Introduction
Gastric cancer has some of the highest morbidity and
mortality rates among cancers of the digestive system (1). In China in 2015, the incidence and
mortality rates of gastric cancer ranked second for all types of
cancer, with an estimated 679,100 new cases and 498,000
cancer-associated mortalities (2).
Stomach adenocarcinoma (STAD) accounts for 80–90% of all gastric
cancer cases and surgery is considered a plausible curative
treatment, particularly in the early stages of disease (3). However, effective biomarkers for
prognosis following surgery are still lacking for patients with
gastric cancer. Thus, the discovery and application of novel
predictive biomarkers are critical to improve therapeutic efficacy
and prediction of clinical response.
ADAM metallopeptidase with thrombospondin type 1
motif 18 (ADAMTS18) is a novel member of the metalloproteinase
family, which plays an essential role in the physiological growth
and development of several organisms (4,5). Loss of
expression, genetic mutation and gene methylation of ADAMTS18 can
lead to abnormal development and disease, such as arthritis, cancer
and cardiovascular disease (6).
Aberrant ADAMTS18 expression has been reported to be closely
associated with the development of the bone, eye and central
nervous system, as well as thrombosis and tumorigenesis (4). Furthermore, a previous study has
demonstrated that abnormal ADAMTS18 expression is associated with
tumor occurrence and development (7). Evidence suggests that ADAMTS18 may be a
tumor suppressor (8); however,
overexpression of ADAMTS18 has been reported to promote the
proliferation and migration of HCC cells (9). These findings suggest that the
biological function of ADAMTS18 varies between different types of
tumor.
To the best of our knowledge, few studies have
focused on the role of ADAMTS18 in STAD, whereby the results are
contradictory. Therefore, the present study aimed to investigate
the prognostic value of ADAMTS18 expression in patients with STAD,
and to determine its association with STAD occurrence and
development.
Materials and methods
Study population and data collection
from the cancer genome atlas (TCGA) database
Data on the survival time, clinicopathological
characteristics and gene expression profiles of patients with STAD
were downloaded from TCGA database (tcga-data.nci.nih.gov/tcga/) (10,11). The
cohort included 191 men and 109 women with an age range of 30 to 90
years old and median age of 67 years. The clinical data included:
Ethnicity, sex, age, tumor stage, lymph node metastasis,
Tumor-Node-Metastasis (TNM) stage (12), survival time and status.
Association between ADAMTS18
expression and survival in TCGA database
Tumor ADAMTS18 expression levels from TCGA were
divided into two groups based on the 50% cut-off values. The
Kaplan-Meier method and log-rank tests were used to assess the
median survival time and OS rate, with adjustment for sex, age,
tumor grade, tumor stage, lymph node metastasis and TNM stage.
Patient information and data
collection
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Guangxi Medical
University (Nanning, China), and written informed consent was
obtained from all patients prior to study commencement. A total of
40 paired tumor and non-tumor tissue samples were collected from 30
male and 10 female subjects at the First Affiliated Hospital of
Guangxi Medical University between October 2016 and February 2017.
The age range of these patients was 34 to 87 and the median age was
63 years. STAD diagnoses were pathologically confirmed by the
Department of Pathology of the First Affiliated Hospital of Guangxi
Medical University following gastrointestinal surgery. The
following patient data were acquired: Name, sex, age, degree of
tumor differentiation, infiltration depth, lymph node metastasis
and clinical stage. Patients were divided into high and low
expression level groups based on their 50% cut-off values of
relative ADAMTS18 expression levels in tumor tissues. The
association between ADAMTS18 mRNA expression and the
clinicopathological characteristics was then assessed.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from tissue samples using
TRIzol® reagent (Aidlab; aidlab.cn). The RNA was then
reverse transcribed into cDNA using the PrimeScript™ RT reagent
kit, with gDNA Eraser (Takara Bio, Inc). qPCR was subsequently
performed using FastStart Universal SYBR Green Master (Rox) (Roche
Life Science) on the ABI PRISM 7500 Sequence Detection System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) (13). All methodologies were performed
according to the manufacturer's protocols. The following primer
sequences (Sangon Biotech Co., Ltd.) were used for qPCR: ADAMTS18
forward, 5′-ACCTTGACCAGAACACCATCGAG-3′ and reverse,
5′-CAGGGTCCAGGTCAGGTGTGTA-3′; and GAPDH forward,
5′-GGAGATTACTGCCCTGGCTCCTA-3′ and reverse,
5′-GACTCATCGTACTCCTGCTTGCTG-3′. The reaction system had a total
volume of 20 µl, and the following thermocycling conditions were
used for qPCR: Initial denaturation at 95°C for 30 sec, followed by
50 cycles of 95°C for 5 sec and 60°C for 30 sec. Relative mRNA
expression levels were measured using the 2−∆∆Cq method
(14) and normalized to the internal
reference gene GAPDH. All experiments were performed in
triplicate.
