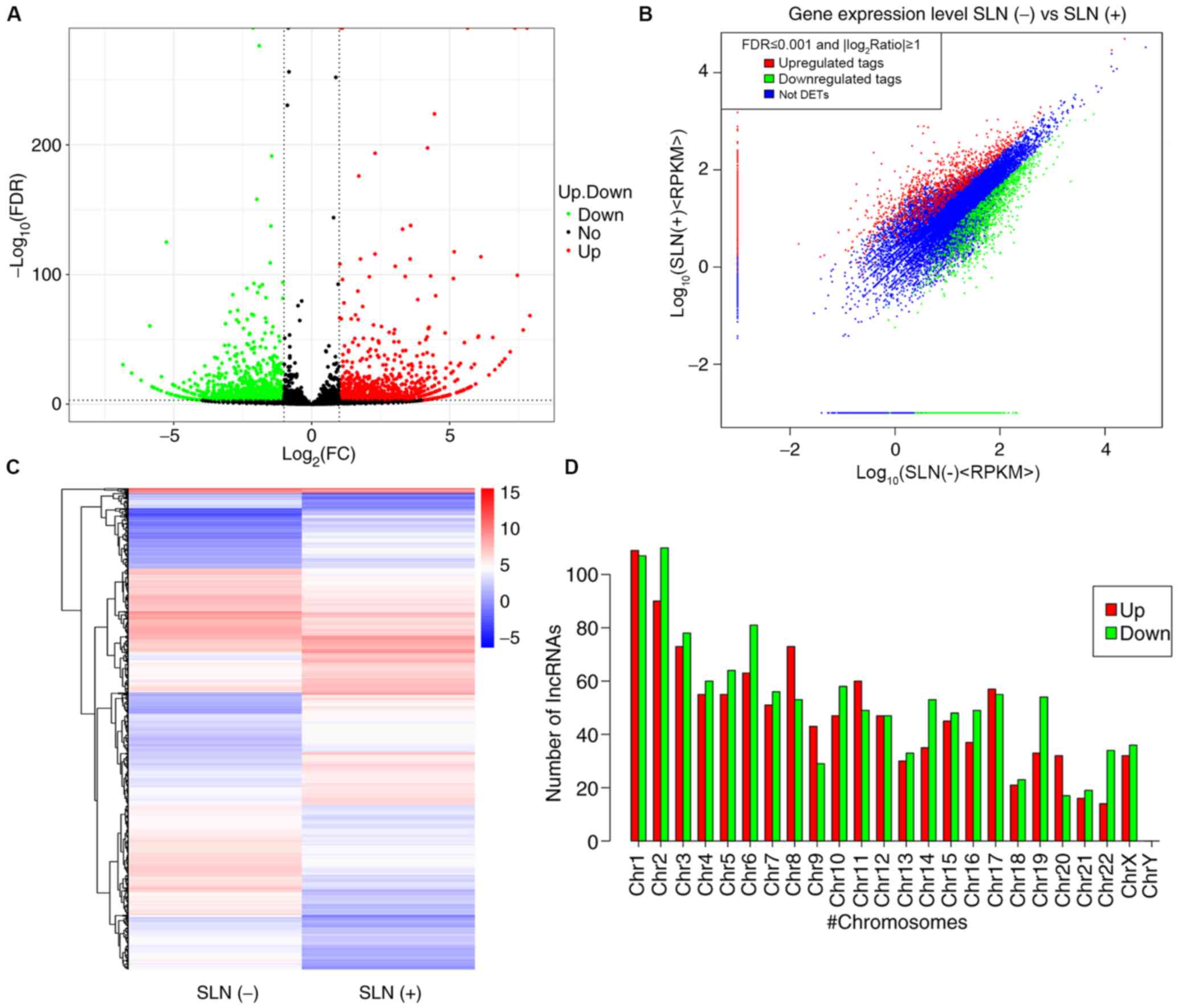
heatmaps. A total of 2,335 differentially expressed lncRNAs were
identified between patients with SLN(+/−) metastasis; of these,
1,120 were upregulated and 1,215 were downregulated (Fig. 1A-C; Table
SI). Furthermore, the expression levels of lncRNAs on different
chromosomes were both upregulated and downregulated (Fig. 1D). A total of 2,335 differentially
expressed mRNAs were found between patients with SLN(+/−)
metastasis; of these, 1,120 were upregulated and 1,215 were
downregulated (Fig. 2A-C and
Table SII). The Reads Per Kilobase
of transcript per Million mapped reads profiles for lncRNAs and
mRNAs were summarized and a fold-change >2 was used as a
selection criterion to screen significantly differentially
expressed lncRNAs and mRNAs. The top ten upregulated and
downregulated lncRNAs and mRNAs are summarized in Tables III and IV, respectively.
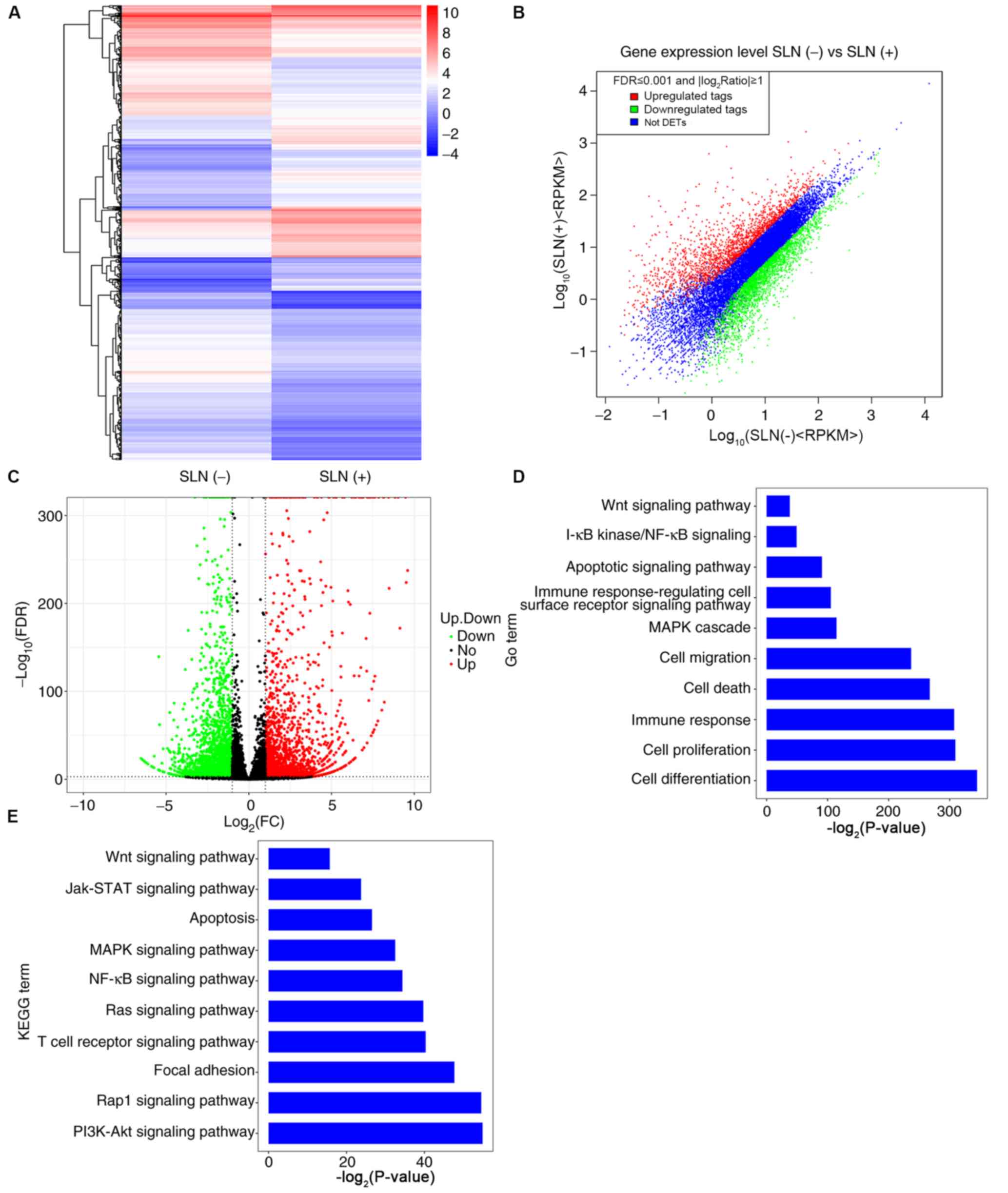 | Figure 2.Differentially expressed mRNAs, and
GO and KEGG analyses in patients with SLN(+) and SLN(−) metastasis.
