Introduction
Influenza is an acute respiratory infectious disease
caused by influenza virus. Humans often suffer from fever, cough,
sore throat, headache, nasal discharge, fatigue and other symptoms
following infection with influenza virus (1). In certain cases, severe complications
induced by the influenza virus, such as pneumonia, respiratory
failure, and multiple organ dysfunction, occur and may lead to
death (2). In recent years, there
have been frequent outbreaks of influenza viruses, and among these,
the novel influenza A (H1N1) virus originating from the influenza
pandemic in North America in 2009 has been identified as a leading
cause of severe pneumonia (3).
Therefore, H1N1 infection represents a critical threat to public
health.
MicroRNAs (miRs/miRNAs) are endogenous
single-stranded, non-coding RNA molecules with a length of 18–25
nucleotides, which are widely involved in the post-transcriptional
regulation of target genes (4).
Several miRNAs have been reported to be implicated in various
biological and pathological processes, including pneumonia
pathogenesis (5,6). Previously, miR-4485 expression has been
demonstrated to be downregulated in gastric cancer (7). Chen et al (8) identified miR-4485 as a potential
biomarker of hepatitis B virus infection persistence. Furthermore,
miR-4485 upregulation could suppress the tumorigenicity of breast
cancer via regulation of mitochondrial functions (9). Recently, decreased serum expression
levels of miR-4485 have been observed in severe pneumonia (10). However, the effects of miR-4485 on
the development of severe H1N1 pneumonia are largely unknown.
In the present study, miR-4485 expression was
detected in patients with severe H1N1 pneumonia. In addition, the
effects of miR-4485 in H1N1-induced A549 cell injury were
investigated by assessing cell viability, apoptosis and
inflammation. Furthermore, the regulatory relationship between
miR-4485 and STAT3 was explored, as well as the association between
miR-4485 and the PI3K/AKT/mTOR signaling pathway. It was
hypothesized that miR-4485 may promote cell viability, and inhibit
cell apoptosis and the production of cytokines by targeting STAT3
and inactivating the PI3K/AKT/mTOR signaling pathway, thus
alleviating H1N1-induced injury in A549 cells.
Materials and methods
Patients and controls
A series of 22 patients with confirmed influenza
virus A (H1N1) pneumonia (18 males and 14 females; age range, 28–67
years; mean age, 45.7 years) and 18 age- and sex-matched healthy
controls (10 males and 8 females; age range, 29–65 years; mean age,
44.6 years) were recruited at Gansu Provincial People's Hospital
(Lanzhou, Gansu, China) between September and December, 2019. The
present study was approved by the Ethics Committee of Gansu
Provincial People's Hospital. Written informed consent was obtained
from all subjects.
A confirmed case of infection was defined as an
acute lower respiratory tract illness with two or more of the
following symptoms or signs: Cough, productive sputum, fever,
chills, dyspnea, pleuritic chest pain, crackles and bronchial
breathing plus an opacity or infiltrate seen on a chest radiography
that was interpreted as pneumonia by the treating physicians, and a
laboratory-confirmed H1N1 virus infection as confirmed by reverse
transcription-quantitative PCR (RT-qPCR) or viral culture. A severe
or critical case was defined as a case that met at least one of the
following criteria on admission, according to the pH1N1 2009
Clinical guidelines (11) released
by Chinese Ministry of Health (MOH): i) Respiratory failure; ii)
Septic shock; iii) Multiple organ insufficiency; and iv) Other
critical clinical conditions requiring intensive care. Hospitalized
patients were excluded if they did not have pneumonia, or if they
had been treated as outpatients or in emergency rooms, had a
duration of hospitalization <24 h, or if there was an incomplete
record of clinical outcome.
All patients in the present study had a body
temperature >100.4°F (>38°C), tachypnea and cough. Other
common symptoms included diarrhea (73%) and sore throat (27%). A
total of 5 ml fasting peripheral blood samples was harvested from
all subjects. Serum was obtained following centrifugation of blood
samples at 4,000 × g at 4°C for 10 min and stored at −80°C.