Prediction analysis of ADAMTS18 gene
and protein interactions
The GeneMANIA database (genemania.org)
was used to identify genes potentially associated with ADAMTS18, in
order to predict ADAMTS18 gene function and determine its role in
cancer development. A gene interaction network was constructed
using Cytoscape software (version 3.6.1) (15). Similarly, The Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) database was used
to construct a protein interaction network (16), depicting proteins associated with
ADAMTS18. Gene Ontology (GO) functional enrichment analysis of the
associated genes and proteins was performed using the Database for
Annotation, Visualization and Integrated Discovery (DAVID)
(david.ncifcrf.gov/).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 22.0; IBM Corp.). The Kaplan-Meier method with
log-rank test was used for median survival time and OS analysis. A
paired Student's t-test was used to evaluate the differences in
relative ADAMTS18 mRNA expression levels between STAD and normal
adjacent tissues. The association between ADAMTS18 expression and
clinicopathological characteristics was assessed using the
χ2 and Fisher's exact probability tests. P<0.05 was
considered to indicate a statistically significant difference.
Results
TCGA patient characteristics
The clinical characteristics of 300 patients from
TCGA database with survival times >2 months are presented in
Table I. The results demonstrated
that lymph node metastasis (P=0.003; HR, 2.02; 95% CI, 1.28–3.19)
and TNM stage (P=0.002; HR, 1.84; 95% CI, 1.25–2.72) were
significantly associated with OS rate.
 | Table I.Clinical characteristics of 300
patients with stomach adenocarcinoma in The Cancer Genome Atlas
database. |
Table I.
Clinical characteristics of 300
patients with stomach adenocarcinoma in The Cancer Genome Atlas
database.
|
|
|
| Overall survival
rate |
|---|
|
|
|
|
|
|---|
| Characteristic | Patient, n | MST, (days) | HR (95% CI) | P-value |
|---|
| Sex |
|
| 0.83 (0.56–1.22) | 0.342 |
| Male | 191 | 1,153 |
|
|
|
Female | 109 | NA |
|
|
| Age, years |
|
| 1.22 (0.84–1.76) | 0.301 |
|
<67 | 148 | 1,407 |
|
|
|
≥67 | 152 |
805 |
|
|
| Ethnicity |
