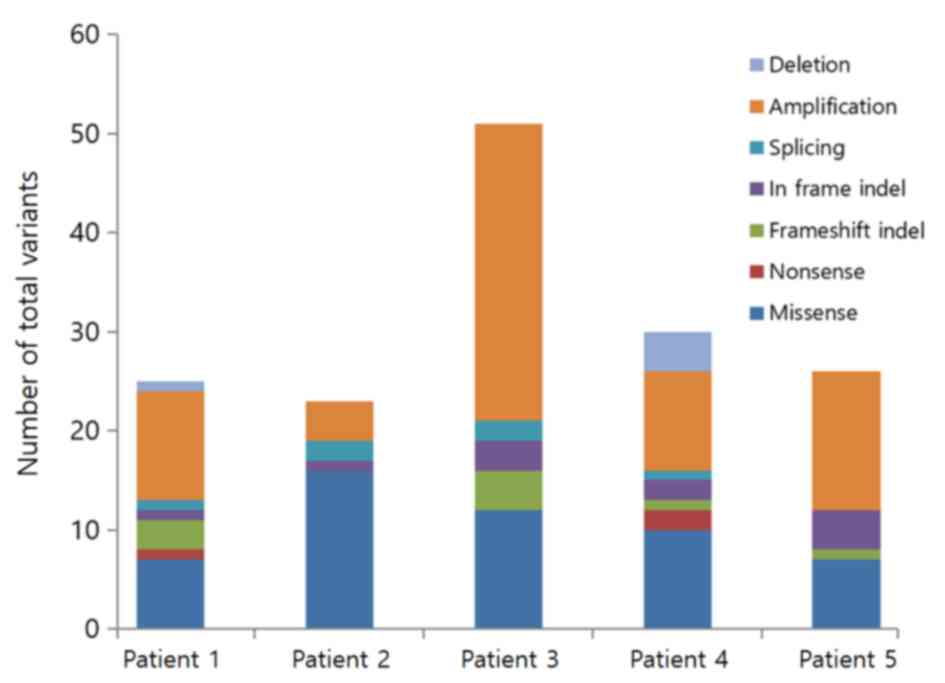
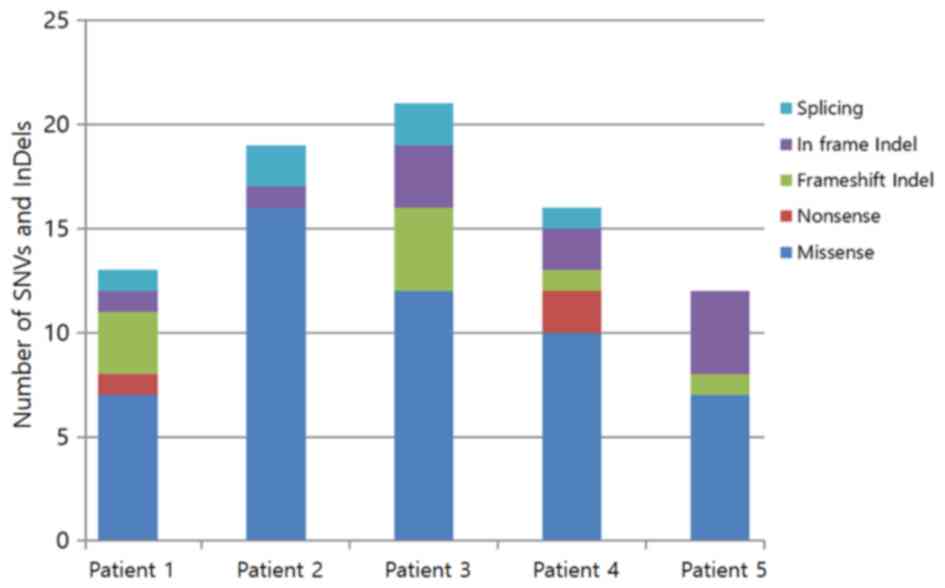
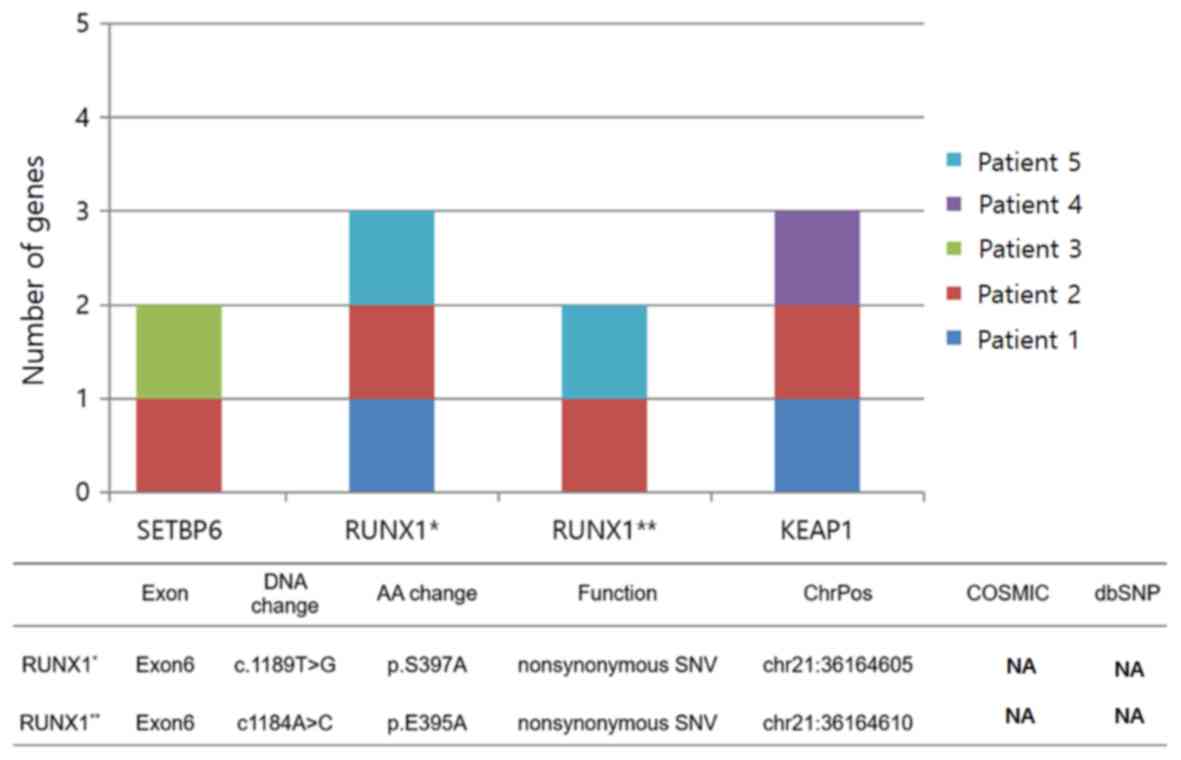
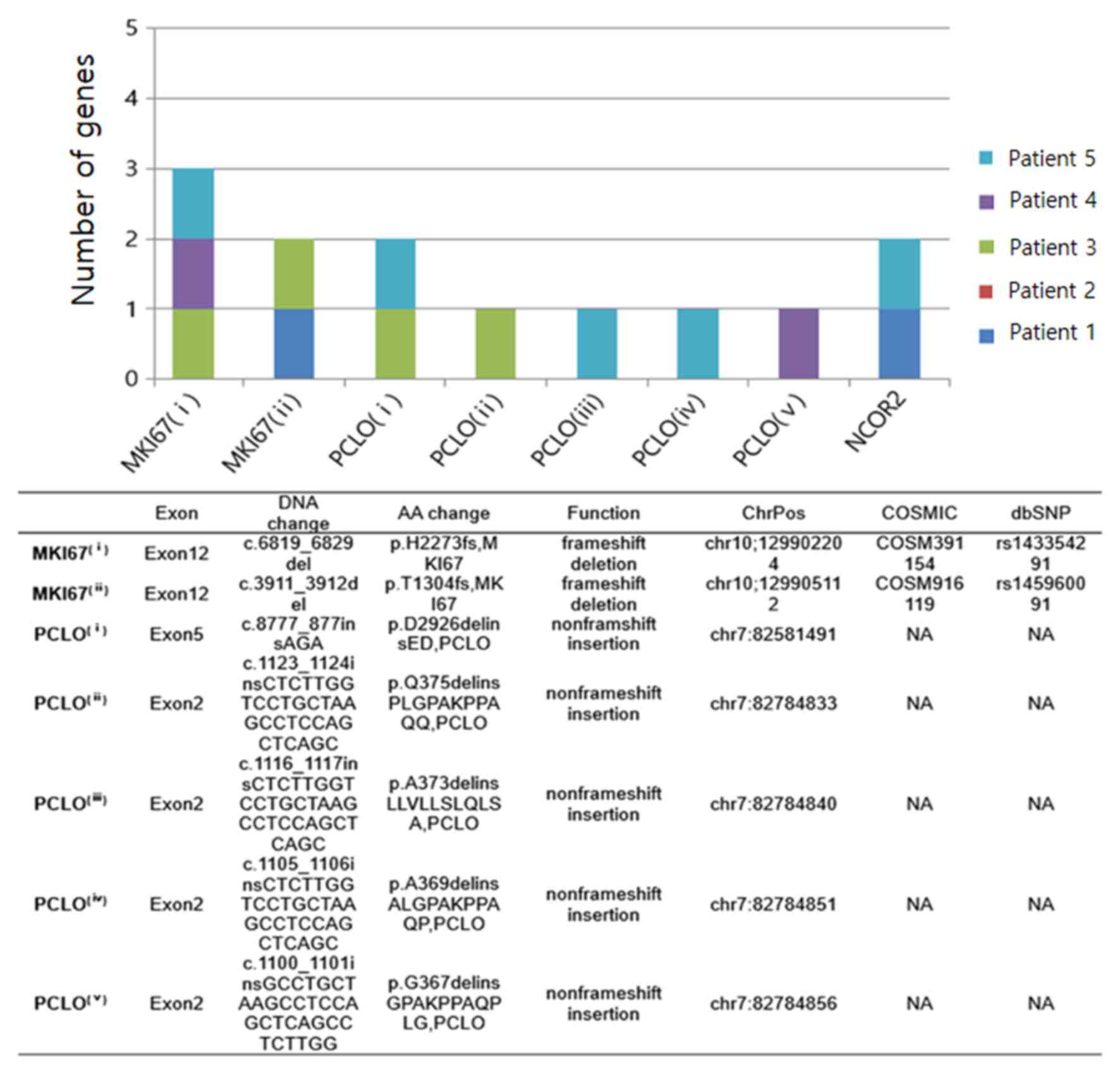
|
1
|
Kahl B and Yang D: Marginal zone
lymphomas: Management of nodal, splenic, and MALT NHL. Hematology
Am Soc Hematol Educ Program. 359–364. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thieblemont C, Berger F, Dumontet C,
Moullet I, Bouafia F, Felman P, Salles G and Coiffier B:
Mucosa-associated lymphoid tissue lymphoma is a disseminated
disease in one third of 158 patients analyzed. Blood. 95:802–806.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Armitage JO: A clinical evaluation of the
International Lymphoma Study Group classification of non-Hodgkins
lymphoma. Blood. 89:3909–3918. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chan JK, Banks PM, Cleary ML, Delsol G, De
Wolf-Peeters C, Falini B, Gatter KC, Grogan TM, Harris NL, Isaacson