Cell culture, transfection and
treatment
Human pulmonary epithelial A549 cells and 293T
cells, purchased from the American Type Culture Collection were
cultured in DMEM supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 100 µg/ml penicillin/streptomycin (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
When cell confluence reached >70%, the miR-4485
mimic (50 nM)/inhibitor (150 nM), mimic/inhibitor negative control
(mimic/inhibitor NC) (50 nM/150 nM), small interfering (si)RNA
against STAT3 or siRNA negative controls (si-NC) (100 nM) were
transfected into cells using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
After 24 h of transfection, cells were infected with or without
H1N1 at a MOI of 2 for another 24 h. Primer sequences were as
follows: miR-4485 mimic sense, 5′-UAACGGCCGCGGUACCCUAA-3′ and mimic
NC antisense, 5′-UGAUCCUCAGUUGUGUGUAA-3′; miR-4485 inhibitor
5′-UAACGGCCGCGGUACCCUAA-3′; si-STAT3: sense,
5′-GGACGACUUUGAUUUCAACtt-3′; antisense,
5′-GUUGAAAUCAAAGUCGUCCtg-3′. si-NC: sense,
5′-UUCUCCGAACGUGUCACGUTT-3′; antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. A total of 48 h following
transfection, the cells were collected for further experiments.
Luciferase reporter assay
The potential binding sites for miR-4485 in STAT3
were predicted using TargetScan Release 7.2 (http://www.targetscan.org/). Wild-type (WT)-STAT3-3′
untranslated region (UTR)-pISo and MUT-STAT3-3′UTR-pISo luciferase
reporter plasmids (50 ng) (Sangon Biotech Co., Ltd.) were
co-transfected with either miR-4485 mimic or mimic NC into 293T
cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h of transfection, the lysate was
analyzed using a Dual-Glo luciferase assay system (Promega
Corporation) and the luciferase activity was measured using a
luminometer. Firefly luciferase activity was normalized to
Renilla luciferase activity.
RT-qPCR
Total RNA was isolated from peripheral blood samples
or A549 cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and purified to obtain the miRNA fraction using
a mirVana miRNA isolation kit (Ambion; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Total RNA was
reverse transcribed to cDNA using a PrimeScript RT reagent kit
(Takara Biotechnology Co., Ltd.). A total of 1 µg of total RNA was
reversely transcribed using oligo(dT) primer at 42°C for 1 h, and 2
µl of the reverse transcription reaction mix was amplified by PCR
with denaturation at 95°C for 2 min, and 50 cycles at 95°C for 30
sec, 55°C for 30 sec, and 72°C for 1 min. The thermocycling
conditions were as follows: Initial denaturation at 95°C for 30
sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec.
The relative expression levels of miR-4485 were normalized to U6
using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) and
calculated with the 2−ΔΔCq method (12). The primer sequences used in qPCR were
as follows: U6, forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and
reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; and miR-4485 Forward,
5′-AGUAACGGCCGCGGUAC-3′ and reverse,
5′-TCCAGTTTTTTTTTTTTTTTTAGG-3′.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 reagent kit (Beyotime Institute of
Biotechnology) was used to analyze the viability rates of A549
cells according to the manufacturer's protocol. At 24, 48 and 72 h
after transfection, cells were treated with 10 µl CCK-8 solution
for an additional 2 h. The optical density was measured at 450
nm.
Apoptosis detection
The TUNEL reaction was performed according to the
manufacturer's protocols (Roche Applied Science). After rinsing
twice with PBS, A549 cells were fixed with 4% paraformaldehyde for
15 min at room temperature, permeabilized in 0.25% Triton X-100 on
ice for 20 min, and incubated in terminal dexynucleotidyl
transferase reaction cocktail for 45 min at 37°C. Cell nuclei were
visualized by DAPI staining (0.3 µg/ml; Sigma-Aldrich; Merck KGaA)
at 4°C for 10 min. The slides were mounted using
DAPI-Fluoromount-GTM, covered with glass slides, and four fields of
view were examined in using a scanning flourescence microscope.