|
| 0.79
(0.49–1.28) | 0.343 |
| White
and black | 223 | 1,043 |
|
|
|
Asian | 77 | NA |
|
|
| Tumor-grade |
|
| 1.45
(0.97–2.16) | 0.069 |
|
G1+G2 | 111 | 1,747 |
|
|
| G3 | 189 |
832 |
|
|
| Tumor stage |
|
| 1.57
(0.96–2.58) | 0.073 |
|
T1+T2 | 72 | 1,811 |
|
|
|
T3+T4 | 228 |
874 |
|
|
| LN metastasis |
|
| 2.02
(1.28–3.19) | 0.003a |
| N0+
NX | 100 | 1,811 |
|
|
|
N1+N2+N3 | 200 |
782 |
|
|
| Metastasis |
|
| 1.26
(0.55–2.89) | 0.579 |
|
M0+MX | 287 | 1,153 |
|
|
| M1 | 13 |
476 |
|
|
| TNM stage |
|
| 1.84
(1.25–2.72) | 0.002a |
|
I+II | 144 | 1,811 |
|
|
|
III+IV | 156 |
766 |
|
|
Association between ADAMTS18 mRNA
expression level and survival
Data from TCGA database was used to divide the
patients into two groups based on the 50% cut-off values of
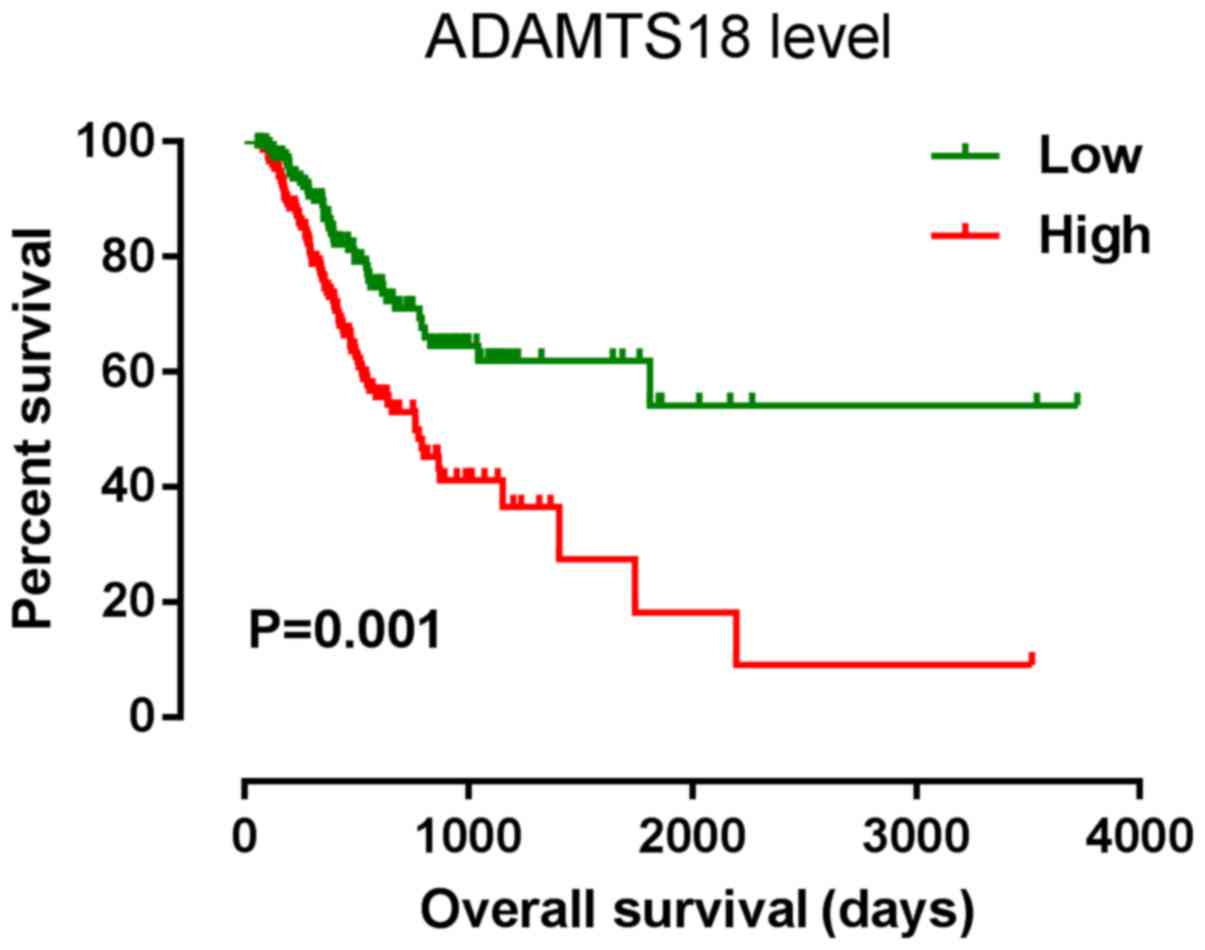
ADAMTS18 mRNA expression. The results indicated that ADAMTS18
expression was significantly associated with OS rate (Fig. 1) (P=0.001; HR, 1.87; 95% CI,
1.27–2.73; Table II), even after
adjusting for age, sex, ethnicity, tumor differentiation degree,
tumor stage, lymph-node metastasis and TNM stage (adjusted P=0.002;
adjusted HR, 1.81; adjusted 95% CI, 1.24–2.65; Table II).
 | Table II.Prognostic survival analysis of
ADAMTS18 gene expression in 300 patients with stomach
adenocarcinoma from The Cancer Genome Atlas database. |
Table II.
Prognostic survival analysis of
ADAMTS18 gene expression in 300 patients with stomach
adenocarcinoma from The Cancer Genome Atlas database.
| Gene | Patient, n | MST, day | Crude HR (95%
CI) | Crude P-value | Adjusted HR (95%
CI) | Adjusted
P-valueb |
|---|
| ADAMTS18 |
|
|
|
|
|
|
|
Low | 150 | 2,281 |
| 0.001a |
| 0.002a |
|
High | 150 | 1,148 | 1.87
(1.27–2.73) |
| 1.81
(1.24–2.65) |
|
Analysis of ADAMTS18 expression in
STAD and non-tumor tissues
In order to determine whether ADAMTS18 was
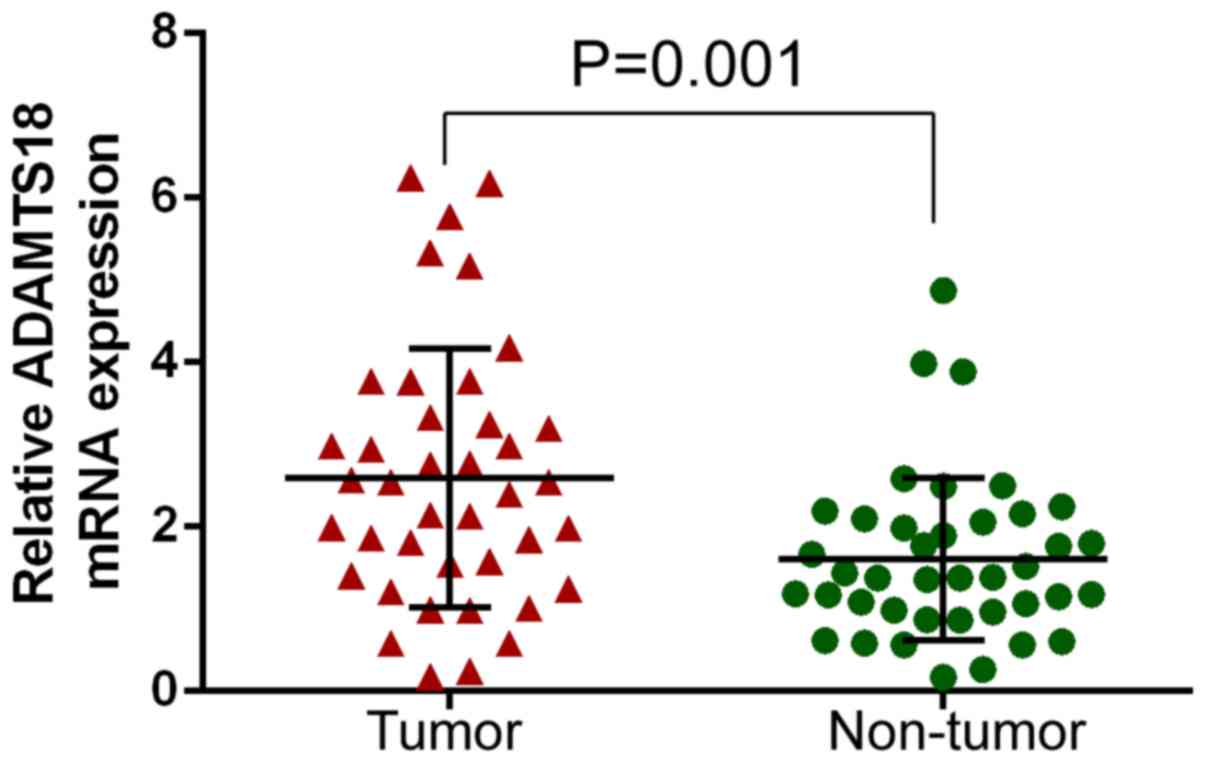
differentially expressed in tumor and non-tumor tissues, relative
ADAMTS18 gene expression was analyzed between 40 paired independent
samples. The results demonstrated that ADAMTS18 expression was
significantly higher in STAD tissues than in normal adjacent
tissues (P=0.001; Fig. 2).