PG, et al: A revised European-American classification of lymphoid
neoplasms proposed by the International Lymphoma Study Group. A
summary version. Am J Clin Pathol. 103:543–560. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Isaacson PG and Du MQ: MALT lymphoma: From
morphology to molecules. Nat Rev Cancer. 4:644–653. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferreri AJ, Govi S and Ponzoni M: Marginal
zone lymphomas and infectious agents. Semin Cancer Biol.
23:431–440. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thieblemont C, Bertoni F, Copie-Bergman C,
Ferreri AJ and Ponzoni M: Chronic inflammation and extra-nodal
marginal-zone lymphomas of MALT-type. Semin Cancer Biol. 24:33–42.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Witkowska M and Smolewski P: Helicobacter
pylori infection, chronic inflammation, and genomic transformations
in gastric MALT lymphoma. Mediators Inflamm. 2013:5231702013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwee I, Rancoita PM, Rinaldi A, Ferreri
AJ, Bhagat G, Gascoyne RD, Canzonieri V, Gaidano G, Doglioni C,
Zucca E, et al: Genomic profiles of MALT lymphomas: Variability
across anatomical sites. Haematologica. 96:1064–1066. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goda JS, Gospodarowicz M, Pintilie M,
Wells W, Hodgson DC, Sun A, Crump M and Tsang RW: Long-term outcome
in localized extranodal mucosa-associated lymphoid tissue lymphomas
treated with radiotherapy. Cancer. 116:3815–3824. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zucca E, Conconi A, Pedrinis E, Cortelazzo
S, Motta T, Gospodarowicz MK, Patterson BJ, Ferreri AJ, Ponzoni M,
Devizzi L, et al: Nongastric marginal zone B-cell lymphoma of
mucosa-associated lymphoid tissue. Blood. 101:2489–2495. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Du MQ: MALT lymphoma: Genetic
abnormalities, immunological stimulation and molecular mechanism.