Western blotting
A549 cells were harvested and lysed using RIPA
buffer (Sigma-Aldrich; Merck KGaA). Protein concentrations were
determined using the BCA protein assay kit (Thermo Fisher
Scientific, Inc.). In total, 30 µg proteins were loaded on a 10%
gel, resolved using SDS-PAGE and transferred onto a nitrocellulose
membrane. The membrane was blocked in 5% skimmed milk at room
temperature for 30 min. Western blotting was performed using
antibodies against Bax (#5023), Bcl-2 (#3498), STAT3 (#12640),
phosphorylated (p)-STAT3 (#9145), PI3K (#4249), p-PI3K (#17366),
AKT (#4685), p-AKT (#4060), mTOR (#2983) and p-mTOR (#5536) (all
1:1,000; Cell Signaling Technology, Inc.) at 4°C overnight.
Membranes were incubated with HRP-coupled IgG antibody (#7074;
1:1,000; Cell Signaling Technology, Inc.) at room temperature for 1
h. Densitometry analysis of the bands was performed using the
ImageJ software (version 1.52; National Institutes of Health).
GAPDH was used as the loading control.
ELISA
Serum samples and supernatants from cultured A549
cells were collected and the concentrations of pro-inflammatory
cytokines IL-6, TNF-α and IL-1β were measured using Human IL-6
Quantikine ELISA Kit (cat. no. D6050), Human TNF-alpha Quantikine
ELISA Kit (cat. no. DTA00D) and Human IL-1 β/IL-1F2 Quantikine
ELISA Kit (cat. no. DLB50) (R&D Systems, Inc.) according to the
manufacturer's protocols.
Statistical analysis
Statistical analysis was performed using SPSS 19.0
statistical software (IBM Corp.). Data are presented as the mean ±
SD from at least three independent experiments. Differences among
groups were compared using Student's t-test or one-way ANOVA with
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-4485 expression is downregulated
in patients with severe H1N1 pneumonia
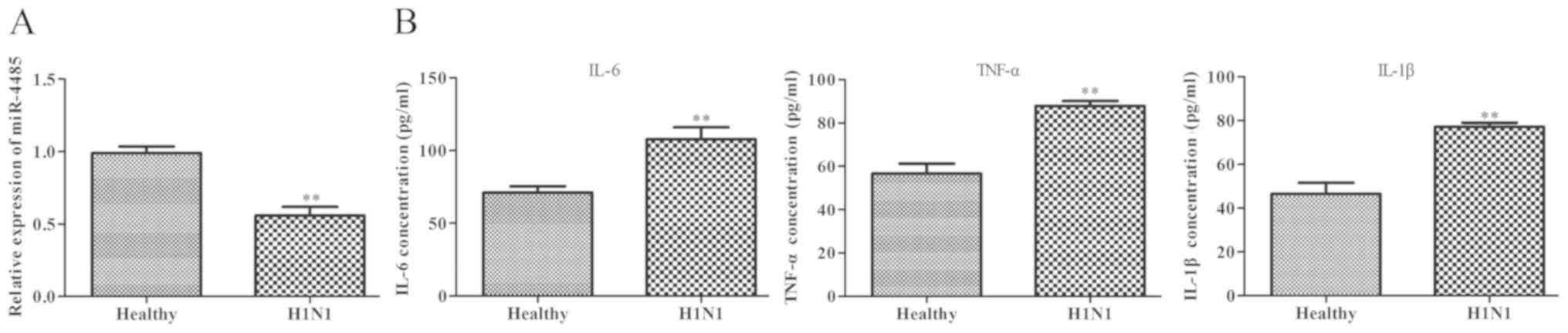
As shown in Fig. 1A,
serum miR-4485 expression was significantly downregulated in
patients with severe H1N1 pneumonia compared with in healthy
subjects. Furthermore, ELISA results demonstrated that patients
with severe H1N1 pneumonia had higher serum levels of IL-6, TNF-α
and IL-1β compared with the healthy controls (Fig. 1B).