Association between ADAMTS18
expression and clinicopathological characteristics in patients with
STAD
ADAMTS18 expression was significantly associated
with tumor differentiation degree (P=0.013; χ2=7.795),
lymph node metastasis (P=0.001; χ2=12.379) and TNM stage
(P=0.001; χ2=12.379). However, ADAMTS18 expression was
not associated with sex, age, ethnicity, tumor size or tumor stage
(Table III).
 | Table III.Association between ADAMTS18
expression and clinicopathological characteristics in patients with
stomach adenocarcinoma (n=40). |
Table III.
Association between ADAMTS18
expression and clinicopathological characteristics in patients with
stomach adenocarcinoma (n=40).
|
|
| ADAMTS18
expression |
|---|
|
|
|
|
|---|
| Characteristic | Patient, n | Low, n=20 | High, n=20 |
χ2-value | P-value |
|---|
| Sex |
|
|
| 0.520 | 0.716 |
|
Male | 30 | 16 | 14 |
|
|
|
Female | 10 | 4 | 6 |
|
|
| Age, years |
|
|
| 0.902 | 0.527 |
|
<63 | 19 | 8 | 11 |
|
|
|
≥63 | 21 | 12 | 9 |
|
|
| Tumor size, cm |
|
|
| 0.921 | 0.337 |
|
<5 | 23 | 13 | 10 |
|
|
| ≥5 | 17 | 7 | 10 |
|
|
|
Differentiation |
|
|
| 7.795 | 0.013a |
|
Poor | 26 | 9 | 17 |
|
|
| Well +
medium | 12 | 10 | 2 |
|
|
|
Missing | 2 | 1 | 1 |
|
|
| Tumor stage |
|
|
| 1.758 | 0.185 |
|
T1+T2 | 14 | 9 | 5 |
|
|
|
T3+T4 | 26 | 11 | 15 |
|
|
| LN metastasis |
|
|
| 12.379 | 0.001a |
|
Yes | 23 | 6 | 17 |
|
|
| No | 17 | 14 | 3 |
|
|
| TNM stage |
|
|
| 12.379 | 0.001a |
|
I+II | 17 | 14 | 3 |
|
|
|
III+IV | 23 | 6 | 17 |
|
|
Prediction analysis of the biological
function of ADAMTS18 and associated signaling pathways
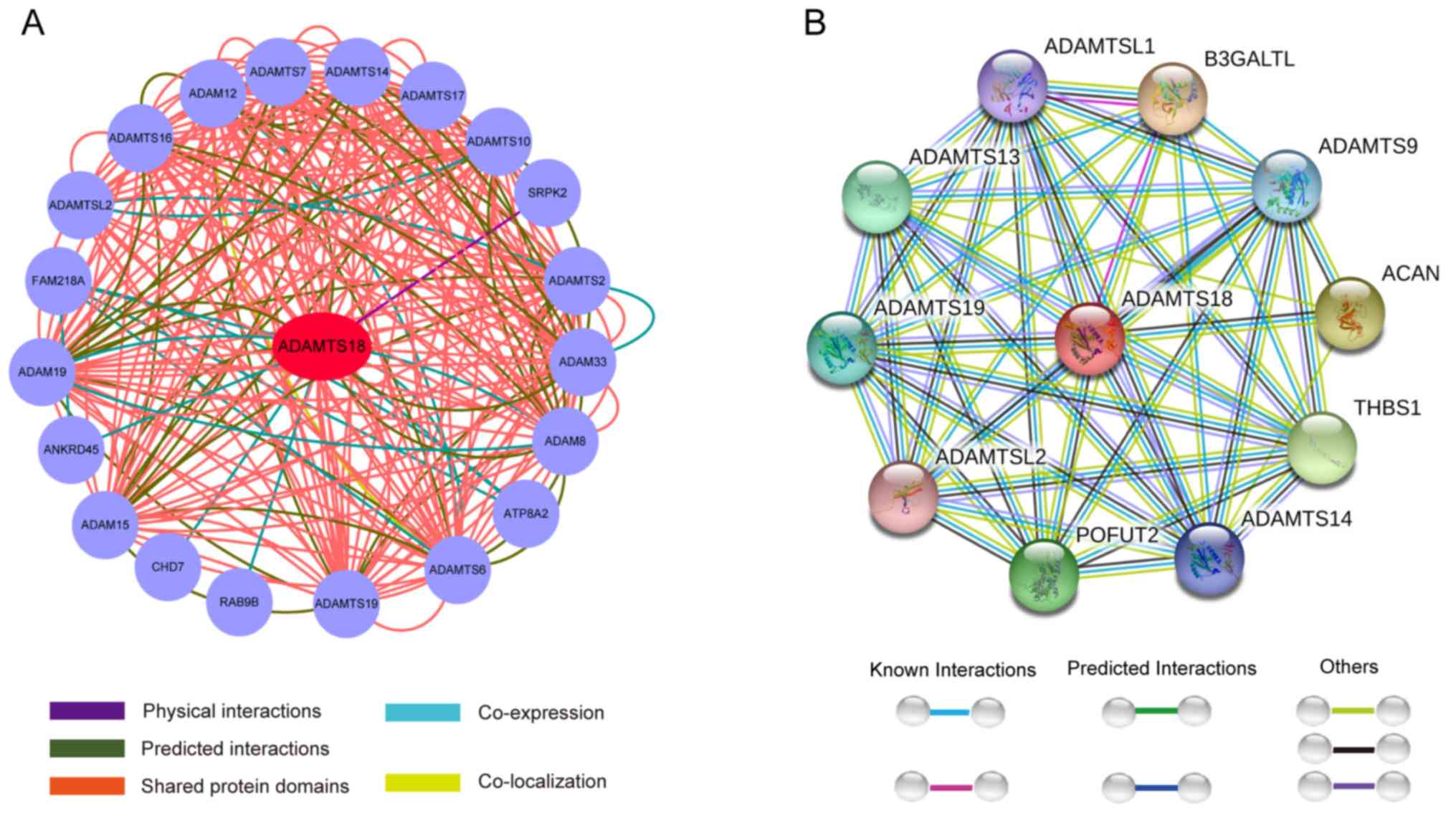
Prediction analysis using GeneMANIA demonstrated
that ADAMTS18 interacts with FAM218A, ATP8A2, ANKRD45, SRPK2, CHD7,
RAB9B and other member of the ADAMTS and ADAM family (Fig. 3A). Protein interaction network