Best Pract Res Clin Haematol. 30:13–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du MQ: MALT lymphoma: A paradigm of NF-κB
dysregulation. Semin Cancer Biol. 39:49–60. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Farinha P and Gascoyne RD: Molecular
pathogenesis of mucosa-associated lymphoid tissue lymphoma. J Clin
Oncol. 23:6370–6378. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morgan JA, Yin Y, Borowsky AD, Kuo F,
Nourmand N, Koontz JI, Reynolds C, Soreng L, Griffin CA,
Graeme-Cook F, et al: Breakpoints of the t(11; 18)(q21; q21) in
mucosa-associated lymphoid tissue (MALT) lymphoma lie within or
near the previously undescribed gene MALT1 in chromosome 18. Cancer
Res. 59:6205–6213. 1999.PubMed/NCBI
|
|
16
|
Akagi T, Motegi M, Tamura A, Suzuki R,
Hosokawa Y, Suzuki H, Ota H, Nakamura S, Morishima Y, Taniwaki M
and Seto M: A novel gene, MALT1 at 18q21, is involved in t (11;
18)(q21; q21) found in low-grade B-cell lymphoma of
mucosa-associated lymphoid tissue. Oncogene. 18:5758–5794. 1999.
View Article : Google Scholar
|
|
17
|
Dierlamm J, Baens M, Wlodarska I,
Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters
C, Hagemeijer A, Van den Berghe H and Marynen P: The apoptosis
inhibitor gene API2 and a novel 18q gene, MLT, are recurrently
rearranged in the t (11; 18)(q21; q21) associated with
mucosa-associated lymphoid tissue lymphomas. Blood. 93:3601–3609.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Auer I, Gascoyne R, Conners J, Cotter FE,
Greiner TC, Sanger WG and Horsman DE: t (11; 18)(q21; q21) is the
most common translocation in MALT lymphomas. Ann Oncol. 8:979–985.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Streubel B, Lamprecht A, Dierlamm J,
Cerroni L, Stolte M, Ott G, Raderer M and Chott A:
T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal
aberration in MALT lymphoma. Blood. 101:2335–2339. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Streubel B, Vinatzer U, Lamprecht A,
Raderer M and Chott A: T(3;14)(p14.1;q32) involving IGH and FOXP1
is a novel recurrent chromosomal aberration in MALT lymphoma.
Leukemia. 19:652–658. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Streubel B, Simonitsch-Klupp I, Müllauer
L, Lamprecht A, Huber D, Siebert R, Stolte M, Trautinger F, Lukas
J, Püspök A, et al: Variable frequencies of MALT
lymphoma-associated genetic aberrations in MALT lymphomas of
different sites. Leukemia. 18:1722–1726. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chanudet E, Ye H, Ferry J, Bacon CM, Adam
P, Müller-Hermelink HK, Radford J, Pileri SA, Ichimura K, Collins
VP, et al: A20 deletion is associated with copy number gain at the
TNFA/B/C locus and occurs preferentially in translocation-negative
MALT lymphoma of the ocular adnexa and salivary glands. J Pathol.
217:420–430. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chanudet E, Huang Y, Ichimura K, Dong G,
Hamoudi RA, Radford J, Wotherspoon AC, Isaacson PG, Ferry J and Du
MQ: A20 is targeted by promoter methylation, deletion and
inactivating mutation in MALT lymphoma. Leukemia. 24:483–487. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Honma K, Tsuzuki S, Nakagawa M, Karnan S,
Aizawa Y, Kim WS, Kim YD, Ko YH and Seto M: TNFAIP3 is the target
gene of chromosome band 6q23. 3-q24. 1 loss in ocular adnexal
marginal zone B cell lymphoma. Genes Chromosomes Cancer. 47:1–7.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Novak U, Rinaldi A, Kwee I, Nandula SV,
Rancoita PM, Compagno M, Cerri M, Rossi D, Murty VV, Zucca E, et
al: The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated
by somatic mutations and genomic deletions in marginal zone
lymphomas. Blood. 113:4918–4921. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martinez-Lopez A, Curiel-Olmo S, Mollejo
M, Cereceda L, Martinez N, Montes-Moreno S, Almaraz C, Revert JB
and Piris MA: MYD88 (L265P) somatic mutation in marginal zone
B-cell lymphoma. Am J Surg Pathol. 39:644–651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ngo VN, Young RM, Schmitz R, Jhavar S,
Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al:
Oncogenically active MYD88 mutations in human lymphoma. Nature.