Overexpression of miR-4485 improves
H1N1-induced A549 cell injury
To determine whether miR-4485 was involved in the
development of severe H1N1 pneumonia, A549 cells were infected with
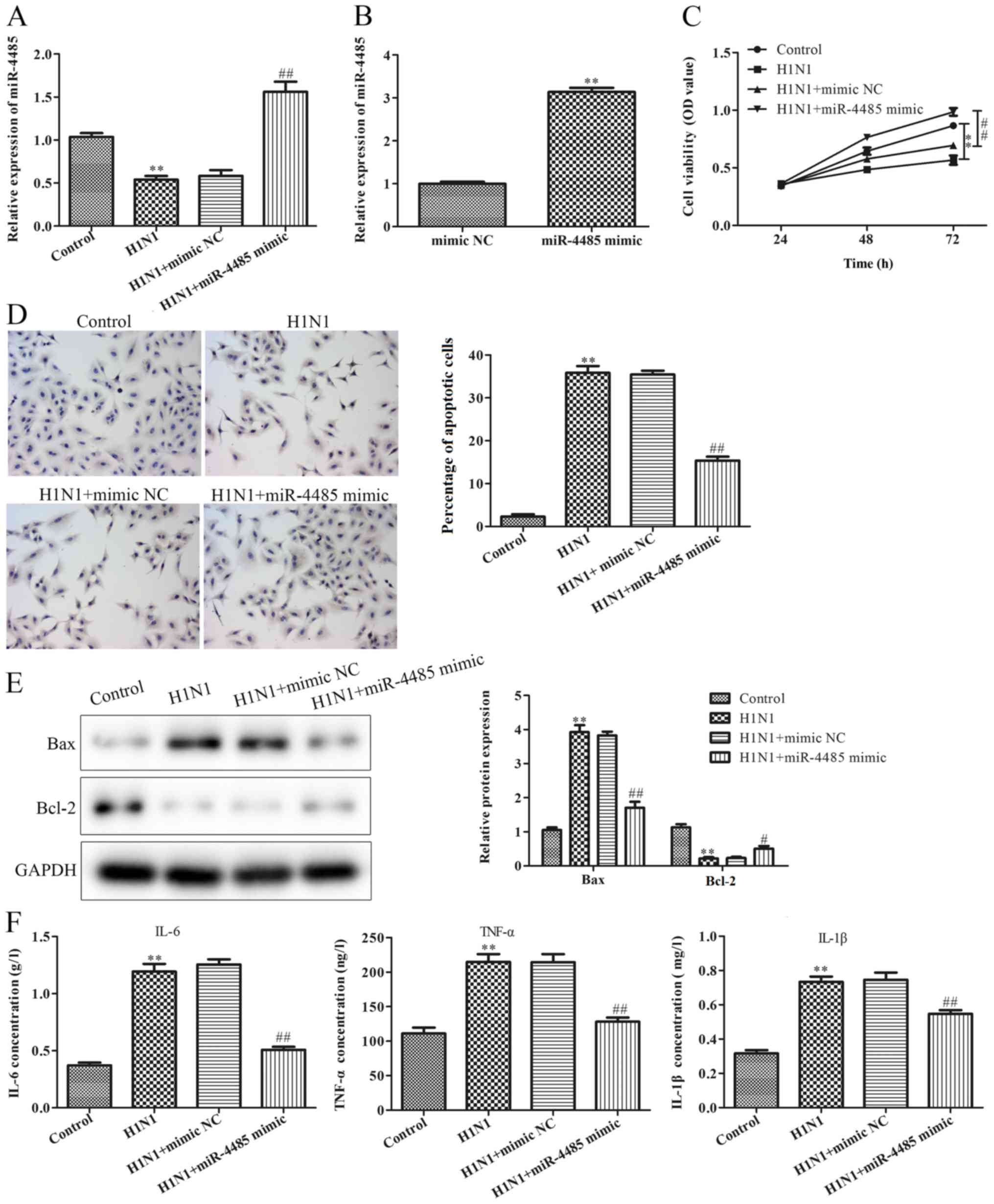
H1N1 to establish a cell model of pneumonia. As shown in Fig. 2A, H1N1 infection decreased miR-4485
expression. To investigate the effects of miR-4485 on H1N1
infection-induced A549 cell injury, A549 cells were transfected
with miR-4485 mimic to overexpress miR-4485 and successful
transfection was confirmed by RT-qPCR (Fig. 2B). Furthermore, the present data
demonstrated that overexpression of miR-4485 markedly ameliorated
H1N1-induced A549 cell injury by promoting cell viability (Fig. 2C), inhibiting apoptosis (Fig. 2D), reversing H1N1-induced expression
changes of apoptosis-related proteins (Fig. 2E), and suppressing the secretion of
IL-6, TNF-α and IL-1β (Fig. 2F).