analysis using the STRING database demonstrated that the ADAMTS18
gene primarily interacts with THBS1, ACAN, B3GALTL, POFUT2 and
other ADAMTS family members (Fig.
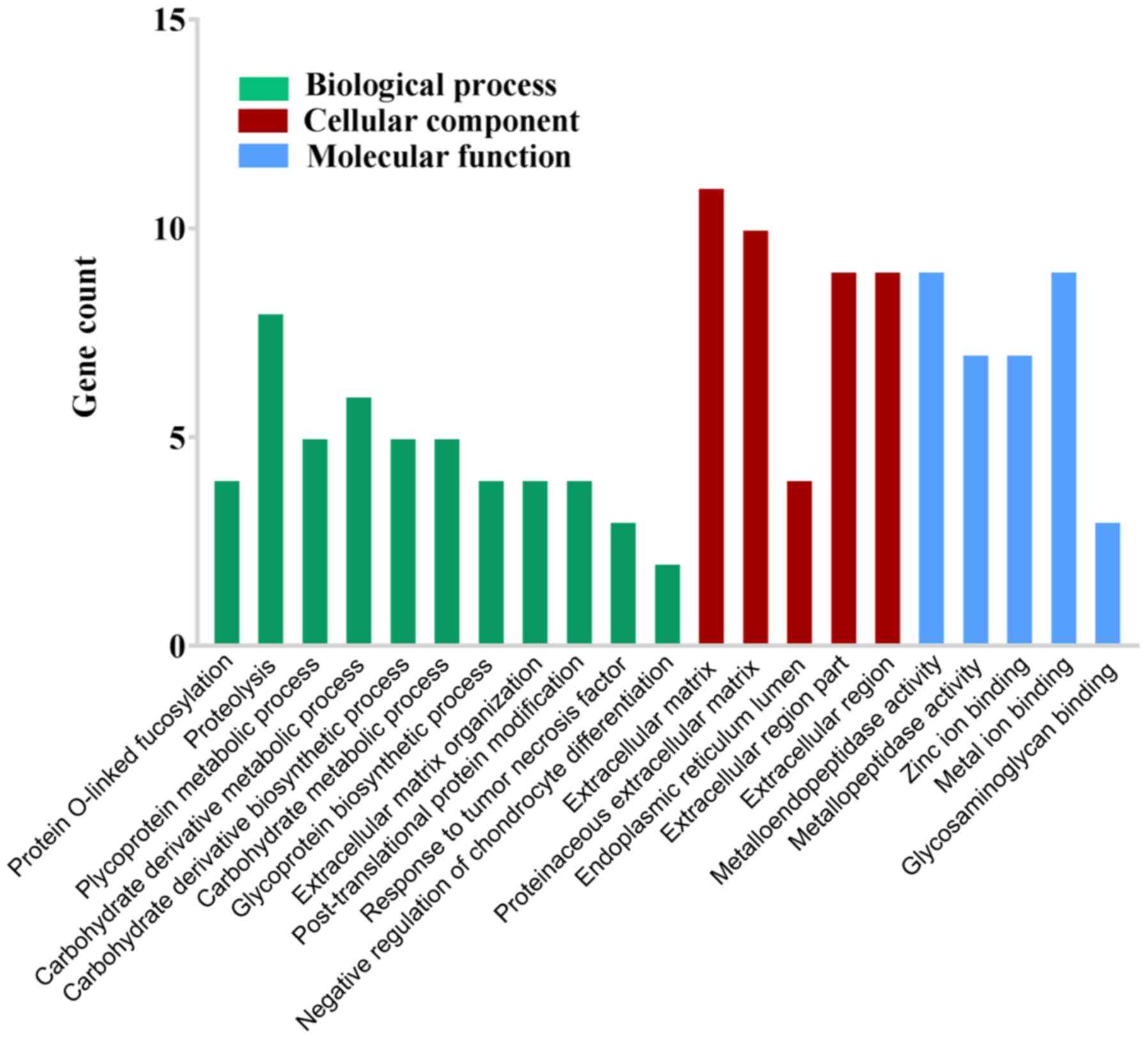
3B). GO functional enrichment analysis suggested that ADAMTS18
is likely to be involved in ‘protein O-linked fucosylation’,
‘proteolysis’, ‘glycoprotein metabolic process’, ‘carbohydrate
derivative metabolic process’, ‘carbohydrate derivative
biosynthetic process’, ‘carbohydrate metabolic process’,
‘glycoprotein biosynthetic process’, ‘extracellular matrix
organization’, ‘post-translational protein modification’, ‘response
to tumor necrosis factor’, ‘negative regulation of chondrocyte
differentiation’, ‘extracellular matrix’, ‘proteinaceous
extracellular matrix’, ‘endoplasmic reticulum lumen’,
‘extracellular region part’, ‘extracellular region’,
‘metalloendopeptidase activity’, ‘metallopeptidase activity’, ‘zinc
ion binding’, ‘metal ion binding’ and ‘glycosaminoglycan binding’
(Fig. 4).
Discussion
ADAMTS18 is a member of the newly discovered ADAMTS
metalloproteinase family (17–19) and
has been associated with tumor occurrence and development in
nasopharyngeal carcinoma, cervical cancer, colorectal cancer,
breast cancer, renal clear cell carcinoma and esophageal
adenocarcinoma (20–23). However, to the best of our knowledge,
few studies have focused on the role of ADAMTS18 in STAD. Thus, the
present study aimed to investigate the association between ADAMTS18
expression and the clinicopathological characteristics and
prognosis of patients with STAD. The results of the present study
demonstrated that ADAMTS18 was upregulated in gastric tumor tissues
and positively associated with tumor differentiation, lymph node
metastasis and TNM stage, compared with normal adjacent tissues.
ADAMTS18 expression was demonstrated to be an adverse prognostic