(A) Heatmap (the color indicates the value after the logarithm of
the expression is taken; the red color indicates higher gene
expression and the blue color indicates lower gene expression), (B)
scatter plot and (C) volcano plot of the differentially expressed
mRNAs in patients with SLN(+) and SLN(−) metastasis. (D) Biological
processes of upregulated and downregulated mRNAs according to GO
enrichment. (E) Signal pathways of upregulated and downregulated
mRNAs according to KEGG pathway enrichment. GO, Gene Ontology;
KEGG, Kyoto Encyclopedia of Genes and Genomes; SLN, sentinel lymph
node; FDR, false discovery rate; FC, fold change; RPKM, Reads Per
Kilobase of transcript per Million mapped reads; DETs,
Differentially expressed tags; Jak, Janus kinase. |
 | Table III.Top ten differentially expressed
lncRNAs. |
Table III.
Top ten differentially expressed
lncRNAs.
| lncRNAs | Length, bp | SLN(−), RPKM | SLN(+), RPKM | log2
ratio, SLN(+)/SLN(−) |
Upregulation/downregulation
(SLN+/SLN-) | FDR |
|---|
| lnc-DDX47-3:1 | 2,740 | 0.391338128 | 225.814620700 | 9.172507504 | Up |
5.90×10−170 |
| LINC01087:1 | 3,516 | 2.744709397 | 779.663719700 | 8.150054784 | Up | 0.00×10° |
|
lnc-SLC39A11-10:48 | 5,836 | 0.183733117 | 50.494217380 | 8.102362680 | Up |
3.28×10−79 |
| TBILA:3 | 1,937 | 0.553570713 | 133.185239200 | 7.910450865 | Up |
5.70×10−69 |
|
lnc-CCDC74A-8:1 | 1,521 | 3.524873343 | 789.454094100 | 7.807140148 | Up | 0.00×10° |
|
lnc-DTNBP1-16:4 | 73,231 | 0.014642248 | 2.978646596 | 7.668376078 | Up |
6.65×10−58 |
| lnc-DHCR24-1:1 | 503 | 4.263484974 | 748.474609300 | 7.455776394 | Up |
4.34×10−100 |
|
lnc-CCDC74A-11:1 | 5,658 | 3.411240099 | 562.345523100 | 7.365016729 | Up | 0.00×10° |
|
lnc-ADPRHL1-5:1 | 509 | 2.106613892 | 309.046682100 | 7.196755050 | Up |
4.72×10−41 |
| lnc-IDNK-10:1 | 5,970 | 0.179609124 | 23.011640110 | 7.001359361 | Up |
1.47×10−35 |
| lnc-MB-6:1 | 1,733 | 69.298237020 | 0.605134684 | −6.839418562 | Down |
3.73×10−31 |
| lnc-P2RX3-4:1 | 2,727 | 35.388332380 | 0.384561206 | −6.523916736 | Down |
1.18×10−24 |
|
lnc-RANBP3L-4:2 | 3,613 | 21.071386510 | 0.290256963 | −6.181810760 | Down |
4.35×10−19 |
| lnc-TFF3-1:1 | 1,328 | 184.093942300 | 3.158730144 | −5.864953654 | Down |
4.06×10−61 |
|
lnc-TNFRSF13C-1:1 | 1,078 | 53.712791690 | 0.972818560 | −5.786951142 | Down |
3.67×10−14 |
|
lnc-NUDT12-11:1 | 549 | 99.609453600 | 1.910197464 | −5.704488982 | Down |
2.58×10−13 |
| lnc-N4BP2-3:4 | 530 | 103.180358500 | 1.978676241 | −5.704488982 | Down |
2.58×10−13 |
| lnc-MMP23B-1:1 | 1,808 | 27.874183710 | 0.580032305 | −5.586652492 | Down |
3.63×10−12 |
|
lnc-NDUFA10-7:1 | 1,024 | 49.215355610 | 1.024119539 | −5.586652492 | Down |
3.63×10−12 |
| lnc-HACL1-2:1 | 1,778 | 25.932203740 | 0.589819127 | −5.458328395 | Down |
4.94×10−11 |
 | Table IV.Top ten differentially expressed
mRNAs. |
Table IV.
Top ten differentially expressed
mRNAs.