470:115–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li ZM, Rinaldi A, Cavalli A, Mensah AA,
Ponzoni M, Gascoyne RD, Bhagat G, Zucca E and Bertoni F: MYD88
somatic mutations in MALT lymphomas. Br J Haematol. 158:662–664.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Treon SP, Xu L, Yang G, Zhou Y, Liu X, Cao
Y, Sheehy P, Manning RJ, Patterson CJ, Tripsas C, et al: MYD88
L265P somatic mutation in Waldenströms macroglobulinemia. N Engl J
Med. 367:826–833. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu L, Hunter ZR, Yang G, Zhou Y, Cao Y,
Liu X, Morra E, Trojani A, Greco A, Arcaini L, et al: MYD88 L265P
in Waldenström macroglobulinemia, immunoglobulin M monoclonal
gammopathy, and other B-cell lymphoproliferative disorders using
conventional and quantitative allele-specific polymerase chain
reaction. Blood. 121:2051–2058. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oh SY, Kwon HC, Kim WS, Park YH, Kim K,
Kim HJ, Kwon JM, Lee J, Ko YH, Ahn YC, et al: Nongastric marginal
zone B-cell lymphoma: A prognostic model from a retrospective
multicenter study. Cancer Lett. 258:90–97. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Olszewski AJ and Castillo JJ: Survival of
patients with marginal zone lymphoma. Cancer. 119:629–638. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cogliatti SB, Schmid U, Schumacher U,
Eckert F, Hansmann ML, Hedderich J, Takahashi H and Lennert K:
Primary B-cell gastric lymphoma: A clinicopathological study of 145
patients. Gastroenterology. 101:1159–1170. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fischbach W: Gastric MALT lymphoma-update
on diagnosis and treatment. Best Pract Res Clin Gastroenterol.
28:1069–1077. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Alderuccio JP, Zhao W, Desai A, Ramdial J,
Gallastegui N, Kimble E, de la Fuente MI, Husnain M, Rosenblatt JD,
Alencar AJ, et al: Short survival and frequent transformation in
extranodal marginal zone lymphoma with multiple mucosal sites
presentation. Am J Hematol. 94:585–596. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheson BD, Fisher RI, Barrington SF,
Cavalli F, Schwartz LH, Zucca E, Lister TA; Alliance Australasian
Leukaemia and Lymphoma Group and Eastern Cooperative Oncology
Group; European Mantle Cell Lymphoma Consortium; Italian Lymphoma
Foundation, ; et al: Recommendations for initial evaluation,
staging, and response assessment of Hodgkin and non-Hodgkin
lymphoma: The Lugano classification. J Clin Oncol. 32:3059–3068.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
International Non-Hodgkins Lymphoma
Prognostic Factors Project, . A predictive model for aggressive
non-Hodgkins lymphoma. N Engl J Med. 329:987–994. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shin HT, Choi YL, Yun JW, Kim NKD, Kim SY,
Jeon HJ, Nam JY, Lee C, Ryu D, Kim SC, et al: Prevalence and
detection of low-allele-fraction variants in clinical cancer
samples. Nat Commun. 8:13772017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Illumina, . Sample Multiplexing Overview.
https://www.illumina.com/techniques/sequencing/ngs-library-prep/multiplexing.htmlAugust
26–2020
|
|
40
|
Locallo A, Prandi D, Fedrizzi T and
Demichelis F: TPES: Tumor purity estimation from SNVs.
Bioinformatics. 35:4433–4435. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oh BY, Shin HT, Yun JW, Kim KT, Kim J, Bae
JS, Cho YB, Lee WY, Yun SH, Park YA, et al: Intratumor
heterogeneity inferred from targeted deep sequencing as a
prognostic indicator. Sci Rep. 9:45422019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Coward J and Harding A: Size does matter:
Why polyploid tumor cells are critical drug targets in the war on
cancer. Front Oncol. 4:1232014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Andor N, Graham TA, Jansen M, Xia LC,
Aktipis CA, Petritsch C, Ji HP and Maley CC: Pan-cancer analysis of
the extent and consequences of intratumor heterogeneity. Nat Med.
22:105–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cibulskis K, Lawrence MS, Carter SL,
Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES
and Getz G: Sensitive detection of somatic point mutations in
impure and heterogeneous cancer samples. Nat Biotechnol.