factor for STAD, which may potentially be used as a prognostic
marker.
ADAMTS18 has been associated with both tumor
suppression and induction, which suggests that the biological
function of ADAMTS18 varies between different types of tumor.
Previous study reported that ADAMTS18 expression levels are
decreased in cervical cancer tissues compared with normal adjacent
tissues, furthermore, low expression levels of ADAMTS18 were
positively associated with high tumor stage, positive lymph node
metastasis and distant metastasis (8). Xu et al (7) reported that ADAMTS18 expression is
lower in breast cancer cell lines and in situ breast cancer
tissue compared with normal breast cells and tissues, and that the
ADAMTS18 promoter is up to 70.8% methylated in breast cancer
tissue. ADAMTS18 can inhibit metastasis and the invasion of breast
cancer both in vivo and in vitro, which was
demonstrated using overexpression and subcutaneous transplantation
experiments in nude mice. However, the results of the present study
suggest that ADAMTS18 is likely to be cancer promoting and a marker
of poor prognosis in patients with STAD. Consistent with previous
findings, this suggests that ADAMTS18 may possess diverse
biological functions during STAD development. In a melanoma study,
genome sequencing led to the discovery of a mutation in the
ADAMTS18 gene, and subsequent in vitro analysis demonstrated
that the mutant promoted the proliferation, migration and
metastasis of melanoma cells (24).
Notably, it has also been reported that ADAMTS18 expression is
associated with tumor stage in STAD (25). The results of the present study
demonstrated that ADAMTS18 expression was closely associated with
tumor grade, lymph node metastasis and TNM stage, and significantly
affected the postoperative survival time of patients with STAD.