| mRNA | Gene ID | SLN(−), RPKM | SLN(+), RPKM | log2
fold-change, SLN(+)/SLN(−) |
Upregulation/downregulation
(SLN+)/SLN-) | FDR |
|---|
| KRT19 | NM_002276 | 0.879993293 | 624.542682700 | 9.471091928 | Up | 0.00×10° |
| AGR2 | NM_006408 | 1.881223855 | 859.243757400 | 8.835252123 | Up | 0.00×10° |
| LRP2 | NM_004525 | 0.070514511 | 13.785703170 | 7.611036966 | Up | 0.00×10° |
| MUC16 | NM_024690 | 0.278551308 | 49.981202550 | 7.487298711 | Up | 0.00×10° |
| PRLR | NR_037910 | 0.237386784 | 38.441857770 | 7.339294629 | Up | 0.00×10° |
| SHANK2 | NR_110766 | 0.176387166 | 27.083658300 | 7.262533218 | Up | 0.00×10° |
| SORD | NR_034039 | 2.297196335 | 328.383682400 | 7.159364465 | Up | 0.00×10° |
| STC2 | NM_003714 | 0.519158633 | 62.454697730 | 6.910490851 | Up | 0.00×10° |
| EPCAM | NM_002354 | 1.184032220 | 135.573309000 | 6.839221026 | Up | 0.00×10° |
| PROM1 | NM_006017 | 0.564720687 | 62.228977230 | 6.783905243 | Up | 0.00×10° |
| SNORD17 | NR_003045 | 380.772408500 | 8.774673935 | −5.439439615 | Down |
4.50×10−140 |
| LRRC55 | NM_001005210 | 7.489998665 | 0.177513285 | −5.398966555 | Down |
7.56×10−63 |
| SDK2 | NM_001144952 | 4.915447389 | 0.179036436 | −4.778997604 | Down |
1.04×10−76 |
| CNR2 | NM_001841 | 18.023608310 | 0.720987293 | −4.643770224 | Down |
5.32×10−46 |
| TIE1 | NM_005424 | 5.619246835 | 0.241038256 | −4.543042733 | Down |
6.06×10−32 |
| MARCO | NM_006770 | 26.302585060 | 1.133295762 | −4.536608274 | Down |
3.77×10−68 |
| FABP4 | NM_001442 | 74.146260690 | 3.245195638 | −4.513996578 | Down |
2.52×10−87 |
| NPIPB3 | NM_130464 | 5.976835270 | 0.265142081 | −4.494544215 | Down |
1.04×10−30 |
| MAST3 | NM_015016 | 6.150545353 | 0.298601665 | −4.364420222 | Down |
4.44×10−50 |
| GRAP | NM_006613 | 21.648787300 | 1.101606876 | −4.296604837 | Down |
4.67×10−60 |
Functional analysis of differentially
expressed genes
GO annotation analysis was used to examine the
processes in which the differentially expressed mRNAs were
involved. The majority of these mRNAs were involved in ‘cell
differentiation’, ‘cell proliferation’, ‘immune response’, ‘cell
death’, ‘cell migration’ and ‘mitogen-activated protein kinase
(MAPK) cascade’, but a few were also involved in the ‘Wnt signaling
pathway’ and the ‘apoptotic signaling pathway’ (Fig. 2D). In order to assess the pathways in
which the mRNAs were involved, KEGG pathway annotation analysis was
conducted; the results revealed that the ‘PI3K/Akt signaling
pathway’, ‘Rhoptry-associated protein1 (Rap1) signaling pathway’
and ‘MAPK signaling pathway’ were the main enriched pathways
(Fig. 2E), suggesting that these
were the primary signaling pathways in which mRNAs were
involved.
Validation of lncRNA expression levels
in patients with SLN(+) and SLN(−) metastasis
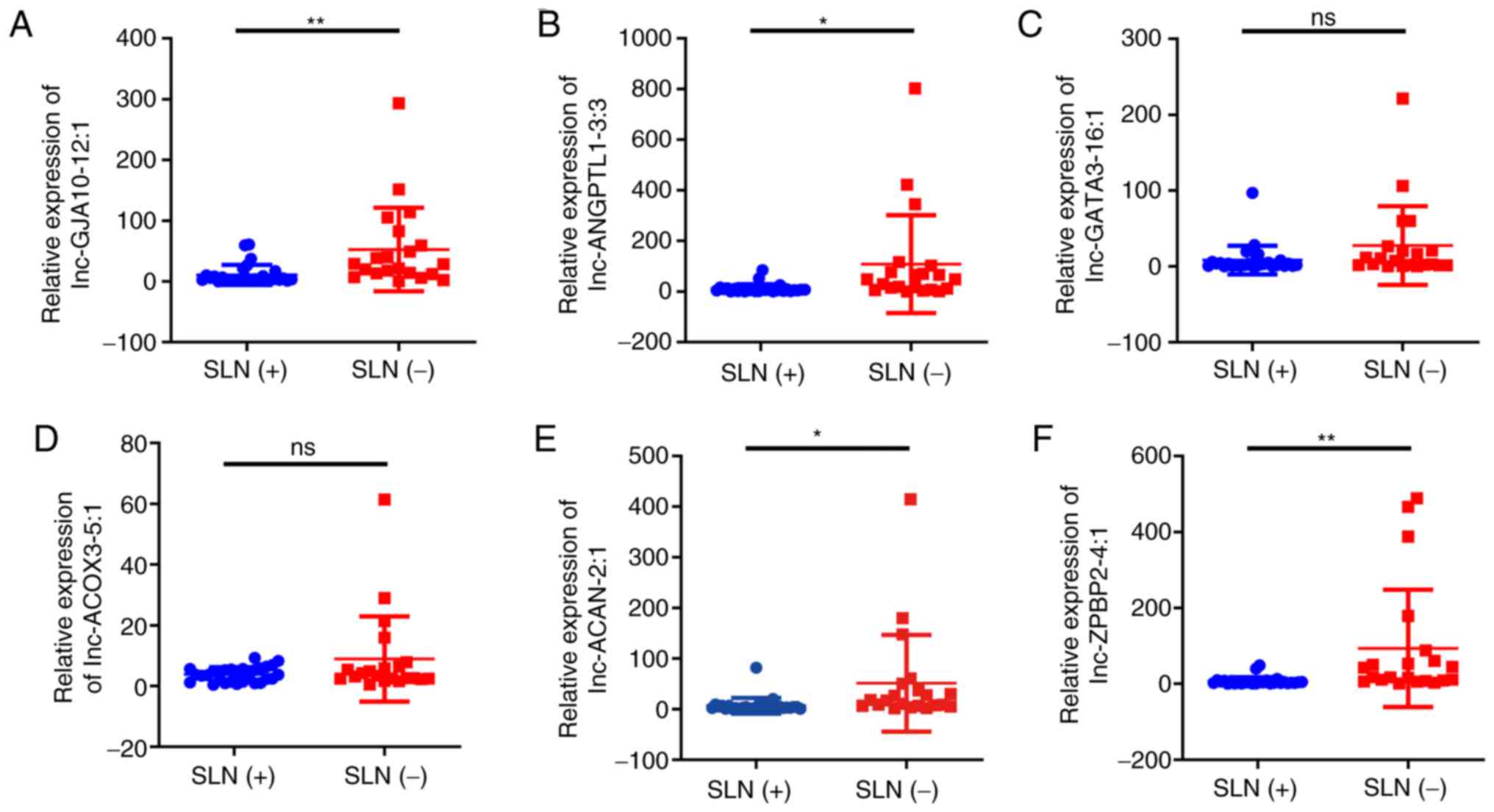
In the present study, the differentially expressed
lncRNAs, including lnc-ANGPTL1-3:3, lnc-GJA10-12:1, lnc-ACAN-2:1,
lnc-ZPBP2-4:1, lnc-GATA3-16:1 and lnc-ACOX3-5:1 were analyzed by
qPCR. As shown in Fig. 3, the
expression levels of lnc-ANGPTL1-3:3, lnc-GJA10-12:1, lnc-ACAN-2:1
and lnc-ZPBP2-4:1 were significantly downregulated in the SLN (+)
group compared with the SLN (−) groups. However, only the
expression results of lnc-ANGPTL1-3:3 and lnc-GJA10-12:1 were
confirmed using RNA-seq, while the results for lnc-ACAN-2:1 and
lnc-ZPBP2-4:1 expression were the opposite to those obtained via
RNA-seq (Table SI). It was also
found that lnc-GATA3-16:1 and lnc-ACOX3-5:1 did not exhibit
significant differential expression levels according to the qPCR
results (Fig. 3).