STAT3 is a direct target gene of
miR-4485
It has been demonstrated that the STAT3-mediated
PI3K/AKT/mTOR signaling pathway may serve a key role in the process
of lung injury (13). In the present
study, H1N1 infection resulted in increased protein phosphorylation
levels of PI3K, AKT and mTOR, as demonstrated by an enhanced ratio
of p-PI3K/PI3K, p-AKT/AKT and p-mTOR/mTOR (Fig. 3A). According to the predicted results
of TargetScan, STAT3 was identified as a potential target of
miR-4485. To confirm the predicted results, a luciferase reporter
assay was performed, and the results revealed that the relative
luciferase activities of STAT3 3′UTR WT were significantly
decreased in the miR-4485 mimic group compared with in the mimic NC
group (Fig. 3B). RT-qPCR analysis
confirmed the knockdown of miR-4485 expression in A549 cells
following transfection with miR-4485 inhibitor (Fig. 3C). Furthermore, the phosphorylated
STAT3, PI3K, AKT and mTOR/total protein ratios were significantly
reduced in the miR-4485 mimic group compared with in the mimic NC
group, whereas they were markedly increased in the miR-4485
inhibitor group compared with in the inhibitor NC group (Fig. 3D).
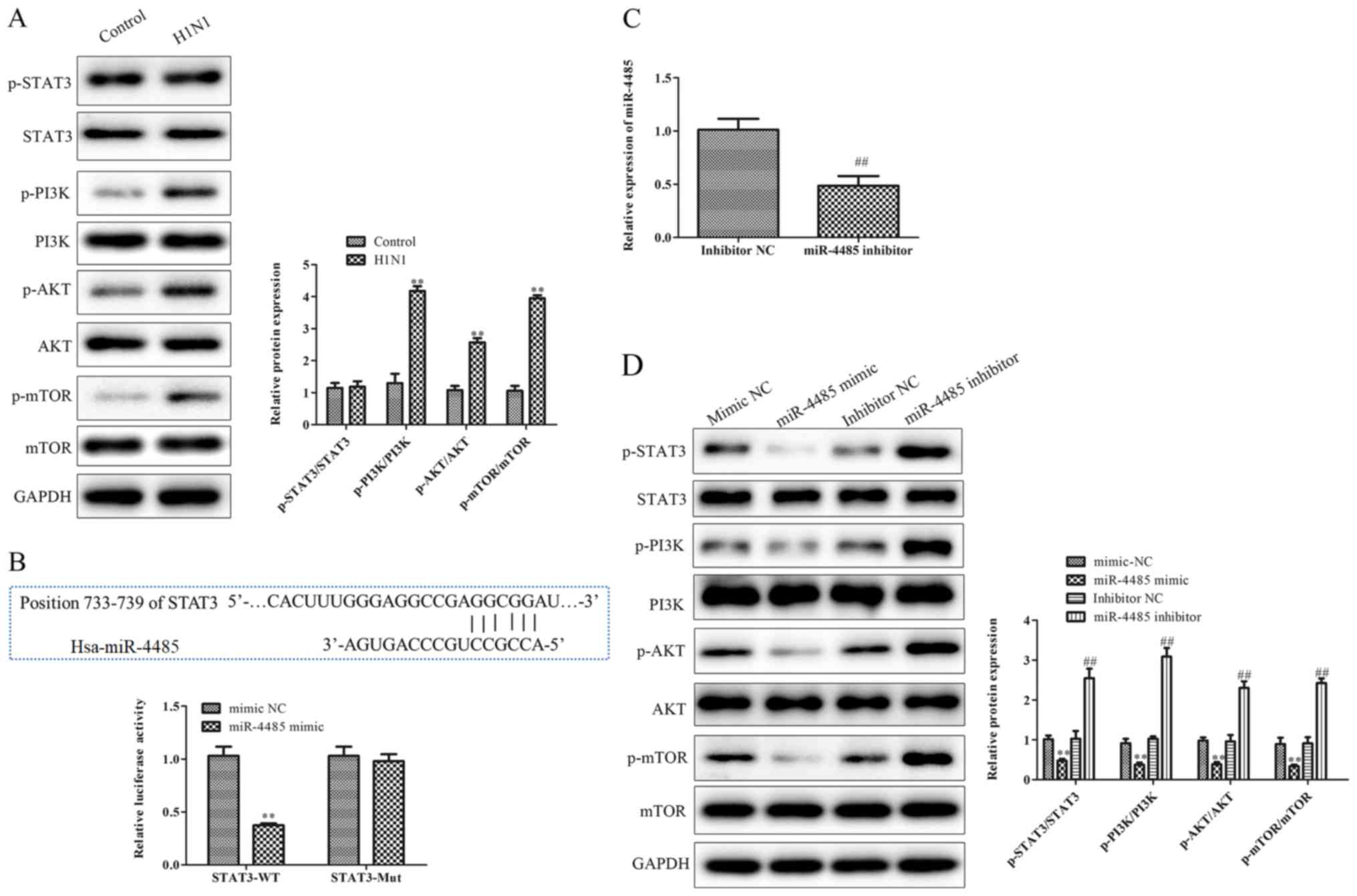 | Figure 3.STAT3 is a direct target gene of
miR-4485. (A) Phosphorylated and total protein levels of STAT3,
PI3K, AKT and mTOR and the ratio of p-STAT3/STAT3, p-PI3K/PI3K,
p-AKT/AKT and p-mTOR/mTOR in A549 cells infected with or without
H1N1. (B) Predicted binding sequences of STAT3 and miR-4485. A
luciferase reporter assay demonstrated that STAT3 3′ untranslated