The ADAMTS family includes 19 members that are
involved in several pathological and physiological processes,
including tumor formation, thrombosis, angiogenesis and cellular
migration (6,26). Different ADAMTS members play varying
roles in different tissue types (4).
For example, ADAMTS1 and ADAMTS8 are expressed at low levels in
breast cancer and can exhibit an antitumor effect through their
platelet reactive protein-1 domain (27,28).
Furthermore, ADAMTS12 can regulate extracellular signals to
modulate kinase signaling pathways and inhibit tumor formation
(29). However, high ADAMTS8 and
ADAMTS18 expression levels have been reported in breast cancer,
suggesting a tumor-inducing role (30). These observations highlight a complex
role of the ADAMTS family members in tumor development. Other
members of the ADAMTS family, such as ADAMTS4, 5, 8, 10 and 17, are
highly expressed in several cancers and cell lines (31–34), and
silencing or overexpressing these genes can inhibit or promote the
proliferation, migration and invasiveness of cancer cells,
indicating that the ADAMTS family serves a role in tumor biology
and progression. Li et al (22) reported that the frequency of ADAMTS18
gene methylation in STAD, colorectal cancer and pancreatic cancer
tissues is significantly higher compared with that in the
respective normal adjacent tissues, suggesting that gene
hypermethylation may cause decreased ADAMTS18 expression in cancer
tissues. A recent report demonstrated that upregulated ADAMTS18
expression is associated with a significantly higher immune
response score in lymph nodes with metastasis, and in gastric
adenocarcinoma tissues compared with normal gastric tissues;
ADAMTS18 expression in STAD tissues was also positively associated
with tumor TNM staging (35).
Consistent with these findings, the results of the present study
demonstrated that ADAMTS18 expression increased in cancer tissues
at both the mRNA and protein levels, indicating that this gene may
promote STAD occurrence and development.
In conclusion, the present study demonstrated that
ADAMTS18 was highly expressed in STAD tissues compared with normal
gastric tissues. Furthermore, ADAMTS18 expression was significantly
associated with STAD prognosis, and thus may potentially be used as
a prognostic biomarker for patients with STAD. However, a
validation study implementing larger sample sizes and a long-term
follow-up period in multiple centers is required to confirm the
findings of the present study and the potential biological function
of the ADAMTS18 gene needs further experimental exploration.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81660511) and Guangxi
Natural Science Foundation of Key Projects (grant no.
2015GXNSFDA227001).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
QX and YX designed and supervised the present study.
KJ and LL performed the literature review, analyzed the data and
drafted the initial manuscript. DX performed RT-qPCR and helped
analyze the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of The First Affiliated Hospital of Guangxi Medical
University (Nanning, China), and written informed consent was
obtained from all patients prior to study commencement.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Okines A, Verheij M, Allum W, Cunningham D
and Cervantes A; ESMO Guidelines Working Group, : Gastric cancer:
ESMO clinical practice guidelines for diagnosis, treatment and
follow-up. Ann Oncol. 21 (Suppl 5):v50–v54. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei J, Liu CJ and Li Z: ADAMTS-18: A
metalloproteinase with multiple functions. Front Biosci (Landmark
Ed). 19:1456–1467. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ataca D, Caikovski M, Piersigilli A,
Moulin A, Benarafa C, Earp SE, Guri Y, Kostic C, Arsenijevic Y,
Soininen R, et al: Adamts18 deletion results in distinct
developmental defects and provides a model for congenital disorders
of lens, lung, and female reproductive tract development. Biol
Open. 5:1585–1594. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kelwick R, Desanlis I, Wheeler GN and
Edwards DR: The ADAMTS (A Disintegrin and Metalloproteinase with
Thrombospondin motifs) family. Genome Biol. 16:1132015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu H, Xiao Q, Fan Y, Xiang T, Li C, Li C,
Li S, Hui T, Zhang L, Li H, et al: Epigenetic silencing of ADAMTS18
promotes cell migration and invasion of breast cancer through AKT
and NF-κB signaling. Cancer Med. 6:1399–1408. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang L, Liu Y and Zheng P: Downregulation
of ADAMTS18 May serve as a poor prognostic biomarker for cervical
cancer patients. Appl Immunohistochem Mol Morphol. 26:670–675.
2018.PubMed/NCBI
|
|
9
|
Jin H, Wang X, Ying J, Wong AH, Li H, Lee
KY, Srivastava G, Chan AT, Yeo W, Ma BB, et al: Epigenetic
identification of ADAMTS18 as a novel 16q23.1 tumor suppressor
frequently silenced in esophageal, nasopharyngeal and multiple
other carcinomas. Oncogene. 26:7490–7498. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
11
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brierley JD, Gospodarowicz MK and
Wittekind C: TNM Classification of Malignant Tumours. 8th. John
Wiley & Sons; Hoboken, NJ, USA: 2017
|
|
13
|
Batey L, Moon JE, Yu Y, Wu B, Hirschhorn
JN, Shen Y and Dauber A: A novel deletion of IGF1 in a patient with
idiopathic short stature provides insight Into IGF1
haploinsufficiency. J Clin Endocrinol Metab. 99:E153–E159. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1): D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
López-Otín C and Matrisian LM: Emerging
roles of proteases in tumour suppression. Nat Rev Cancer.