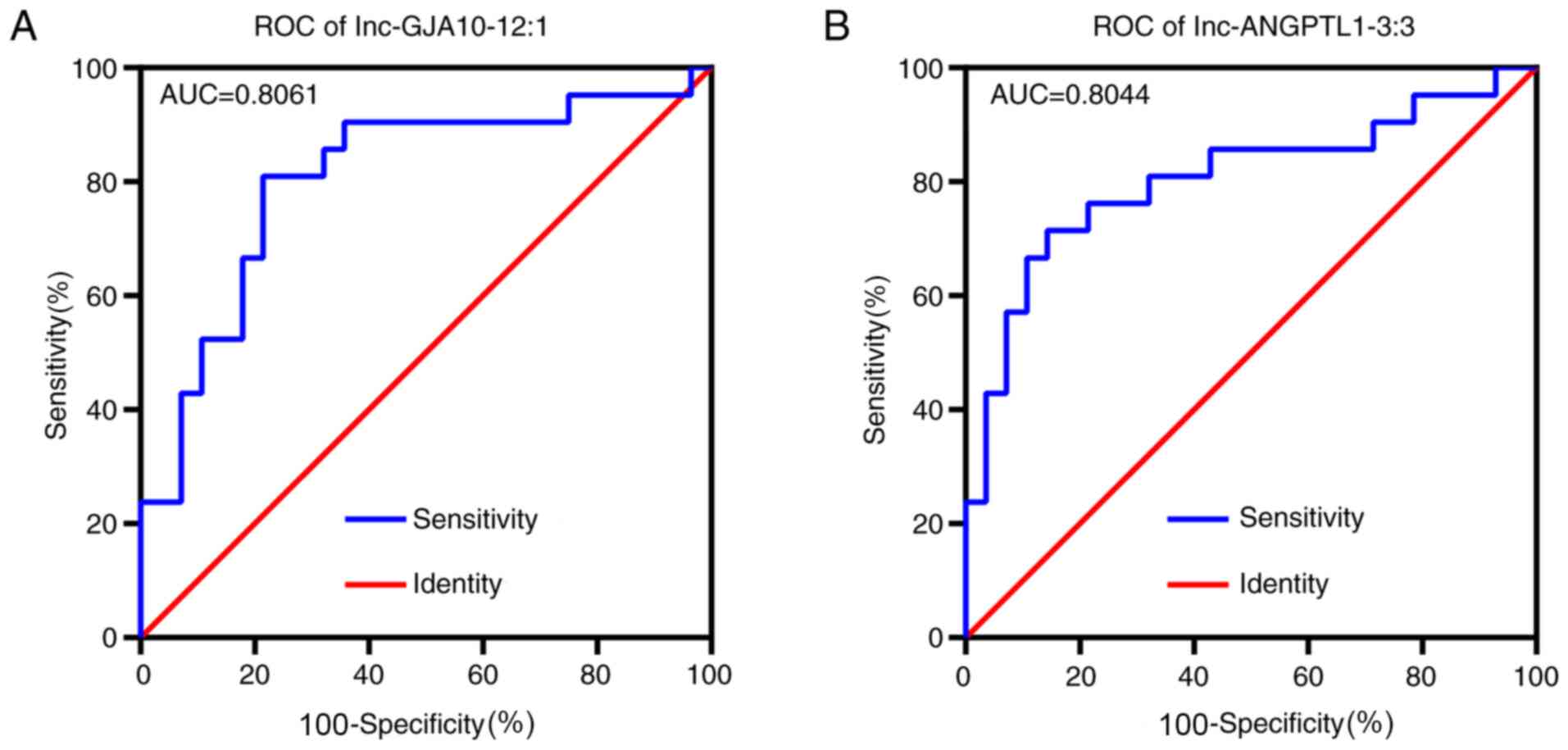
Receiver operating characteristic (ROC) curve
analysis identified that both lnc-ANGPTL1-3:3 and lnc-GJA10-12:1
had high area under the curve values (>0.8), which indicated
that both lncRNAs were closely associated with SLN metastasis and
may be suitable biomarkers for SLN diagnosis (Fig. 4). According to associations of
lnc-ANGPTL1-3:3 and lnc-GJA10-12-1 expression levels with
pathological features, lnc-ANGPTL1-3:3 and lnc-GJA10-12:1
expression levels were highly associated with patients with
axillary lymph node metastasis or SLN metastasis diagnosis
(Table V).
 | Table V.Association of lnc-ANGPTL1-3:3 and
lnc-GJA10-12:1 with patient clinicopathological features. |
Table V.
Association of lnc-ANGPTL1-3:3 and
lnc-GJA10-12:1 with patient clinicopathological features.