region-WT was targeted by miR-4485 in 293T cells. (C) Reverse
transcription-quantitative PCR confirmation of miR-4485 knockdown
in A549 cells following transfection with miR-4485 inhibitor. (D)
Phosphorylated and total protein levels of STAT3, PI3K, AKT and
mTOR and the ratio of p-STAT3/STAT3, p-PI3K/PI3K, p-AKT/AKT and
p-mTOR/mTOR in A549 cells transfected with miR-4485 mimic/inhibitor
or mimic/inhibitor NC. The experiments were performed three times
and the data are presented as the mean ± SD. **P<0.01 vs.
control or mimic NC group; ##P<0.01 vs. inhibitor NC
group. miR-4485, microRNA-4485; Mut, mutant; NC, negative control;
p-, phosphorylated; WT, wild type. |
STAT3 knockdown alleviates the effects
of miR-4485 inhibition on H1N1-induced A549 cell injury
To explore the regulatory relationship between
miR-4485 and STAT3, STAT3 expression was knocked down by
transfection with si-STAT3 in A549 cells following miR-4485
downregulation by transfection with miR-4485 inhibitor. As shown in
Fig. 4A, si-STAT3 transfection
decreased the total and phosphorylated protein levels of STAT3.
Furthermore, the suppression of miR-4485 expression deteriorated
H1N1-induced PI3K/AKT/mTOR phosphorylation (Fig. 4B) with no change in STAT3 expression,
inhibited cell viability (Fig. 4C),
promoted apoptosis (Fig. 4D) via
upregulation of Bax expression and downregulation of the expression
levels of Bcl-2 (Fig. 4E), and
increased production of cytokines (Fig.
4F), and these effects were partly reversed by si-STAT3
transfection.