7:800–808. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kumar S, Rao N and Ge R: Emerging roles of
ADAMTSs in angiogenesis and cancer. Cancers (Basel). 4:1252–1299.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wagstaff L, Kelwick R, Decock J and
Edwards DR: The roles of ADAMTS metalloproteinases in tumorigenesis
and metastasis. Front Biosci (Landmark Ed). 16:1861–1872. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zeng W, Corcoran C, Collins-Racie LA,
Lavallie ER, Morris EA and Flannery CR: Glycosaminoglycan-binding
properties and aggrecanase activities of truncated ADAMTSs:
Comparative analyses with ADAMTS-5, −9, −16 and −18. Biochim
Biophys Acta. 1760:517–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Z, Nardi MA, Li YS, Zhang W, Pan R,
Dang S, Yee H, Quartermain D, Jonas S and Karpatkin S: C-terminal
ADAMTS-18 fragment induces oxidative platelet fragmentation,
dissolves platelet aggregates, and protects against carotid artery
occlusion and cerebral stroke. Blood. 113:6051–6060. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Z, Zhang W, Shao Y, Zhang C, Wu Q, Yang
H, Wan X, Zhang J, Guan M, Wan J and Yu B: High-resolution melting
analysis of ADAMTS18 methylation levels in gastric, colorectal and
pancreatic cancers. Med Oncol. 27:998–1004. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wei X, Prickett TD, Viloria CG, Molinolo
A, Lin JC, Cardenas-Navia I, Cruz P; NISC Comparative Sequencing
Program, ; Rosenberg SA, Davies MA, et al: Mutational and
functional analysis reveals ADAMTS18 metalloproteinase as a novel
driver in melanoma. Mol Cancer Res. 8:1513–1525. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malik R, Lelkes PI and Cukierman E:
Biomechanical and biochemical remodeling of stromal extracellular
matrix in cancer. Trends Biotechnol. 33:230–236. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun Y, Huang J and Yang Z: The roles of
ADAMTS in angiogenesis and cancer. Tumour Biol. 36:4039–4051. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Martino-Echarri E, Fernández-Rodríguez R,
Rodríguez-Baena FJ, Barrientos-Durán A, Torres-Collado AX,
Plaza-Calonge Mdel C, Amador-Cubero S, Cortés J, Reynolds LE,
Hodivala-Dilke KM, et al: Contribution of ADAMTS1 as a tumor
suppressor gene in human breast carcinoma. Linking its tumor
inhibitory properties to its proteolytic activity on nidogen-1 and
nidogen-2. Int J Cancer. 133:2315–2324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ocak Z, Acar M, Gunduz E, Gunduz M,
Demircan K, Uyeturk U and Ozlü T: Effect of hypericin on the
ADAMTS-9 and ADAMTS-8 gene expression in MCF7 breast cancer cells.
Eur Rev Med Pharmacol Sci. 17:1185–1190. 2013.PubMed/NCBI
|
|
29
|
Fontanil T, Rúa S, Llamazares M,
Moncada-Pazos A, Quirós PM, García-Suárez O, Vega JA, Sasaki T,
Mohamedi Y, Esteban MM, et al: Interaction between the ADAMTS-12
metalloprotease and fibulin-2 induces tumor-suppressive effects in
breast cancer cells. Oncotarget. 5:1253–1264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo X, Li J, Zhang H, Liu H, Liu Z and Wei
X: Relationship between ADAMTS8, ADAMTS18, and ADAMTS20 (A
Disintegrin and Metalloproteinase with Thrombospondin Motifs)
expressions and tumor molecular classification, clinical
pathological parameters, and prognosis in breast invasive ductal
carcinoma. Med Sci Monit. 24:3726–3735. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Held-Feindt J, Paredes EB, Blömer U,
Seidenbecher C, Stark AM, Mehdorn HM and Mentlein R:
Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and
metalloproteinases with thrombospondin motifs 4 and 5) are
expressed in human glioblastomas. Int J Cancer. 118:55–61. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ishikawa N, Daigo Y, Yasui W, Inai K,
Nishimura H, Tsuchiya E, Kohno N and Nakamura Y: ADAM8 as a novel
serological and histochemical marker for lung cancer. Clin Cancer
Res. 10:8363–8370. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ko SY, Lin SC, Wong YK, Liu CJ, Chang KW
and Liu TY: Increase of disintergin metalloprotease 10 (ADAM10)
expression in oral squamous cell carcinoma. Cancer Lett. 245:33–43.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lendeckel U, Kohl J, Arndt M, Carl-McGrath
S, Donat H and Röcken C: Increased expression of ADAM family
members in human breast cancer and breast cancer cell lines. J
Cancer Res Clin Oncol. 131:41–48. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kilic MO, Aynekin B, Kara A, Icen D and
Demircan K: Differentially regulated ADAMTS1, 8, and 18 in gastric
adenocarcinoma. Bratisl Lek Listy. 118:71–76. 2017.PubMed/NCBI
|