| Groups | Cases | lnc-ANGPTL1-3:3
expression |
χ2-value | P-value | lnc-GJA10-12:1
expression |
χ2-value | P-value |
|---|
| Age, years |
|
|
|
|
| 1.525 | 0.1345 |
|
<45 | 22 | 69.65±37.93 | 0.8166 | 0.4185 | 42.19±14.43 |
|
|
|
≥45 | 24 | 40.52±15.60 |
|
| 19.23±5.765 |
|
|
| SLN metastasis |
|
|
|
|
| 2.812 | 0.0074 |
| + | 26 | 21.32±5.312 | 3.574 | 0.0009 | 19.61±4.827 |
|
|
| − | 20 | 188.7±93.27 |
|
| 71.16±31.90 |
|
|
| Pattern of the
axillary |
|
|
|
|
| 4.215 | 0.0001 |
| lymph nodes |
|
|
|
|
|
|
|
|
Non-suspicious | 13 | 149.2±67.41 | 2.944 | 0.0052 | 77.07±23.62 |
|
|
|
Suspicious | 33 | 21.16±10.50 |
|
| 13.92±3.269 |
|
|
| Tumor size, cm |
|
|
|
|
| 0.898 | 0.3741 |
|
<2 | 6 | 24.51±11.61 | 0.5866 | 0.5605 | 12.47±5.894 |
|
|
| ≥2 | 40 | 60.29±23.34 |
|
| 32.87±8.677 |
|
|
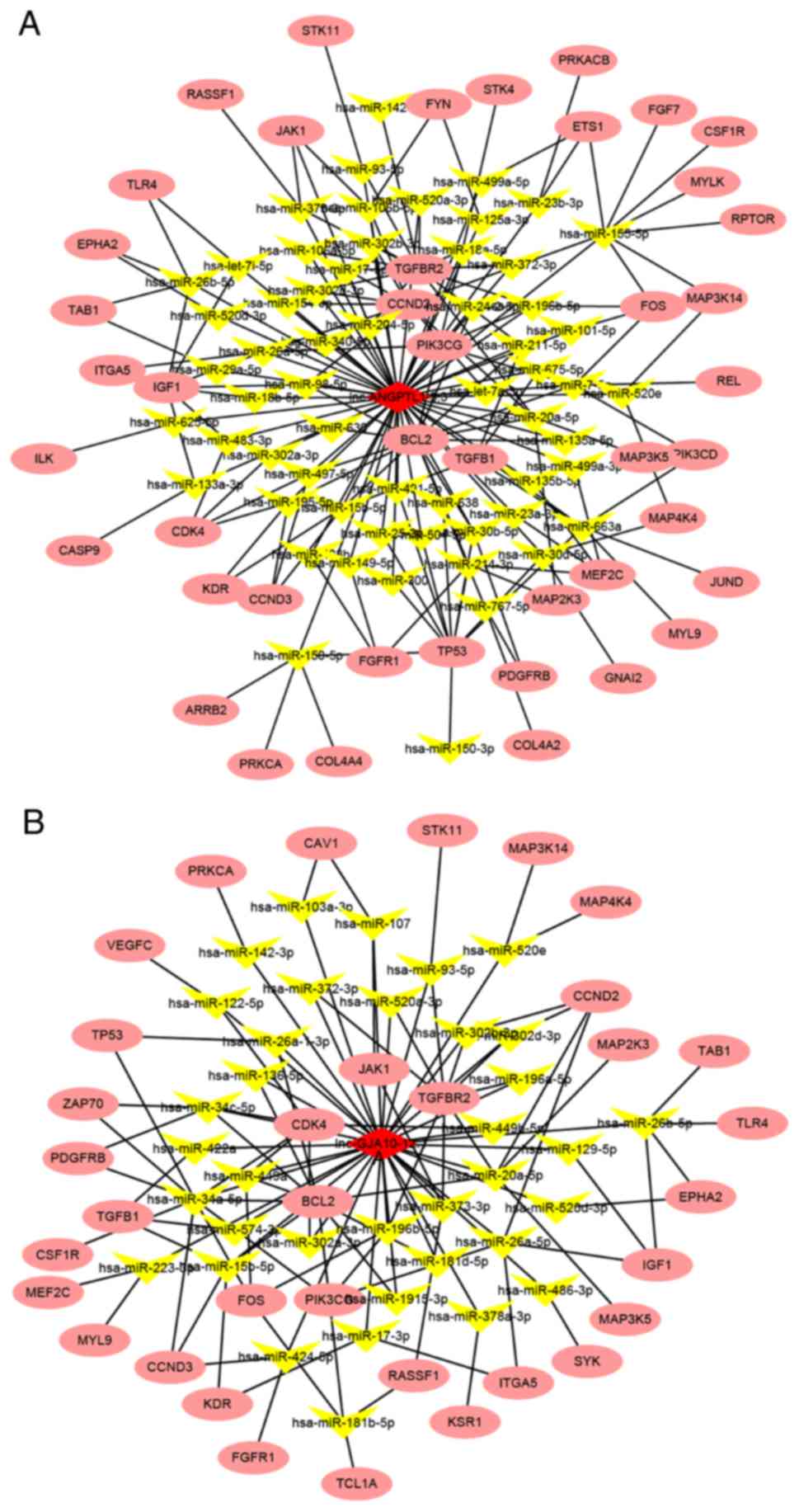
Co-expression network analysis of
lncRNAs and mRNAs
After ROC analysis and verification using qPCR,
lnc-ANGPTL1-3:3 and lnc-GJA10-12:1 were selected for functional
prediction analysis, in which the targets of these lncRNAs were
predicted based on the competing endogenous RNA (ceRNA) principle
(32). The target genes of lncRNAs,
which were significantly downregulated in the SLN (+) group in mRNA
sequencing compared with the SLN (−) group, in the top five
pathways in the KEGG pathway analysis based on the ceRNA principle
were used to establish network regulatory maps. The miRNAs that
interacted with lncRNAs were predicted based on the sequences of
lncRNAs, and the targets of miRNAs were thus also predicted. Under
this principle, lncRNA-miRNA-mRNA cascades were identified. The top
ten miRNAs and their corresponding targeted mRNAs associated with
either lnc-ANGPTL1-3:3 or lnc-GJA10-12:1 are presented in Tables VI and VII, respectively. Moreover, the
interactions are summarized in the regulation network illustrated
in Fig. 5. The results demonstrated
that the miRNAs associated with lnc-GJA10-12:1 and lnc-ANGPTL1-3:3
were commonly involved in regulating the miR-302 family, including
miR-302d-3p and miR-302c-3p, which together targeted AKT1.
Specifically, lnc-ANGPTL1-3:3 was predicted to target miR-520b to
regulate MAP3K2 expression. In addition, lnc-GJA10-12:1 was
predicted to target miR-34a-5p to regulate MAP2K1 and MAP3K9
expression, as well as miR-449a to regulate MAP2K1 expression.
Therefore, the results indicated that lnc-GJA10-12:1 and
lnc-ANGPTL1-3:3 may be involved in signal conditioning networks to
control SLN metastasis in breast cancer.
 | Table VI.Top ten miRNAs and corresponding
targeted mRNAs associated with lnc-ANGPTL1-3:3. |
Table VI.
Top ten miRNAs and corresponding
targeted mRNAs associated with lnc-ANGPTL1-3:3.
| miRNA | Specific binding
sites predicted | mRNA |
|---|
|
hsa-miR-302c-3p | 306, 1284, 1993,
2215 | ESR1, CCND1, BMPR2,
AKT1, MICA |
|
hsa-miR-548d-5p | 960, 1053, 1491,
2264 | PPARA |
|
hsa-miR-302a-3p | 306, 1281,
2215 | CCND1, CCND1, CDK4,
CDKN1A, LEFTY1, LEFTY2, DAZAP2, SLAIN1, TOB2, NR2F2, AKT1, TAC1,
CDK2, BMI1, CDK1, MBD2, NANOG |
|
hsa-miR-302b-3p | 307, 1284,
2215 | CCND2, BMI1,
TGFBR2, RHOC, AKT1, HDAC4, EGFR, ERBB4, AKT2 |
| hsa-miR-373-3p | 304, 1281,
2215 | RAD52, RAD23B, XPA,
MRE11A, CD44, CD44, LATS2, LATS2, LATS2, LATS2, RECK, VEGFA, TXNIP,
RABEP1, MYC, MBD2, RASSF1, MTOR, SIRT1, NFIB, DKK1, TGFBR2, BTG1,
LEFTY1, TNFAIP1, LEFTY2 |
|
hsa-miR-520c-3p | 307, 1282,
2218 | APP, CD44, CD44,
MTOR, SIRT1, GPC3, GPC3, MICA, EIF4G1 |
|
hsa-miR-302d-3p | 306, 1284,
2218 | TRPS1, KLF13,
VEGFA, MBNL2, NR4A2, ERBB4, CDK2, CCND2, LEFTY1, LEFTY2, AKT1 |
| hsa-miR-520b | 308, 1283,
2219 | CDKN1A, MICA,
LAMTOR5, IL8, MAP3K2, CCND1, CD46, PFKP |
|
hsa-miR-520a-3p | 304, 1282,
2216 | CDKN1A, PFKP |
|
hsa-miR-520d-3p | 304, 1282,
2216 | EPHA2, EPHA2 |
 | Table VII.Top ten miRNAs and corresponding
targeted mRNAs associated with lnc-GJA10-12:1. |
Table VII.