31:213–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH,
Wong CH, Khor CC, Petric R, Hibberd ML and Nagarajan N: LoFreq: A
sequence-quality aware, ultra-sensitive variant caller for
uncovering cell-population heterogeneity from high-throughput
sequencing datasets. Nucleic Acids Res. 40:11189–11201. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ye H, Gong L, Liu H, Hamoudi RA, Shirali
S, Ho L, Chott A, Streubel B, Siebert R, Gesk S, et al: MALT
lymphoma with t(14;18)(q32;q21)/IGH-MALT1 is characterized by
strong cytoplasmic MALT1 and BCL10 expression. J Pathol.
205:293–301. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Oetting WS, Brookes AJ, Béroud C and
Taschner PE: Clinical interpretation of variants from
next-generation sequencing: The 2016 scientific meeting of the
human genome variation society. Hum Mutat. 37:1110–1113. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Morin RD, Johnson NA, Severson TM, Mungall
AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et
al: Somatic mutations altering EZH2 (Tyr641) in follicular and
diffuse large B-cell lymphomas of germinal-center origin. Nat
Genet. 42:181–185. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
49
|
Puente XS, Pinyol M, Quesada V, Conde L,
Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz
M, Bassaganyas L, et al: Whole-genome sequencing identifies
recurrent mutations in chronic lymphocytic leukaemia. Nature.
475:101–105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Spina V, Khiabanian H, Messina M, Monti S,
Cascione L, Bruscaggin A, Spaccarotella E, Holmes AB, Arcaini L,
Lucioni M, et al: The genetics of nodal marginal zone lymphoma.
Blood. 128:1392–1373. 2016. View Article : Google Scholar
|
|
51
|
Kiel MJ, Velusamy T, Betz BL, Zhao L,
Weigelin HG, Chiang MY, Huebner-Chan DR, Bailey NG, Yang DT, Bhagat
G, et al: Whole-genome sequencing identifies recurrent somatic
NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med.
209:1553–1565. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kridel R, Meissner B, Rogic S, Boyle M,
Telenius A, Woolcock B, Gunawardana J, Jenkins C, Cochrane C,
Ben-Neriah S, et al: Whole transcriptome sequencing reveals
recurrent NOTCH1 mutations in mantle cell lymphoma. Blood.
119:1963–1971. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Feldman AL, Dogan A, Smith DI, Law ME,
Ansell SM, Johnson SH, Porcher JC, Ozsan N, Wieben ED, Eckloff BW
and Vasmatzis G: Discovery of recurrent t (6; 7)(p25. 3; q32. 3)
translocations in ALK-negative anaplastic large cell lymphomas by
massively parallel genomic sequencing. Blood. 117:915–919. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hyeon J, Lee B, Shin SH, Yoo HY, Kim SJ,
Kim WS, Park WY and Ko YH: Targeted deep sequencing of gastric
marginal zone lymphoma identified alterations of TRAF3 and TNFAIP3
that were mutually exclusive for MALT1 rearrangement. Mod Pathol.
31:1418–1428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cascione L, Rinaldi A, Bruscaggin A,
Tarantelli C, Arribas AJ, Kwee I, Pecciarini L, Mensah AA, Spina V,
Chung EYL, et al: Novel insights into the genetics and epigenetics
of MALT lymphoma unveiled by next generation sequencing analyses.
Haematologica. 104:e558–e561. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bonifer C, Levantini E, Kouskoff V and
Lacaud G: Runx1 Structure and Function in Blood Cell Development.
Adv Exp Med Biol. 962:65–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Maki K, Yamagata T and Mitani K: Role of
the RUNX1-EVI1 fusion gene in leukemogenesis. Cancer Sci.
99:1878–1883. 2008.PubMed/NCBI
|
|
58
|
Gaidzik VI, Teleanu V, Papaemmanuil E,
Weber D, Paschka P, Hahn J, Wallrabenstein T, Kolbinger B, Köhne
CH, Horst HA, et al: RUNX1 mutations in acute myeloid leukemia are
associated with distinct clinico-pathologic and genetic features.
Leukemia. 30:22822016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ichikawa M, Yoshimi A, Nakagawa M,
Nishimoto N, Watanabe-Okochi N and Kurokawa M: A role for RUNX1 in
hematopoiesis and myeloid leukemia. Int J Hematol. 97:726–734.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Klaunig JE: Oxidative stress and cancer.