Top ten miRNAs and corresponding
targeted mRNAs associated with lnc-GJA10-12:1.
| miRNA | Specific binding
sites predicted | mRNA |
|---|
| hsa-miR-223-3p | 466, 539 | RHOB, RHOB, RHOB,
NFIX, E2F1, MEF2C, MEF2C, NFIA, NFIA, STMN1, STMN1, STMN1, LMO2,
LMO2, Arid4b, Il6, Lpin2, CHUK, FBXW7, FBXW7, FBXW7, FBXW7, IGF1R,
IGF1R, IGF1R, IGF1R, EPB41L3, SLC2A4, LIF, SP3, ARTN, FOXO1, FOXO1,
HSP90B1, SCARB1, PARP1, CDK2, ECT2, PTBP2, TAL1, ATM, CYB5A, TOX,
PRDM1, STAT5A, CARM1, POLR3G, FOXO3, CDC27, SP1, CCL3, IL6, CXCL2,
ABCB1, CAPRIN1, PAX6, CFTR |
| hsa-miR-136-5p | 437, 642 | MTDH, BCL2 |
| hsa-miR-34a-5p | 619 | SIRT1, SIRT1,
SIRT1, SIRT1, SIRT1, SIRT1, SIRT1, SIRT1, SIRT1, SIRT1, VAMP2,
NOTCH1, NOTCH1, NOTCH1, NOTCH1, NOTCH1, NOTCH1, NOTCH1, NOTCH1,
NOTCH1, NOTCH1, NOTCH1, DLL1, DLL1, DLL1, DLL1, BCL2, BCL2, BCL2,
BCL2, BCL2, BCL2, BCL2, BCL2, BCL2, BCL2, YY1, YY1, YY1, MAGEA12,
MAGEA6, MAGEA3, MAGEA2, AXIN2, WNT1, WNT1, CCND1, CCND1, CCND1,
CDK6, CDK6, CDK6, CDK6, CDK6, CDK6, CDK6, CDC25A, CCND3, MYCN,
MYCN, MYCN, MYCN, MYCN, MYCN, E2F3, E2F3, E2F3, E2F3, E2F3, E2F3,
CCNE2, CCNE2, CDK4, CDK4, CDK4, MET, MET, MET, MET, MET, MET, MET,
MET, MET, MET, MYB, MYC, MYC, MYC, MYC, MAP2K1, JAG1, JAG1, JAG1,
PEA15, VEGFA, IFNB1, HNF4A, HNF4A, HNF4A, Sirt1, Mycn, E2f3,
NOTCH2, NOTCH2, CD44, CD44, FOXP1, FOXP1, MAP3K9, CEBPB, SPI1,
ZAP70, PDGFRA, PDGFRA, PDGFRA, NANOG, SOX2, IMPA1, IMPDH2, ULBP2,
SYT1, STX1A, FOSL1, FOSL1, EPHA5, AXL, AXL, CCL22, KLB, PPP1R10,
BMP7, LEF1, ACSL1, GRM7, LDHA, MTA2, CDKN2A, HDAC1, HDAC1, E2F1,
ACSL4, CDKN2C, SNAI1, CD24, AR, NAMPT, PDGFRB, PDGFRB, BIRC5,
BIRC5, SRC, SRC, PCBP2, KDM4A, KCNH1, KLF4, MDM4, MDM4, INHBB,
RICTOR, POU5F1, ARHGDIB, CYBB, HOTAIR, DGUOK, GAS1, CSF1R, KIT,
SIRT7, GALNT7, IL6R, FUT8, L1CAM |
|
hsa-miR-449b-5p | 618 | SIRT1, CCNE2, MET,
GMNN, HDAC1, CDK4, CDC25A, CDK6 |
| hsa-miR-449a | 618 | HDAC8, HDAC1,
HDAC1, CDK6, CDK6, CDK6, CDC25A, CDC25A, CCND1, CCND1, HNF4A,
HNF4A, GMNN, MET, MET, CCNE2, SIRT1, BCL2, BCL2, BCL2, BCL2,
NOTCH1, NOTCH1, WISP2, CDK4, LEF1, E2F3, E2F3, ITPR1, MAP2K1 |
|
hsa-miR-302d-3p | 420 | TRPS1, KLF13,
VEGFA, MBNL2, NR4A2, ERBB4, CDK2, CCND2, LEFTY1, LEFTY2, AKT1 |
|
hsa-miR-302c-3p | 420 | ESR1, CCND1, BMPR2,
AKT1, MICA |
| hsa-miR-561-3p | 98 | NR0B1 |
|
hsa-miR-196b-5p | 665 | HOXB8, HOXC8, CD8A,
HOXA9, HOXA9, MEIS1, FAS, ETS2, RDX, HOXB7 |
|
hsa-miR-181d-5p | 679 | BCL2, BCL2, HRAS,
MGMT, MGMT, RAP1B |
Discussion
In the present study, RNA-seq was performed to
identify the lncRNAs involved in the SLN metastasis processes in
breast cancer. According to the qPCR results, lnc-ANGPTL1-3:3 and
lnc-GJA10-12:1 were downregulated in SLN metastasis specimens. The
first identified lncRNA, lnc-ANGPTL1-3:3, is derived from the
angiopoietin-like 1 (ANGPTL1) gene, which is a potent
regulator of angiogenesis and can interact with integrin α1β1 to
suppress hepatocellular carcinoma angiogenesis and metastasis via
inhibition of Janus kinase 2/STAT3 signaling (33). Furthermore, the second lncRNA,
lnc-GJA10-12:1, is derived from the gap junction protein α10
(GJA10) gene (34). Thus, the
results of the current study indicated that both lncRNAs may
potentially serve important roles in the regulation of SLN
metastasis in patients with breast cancer.
lncRNAs interact with miRNAs and can activate or
suppress their functions, resulting in increased or decreased
expression of their downstream targeted mRNAs (35). The GO and KEGG analyses in the
present study revealed that the MAPK and PI3K/Akt signaling
pathways may serve critical roles in SLN metastasis in breast
cancer. Additionally, bioinformatics analysis predicted the
targeted miRNAs, as well as the mRNAs. It was found that
miR-302c-3p and miR-302d-3p were each targeted by both candidate
lncRNAs, lnc-ANGPTL1-3:3 and lnc-GJA10-12:1, whereas miR-302a-3p
and miR-302b-3p were targeted by lnc-ANGPTL1-3:3 alone to regulate
the SLN metastasis processes of breast cancer; therefore, the
miR-302 family may serve an important role in SLN metastasis of
lncRNA-regulated breast cancer. A previous study reported that
miR-302c-3p suppressed tumorigenesis and development in
hepatocellular carcinoma by targeting tumor necrosis factor
receptor-associated factor 4 (36).
Furthermore, AKT1 serves a crucial role as a regulator of cell
invasion and proliferation in breast cancer (37–39). It
has also been shown that the inhibition of AKT1 induces breast
cancer metastasis via β-catenin nuclear accumulation mediated by
the epidermal growth factor receptor (40). Based on the present results, it was
speculated that lnc-ANGPTL1-3:3 and lnc-GJA10-12:1 may regulate two
AKT1-targeting miRNAs, miR-302c-3p and miR-302d-3p, and thus
control SLN metastasis in breast cancer.
MAPK, which is an important transmitter of signals
from the cell surface to the nucleus, serves an important role in
the development and metastasis of cancer, and it has been reported
that chemokine ligand 28 promotes the growth and metastasis of
breast cancer via MAPK-mediated pro-metastatic and anti-apoptotic
mechanisms (41). A previous study
revealed that miR-34c-3p regulated cancerous development and
epithelial-mesenchymal transition of triple-negative breast cancer
cells by regulating MAP3K2 signaling (42). Furthermore, in non-small cell lung
cancer, miR-520a-3p suppresses cell proliferation, metastasis and
apoptosis by targeting MAP3K2 (43),
and miR-449a inhibits cell invasion by suppressing MAP2K1 (44). Based on the aforementioned studies
and the present results, it was indicated that in SLN metastasis of
breast cancer lnc-ANGPTL1-3:3 may target miR-520b to regulate
MAP3K2 expression, and that lnc-GJA10-12:1 may target miR-34a-5p to
regulate MAP2K1 and MAP3K9 expression, as well as miR-449a to
regulate MAP2K1 expression.
At present, there are no mature animal models in
vivo and cell models in vitro for the study of SLN
metastasis. Therefore, the specific mechanism of lnc-ANGPTL1-3:3
and lnc-GJA10-12:1 regulating SLN metastasis were not investigated
in the present study. In addition, the expression levels of
lnc-ANGPTL1-3:3 and lnc-GJA10-12:1 in breast cancer in situ
were not investigated in the present study. The aforementioned
limitations should be investigated by future studies.
In conclusion, lnc-ANGPTL1-3:3 and lnc-GJA10-12:1
may be important regulators of SLN metastasis in breast cancer via
their downregulated expression. Moreover, the present results
suggested that lnc-ANGPTL1-3:3 and lnc-GJA10-12:1 may regulate the
PI3K/Akt and MAPK signaling pathways by targeting the miR-302
family, miR-520a-3p, miR-34a-5p and miR-449a, which may serve a
crucial role in SLN metastasis of breast cancer. It was also
demonstrated that lnc-ANGPTL1-3:3 and lnc-GJA10-12:1 in SLN may
serve as potential markers of breast cancer metastasis. Hence,
future studies should further investigate lnc-ANGPTL1-3:3 and
lnc-GJA10-12:1 expression in breast cancer tissues to provide a
basis for the surgical treatment of breast cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Guangzhou Forevergen
Biosciences Co., Ltd. for providing technical help.
Funding
The present study was supported by the Health and
Family Planning Commission of Shenzhen Municipality (grant no.
SZXJ2017026), the Shenzhen ‘Sanming’ Project of Medicine (grant no.
SZSM201512026) and Shenzhen Key Medical Discipline Construction
Fund.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DS and JZ conceived and designed the experiments.
DS, JZ, and WW performed the experiments. LL, JL, and XL analyzed
the experimental data. All authors reviewed and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institute
Research Medical Ethics Committee of the Peking University Shenzhen
Hospital (Shenzhen, China), and informed consent was provided
orally and in writing by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Choi HY, Park M, Seo M, Song E, Shin SY
and Sohn YM: Preoperative axillary lymph node evaluation in breast
cancer: Current issues and literature review. Ultrasound Q.
33:6–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Promish DI: Prediction of axillary lymph
node involvement of women with invasive breast carcinoma: A
multivariate analysis. Cancer. 85:1201–1203. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fein DA: Identification of women with
T1-T2 breast cancer at low risk of positive axillary nodes. J Surg
Oncol. 65:34–39. 2015. View Article : Google Scholar
|
|
5
|
Gajdos C, Tartter PI and Bleiweiss IJ:
Lymphatic invasion, tumor size, and age are independent predictors
of axillary lymph node metastases in women with T1 breast cancers.
Ann Surg. 230:692–696. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gann PH, Colilla SA, Gapstur SM,
Winchester DJ and Winchester DP: Factors associated with axillary
lymph node metastasis from breast carcinoma: Descriptive and
predictive analyses. Cancer. 86:1511–1519. 2015. View Article : Google Scholar
|
|
7
|
Rivadeneira DE, Simmons RM, Christos PJ,
Hanna K, Daly JM and Osborne MP: Predictive factors associated with
axillary lymph node metastases in T1a and T1b breast carcinomas:
Analysis in more than 900 patients. J Am Coll Surg. 191:1–6,
Discussion 6–8. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tabár L, Chen HH, Duffy SW, Yen MF, Chiang
CF, Dean PB and Smith RA: A novel method for prediction of
long-term outcome of women with T1a, T1b, and 10–14 mm invasive
breast cancers: A prospective study. Lancet. 355:429–433. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wong JH: Sentinel lymphadenectomy in
breast cancer: University research tool or community practice? Surg
Clin North Am. 80:1821–1830. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cserni G, Boross G and Baltás B: The role
of the histopathological analysis of sentinel lymph nodes in breast
cancer. Preliminary findings. Orv Hetil. 139:1899–1903. 1998.(In
Hungarian). PubMed/NCBI
|
|
11
|
Lucci A, McCall LM, Beitsch PD, Whitworth
PW, Reintgen DS, Blumencranz PW, Leitch AM, Saha S, Hunt KK and
Giuliano AE; American College of Surgeons Oncology Group, :
Surgical complications associated with sentinel lymph node
dissection (SLND) plus axillary lymph node dissection compared with
SLND alone in the American College of Surgeons Oncology Group Trial
Z0011. J Clin Oncol. 25:3657–3663. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Veronesi U: A randomized comparison of
sentinel-node biopsy with routine axillary dissection in breast
cancer. N Engl J Med. 349:546–553. 2005. View Article : Google Scholar
|
|
13
|
Giuliano AE, Haigh PI, Brennan MB, Hansen
NM, Kelley MC, Ye W, Glass EC and Turner RR: Prospective
observational study of sentinel lymphadenectomy without further
axillary dissection in patients with sentinel node-negative breast
cancer. J Clin Oncol. 18:2553–2559. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Burak WE, Hollenbeck ST, Zervos EE, Hock
KL, Kemp LC and Young DC: Sentinel lymph node biopsy results in
less postoperative morbidity compared with axillary lymph node
dissection for breast cancer. Am J Surg. 183:23–27. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Haid A, Kuehn T, Konstantiniuk P,
Köberle-Wührer R, Knauer M, Kreienberg R and Zimmermann G:
Shoulder-arm morbidity following axillary dissection and sentinel
node only biopsy for breast cancer. Eur J Surg Oncol. 28:705–710.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schrenk P, Rieger R, Shamiyeh A and Wayand
W: Morbidity following sentinel lymph node biopsy versus axillary
lymph node dissection for patients with breast carcinoma. Cancer.