Curr Pharm Des. 24:4771–4778. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Reuter S, Gupta SC, Chaturvedi MM and
Aggarwal BB: Oxidative stress, inflammation, and cancer: How are
they linked? Free Radic Biol Med. 49:1603–1616. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hartikainen JM, Tengström M, Winqvist R,
Jukkola-Vuorinen A, Pylkäs K, Kosma VM, Soini Y and Mannermaa A:
KEAP1 genetic polymorphisms associate with breast cancer risk and
survival outcomes. Clin Cancer Res. 21:1591–1601. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Piazza R, Magistroni V, Redaelli S, Mauri
M, Massimino L, Sessa A, Peronaci M, Lalowski M, Soliymani R,
Mezzatesta C, et al: SETBP1 induces transcription of a network of
development genes by acting as an epigenetic hub. Nat Commun.
9:21922018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Montgomery SB, Goode DL, Kvikstad E,
Albers CA, Zhang ZD, Mu XJ, Ananda G, Howie B, Karczewski KJ, Smith
K, et al: The origin, evolution, and functional impact of short
insertion-deletion variants identified in 179 human genomes. Genome
Res. 23:749–761. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Stenson PD, Mort M, Ball EV, Howells K,
Phillips AD, Thomas NS and Cooper DN: The human gene mutation
database: 2008 update. Genome Med. 1:132009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Fisher G, Yang ZH, Kudahetti S, Møller H,
Scardino P, Cuzick J and Berney DM; Transatlantic Prostate Group, :
Prognostic value of Ki-67 for prostate cancer death in a
conservatively managed cohort. Br J Cancer. 108:271–277. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Niemiec J: Ki-67 labelling index in human
brain tumours. Folia Histochem Cytobiol. 39:259–262.
2001.PubMed/NCBI
|
|
68
|
Inwald EC, Klinkhammer-Schalke M,
Hofstädter F, Zeman F, Koller M, Gerstenhauer M and Ortmann O:
Ki-67 is a prognostic parameter in breast cancer patients: Results
of a large population-based cohort of a cancer registry. Breast
Cancer Res Treat. 139:539–552. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Jurić I, Pogorelić Z, Kuzmić-Prusac I,
Biocić M, Jakovljević G, Stepan J, Zupancić B, Culić S and Kruslin
B: Expression and prognostic value of the Ki-67 in Wilms tumor:
Experience with 48 cases. Pediatr Surg Int. 26:487–493. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Nadler A, Cukier M, Rowsell C, Kamali S,
Feinberg Y, Singh S and Law CH: Ki-67 is a reliable pathological
grading marker for neuroendocrine tumors. Virchows Arch.
462:501–505. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Broyde A, Boycov O, Strenov Y, Okon E,
Shpilberg O and Bairey O: Role and prognostic significance of the
Ki-67 index in non-Hodgkins lymphoma. Am J Hematol. 84:338–343.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bubán T, Schmidt M, Broll R, Antal-Szalmás
P and Duchrow M: Detection of mutations in the cDNA of the
proliferation marker Ki-67 protein in four tumor cell lines. Cancer
Genet Cytogenet. 149:81–84. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Oh SY, Ryoo BY, Kim WS, Park YH, Kim K,
Kim HJ, Kwon JM, Lee J, Ko YH, Ahn YC, et al: Nongastric marginal
zone B-cell lymphoma: Analysis of 247 cases. Am J Hematol.
82:446–452. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Thieblemont C: Clinical presentation and
management of marginal zone lymphomas. Hematology Am Soc Hematol
Educ Program. 307–313. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Riva V and Maga G: From the magic bullet
to the magic target: Exploiting the diverse roles of DDX3X in viral
infections and tumorigenesis. Future Med Chem. 11:1357–1381. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ojha J, Secreto CR, Rabe KG, Van Dyke DL,
Kortum KM, Slager SL, Shanafelt TD, Fonseca R, Kay NE and Braggio
E: Identification of recurrent truncated DDX3X mutations in chronic
lymphocytic leukaemia. Br J Haematol. 169:445–448. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Phung B, Cieśla M, Sanna A, Guzzi N,
Beneventi G, Cao Thi Ngoc P, Lauss M, Cabrita R, Cordero E, Bosch
A, et al: The X-Linked DDX3X RNA helicase dictates translation
reprogramming and metastasis in melanoma. Cell Rep. 27:3573–3586
e3577. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Patmore DM, Jassim A, Nathan E, Gilbertson
RJ, Tahan D, Hoffmann N, Tong Y, Smith KS, Kanneganti TD, Suzuki H,
et al: DDX3X suppresses the susceptibility of hindbrain lineages to
medulloblastoma. Dev Cell. S1534-5807(20)30416-0. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Fenster SD and Garner CC: Gene structure
and genetic localization of the PCLO gene encoding the presynaptic
active zone protein Piccolo. Int J Dev Neurosci. 20:161–171. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lohr JG, Stojanov P, Lawrence MS, Auclair
D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW,
Slager SL, et al: Discovery and prioritization of somatic mutations
in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing.