88:608–614. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Temple LK, Baron R, Cody HS III, Fey JV,
Thaler HT, Borgen PI, Heerdt AS, Montgomery LL, Petrek JA and Van
Zee KJ: Sensory morbidity after sentinel lymph node biopsy and
axillary dissection: A prospective study of 233 women. Ann Surg
Oncol. 9:654–662. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y,
Yan F, Zhu P, Wu J, Huang G, et al: Mesenchymal stem cells promote
hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and
miR-34a. Cancer Res. 77:6704–6716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yin J, Luo W, Zeng X, Zeng L, Li Z, Deng
X, Tan X and Hu W: UXT-AS1-induced alternative splicing of UXT is
associated with tumor progression in colorectal cancer. Am J Cancer
Res. 7:462–472. 2017.PubMed/NCBI
|
|
22
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ellinger J, Alam J, Rothenburg J, Deng M,
Schmidt D, Syring I, Miersch H, Perner S and Müller SC: The long
non-coding RNA lnc-ZNF180-2 is a prognostic biomarker in patients
with clear cell renal cell carcinoma. Am J Cancer Res.
5:27992015.PubMed/NCBI
|
|
24
|
Shi J, Li X, Zhang F, Zhang C, Guan Q, Cao
X, Zhu W, Zhang X, Cheng Y, Ou K, et al: Circulating lncRNAs
associated with occurrence of colorectal cancer progression. Am J
Cancer Res. 5:2258–2265. 2015.PubMed/NCBI
|
|
25
|
Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y,
Liang W, Hu C, Liu Y, Li J, et al: Genome-wide lncRNA microarray
profiling identifies novel circulating lncRNAs for detection of
gastric cancer. Theranostics. 7:213–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen X, Bai Y, Luo B and Zhou X:
Upregulation of lncRNA BANCR associated with the lymph node
metastasis and poor prognosis in colorectal cancer. Biol Res.
50:322017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kong Q and Qiu M: Long noncoding RNA
SNHG15 promotes human breast cancer proliferation, migration and
invasion by sponging miR-211-3p. Biochem Biophys Res Commun.
495:1594–1600. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan Y, Pan Y, Cheng Y, Yang F, Yao Z and
Wang O: Knockdown of LncRNA MAPT-AS1 inhibites proliferation and
migration and sensitizes cancer cells to paclitaxel by regulating
MAPT expression in ER-negative breast cancers. Cell Biosci.
8:72018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gershenwald JE, Scolyer RA, Hess KR,
Sondak VK, Long GV, Ross MI, Lazar AJ, Faries MB, Kirkwood JM,
McArthur GA, et al: Melanoma staging: Evidence-based changes in the
American Joint Committee on Cancer eighth edition cancer staging
manual. CA Cancer J Clin. 67:472–492. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Liu J, Zhou G, Guo J, Yan H, Niu
Y, Li Y, Yuan C, Geng R, Lan X, et al: Whole-genome sequencing of
eight goat populations for the detection of selection signatures
underlying production and adaptive traits. Sci Rep. 6:389322016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao
WZ, Peng H, Hong WW, Yin LH, Pu YP and Liang GY: Integrated
analysis of long non-coding RNA-associated ceRNA network reveals
potential lncRNA biomarkers in human lung adenocarcinoma. Int J
Oncol. 49:2023–2036. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan Q, Jiang L, Liu M, Yu D, Zhang Y, Li
Y, Fang S, Li Y, Zhu YH, Yuan YF and Guan XY: ANGPTL1 interacts
with integrin α1β1 to suppress HCC angiogenesis and metastasis by
inhibiting JAK2/STAT3 signaling. Cancer Res. 77:5831–5845. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Söhl G, Nielsen PA, Eiberger J and
Willecke K: Expression profiles of the novel human connexin genes
hCx30.2, hCx40.1, and hCx62 differ from their putative mouse
orthologues. Cell Commun Adhes. 10:27–36. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiao B, Zhang W, Chen L, Hang J, Wang L,
Zhang R, Liao Y, Chen J, Ma Q, Sun Z and Li L: Analysis of the
miRNA-mRNA-lncRNA network in human estrogen receptor-positive and
estrogen receptor--negative breast cancer based on TCGA data. Gene.
658:28–35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang L, Guo Y, Liu X, Wang T, Tong X, Lei
K, Wang J, Huang D and Xu Q: The tumor suppressive miR-302c-3p
inhibits migration and invasion of hepatocellular carcinoma cells
by targeting TRAF4. J Cancer. 9:2693–2701. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cejalvo JM, Pérez-Fidalgo JA, Ribas G,
Burgués O, Mongort C, Alonso E, Ibarrola-Villava M, Bermejo B,
Martínez MT, Cervantes A and Lluch A: Clinical implications of
routine genomic mutation sequencing in PIK3CA/AKT1 and
KRAS/NRAS/BRAF in metastatic breast cancer. Breast Cancer Res
Treat. 160:69–77. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chin YR and Toker A: The actin-bundling
protein palladin is an Akt1-specific substrate that regulates
breast cancer cell migration. Mol Cell. 38:333–344. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu H, Radisky DC, Nelson CM, Zhang H,
Fata JE, Roth RA and Bissell MJ: Mechanism of Akt1 inhibition of
breast cancer cell invasion reveals a protumorigenic role for TSC2.
Proc Natl Acad Sci USA. 103:4134–4139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li W, Hou JZ, Niu J, Xi ZQ, Ma C, Sun H,
Wang CJ, Fang D, Li Q and Xie SQ: Akt1 inhibition promotes breast
cancer metastasis through EGFR-mediated beta-catenin nuclear
accumulation. Cell Commun Signal. 16:822018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang XL, Liu KY, Lin FJ, Shi HM and Ou ZL:
CCL28 promotes breast cancer growth and metastasis through
MAPK-mediated cellular anti-apoptosis and pro-metastasis. Oncol
Rep. 38:1393–1401. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu J, Li WZ, Huang ML, Wei HL, Wang T, Fan
J, Li NL and Ling R: Regulation of cancerous progression and
epithelial-mesenchymal transition by miR-34c-3p via modulation of
MAP3K2 signaling in triple-negative breast cancer cells. Biochem
Biophys Res Commun. 483:10–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu J, Tan Q, Deng B, Fang C, Qi D and Wang
R: The microRNA-520a-3p inhibits proliferation, apoptosis and
metastasis by targeting MAP3K2 in non-small cell lung cancer. Am J
Cancer Res. 5:802–811. 2015.PubMed/NCBI
|
|
44
|
You J, Zhang Y, Li Y, Fang N, Liu B, Zu L
and Zhou Q: MiR-449a suppresses cell invasion by inhibiting MAP2K1
in non-small cell lung cancer. Am J Cancer Res. 5:2730–2744.
2015.PubMed/NCBI
|