Proc Natl Acad Sci USA. 109:3879–3884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chen JD and Evans RM: A transcriptional
co-repressor that interacts with nuclear hormone receptors. Nature.
377:454–457. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Battaglia S, Maguire O and Campbell MJ:
Transcription factor co-repressors in cancer biology: Roles and
targeting. Int J Cancer. 126:2511–2519. 2010.PubMed/NCBI
|
|
83
|
Rosenfeld JA, Coe BP, Eichler EE, Cuckle H
and Shaffer L: Estimates of penetrance for recurrent pathogenic
copy-number variations. Genet Med. 15:478–481. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Nakamura M, Choe SK, Runko AP, Gardner PD
and Sagerström C: Nlz1/Znf703 acts as a repressor of transcription.
BMC Dev Biol. 8:1082008. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Pereira-Castro I, Costa AM, Oliveira MJ,
Barbosa I, Rocha AS, Azevedo L and da Costa LT: Characterization of
human NLZ1/ZNF703 identifies conserved domains essential for proper
subcellular localization and transcriptional repression. J Cell
Biochem. 114:120–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Reynisdottir I, Arason A, Einarsdottir BO,
Gunnarsson H, Staaf J, Vallon-Christersson J, Jonsson G, Ringnér M,
Agnarsson BA, Olafsdottir K, et al: High expression of ZNF703
independent of amplification indicates worse prognosis in patients
with luminal B breast cancer. Cancer Med. 2:437–446. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Yang G, Ma F, Zhong M, Fang L, Peng Y, Xin
X, Zhong J, Yuan F, Gu H, Zhu W and Zhang Y: ZNF703 acts as an
oncogene that promotes progression in gastric cancer. Oncol Rep.
31:1877–1882. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ma F, Bi L, Yang G, Zhang M, Liu C, Zhao
Y, Wang Y, Wang J, Bai Y and Zhang Y: ZNF703 promotes tumor cell
proliferation and invasion and predicts poor prognosis in patients
with colorectal cancer. Oncol Rep. 32:1071–1077. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Baykara O, Dalay N, Kaynak K and Buyru N:
ZNF703 Overexpression may act as an oncogene in non-small cell lung
cancer. Cancer Med. 5:2873–2878. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Previs RA, Coleman RL, Harris AL and Sood
AK: Molecular pathways: Translational and therapeutic implications
of the Notch signaling pathway in cancer. Clin Cancer Res.
21:955–961. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Ntziachristos P, Lim JS, Sage J and
Aifantis I: From fly wings to targeted cancer therapies: A
centennial for notch signaling. Cancer Cell. 25:318–334. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Mirandola L, Comi P, Cobos E, Kast WM,
Chiriva-Internati M and Chiaramonte R: Notchoing from T-cell to
B-cell lymphoid malignancies. Cancer Lett. 308:1–13. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Hozumi K, Negishi N, Suzuki D, Abe N,
Sotomaru Y, Tamaoki N, Mailhos C, Ish-Horowicz D, Habu S and Owen
MJ: Delta-like 1 is necessary for the generation of marginal zone B
cells but not T cells in vivo. Nat Immunol. 5:638–644. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Santos MA, Sarmento LM, Rebelo M, Doce AA,
Maillard I, Dumortier A, Neves H, Radtke F, Pear WS, Parreira L and
Demengeot J: Notch1 engagement by Delta-like-1 promotes
differentiation of B lymphocytes to antibody-secreting cells. Proc
Natl Acad Sci USA. 104:15454–15459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Mensah AA, Rinaldi A, Ponzoni M,
Canzonieri V, Uccella S, Rossi D, Bhagat G, Gaidano G, Zucca E and
Bertoni F: Absence of NOTCH1 gene mutations in MALT lymphomas. Br J
Haematol. 157:382–384. 2012. View Article : Google Scholar : PubMed/NCBI
|