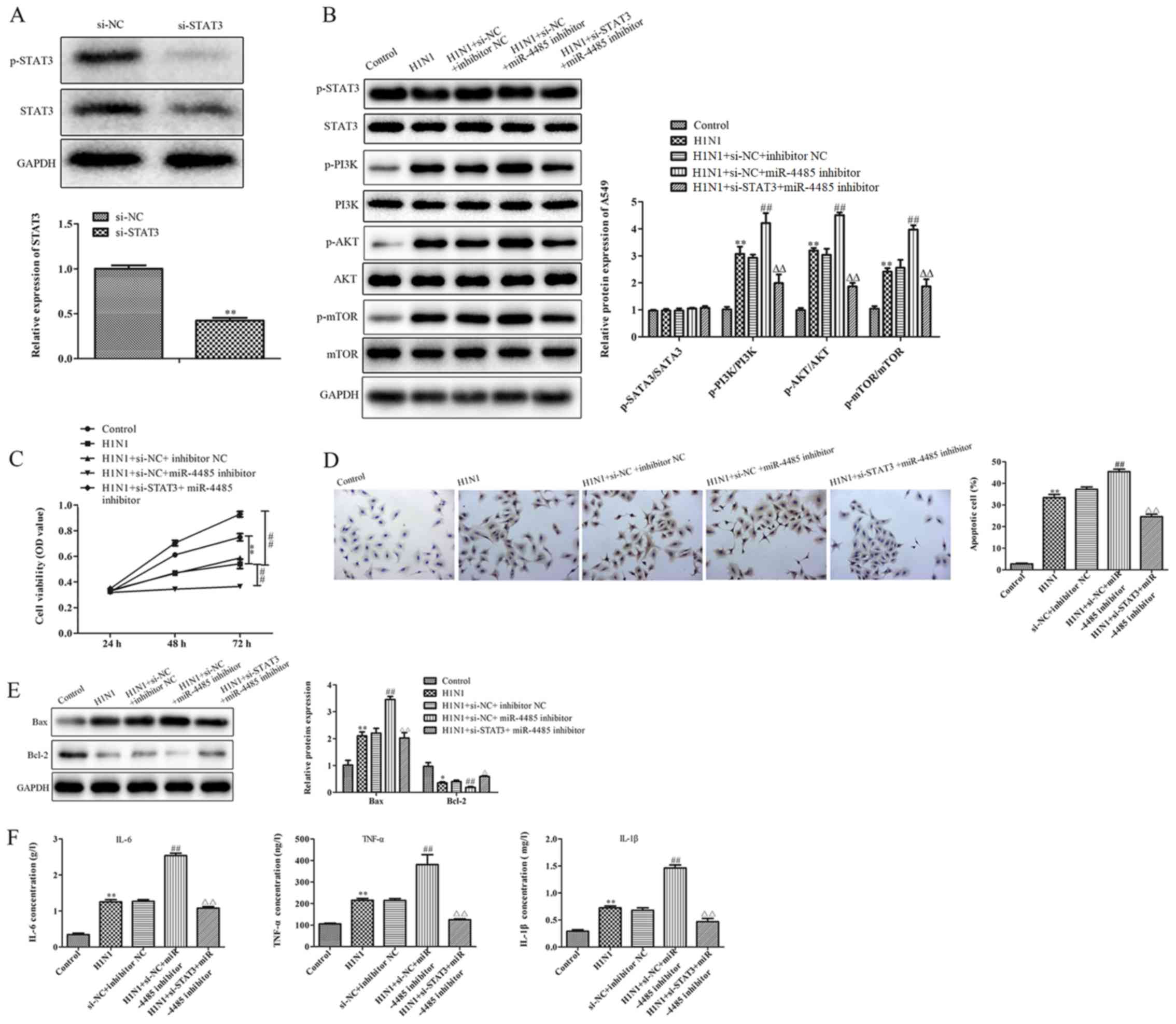 | Figure 4.STAT3 knockdown alleviates the effects
of miR-4485 inhibition on H1N1-induced A549 cell injury. (A)
Western blotting demonstrated STAT3 knockdown in A549 cells
following transfection with si-STAT3. (B) Phosphorylated and total
protein levels of STAT3, PI3K, AKT and mTOR and the ratio of
p-STAT3/STAT3, p-PI3K/PI3K, p-AKT/AKT and p-mTOR/mTOR in A549
cells. (C) Cell Counting Kit-8 cell viability assay, (D) TUNEL
apoptosis detection (magnification ×200), (E) protein expression
levels of Bax and Bcl-2, and (F) production of IL-6, TNF-α and
IL-1β in A549 cells infected with or without H1N1 following
co-transfection with miR-4485 inhibitor and/or si-STAT3. The
experiments were performed three times and the data are presented
as the mean ± SD. **P<0.05 vs. control group;
##P<0.05 vs. H1N1 + si-NC + inhibitor NC group;
ΔΔP<0.01 vs. H1N1 + si-NC + miR-4485 inhibitor group.
miR-4485, microRNA-4485; Mut, mutant; NC, negative control; OD,
optical density; p-, phosphorylated; si, small interfering RNA. |
Discussion
Maintaining epithelial integrity in the lung is
critical in fighting infection and allowing optimal gas exchange.
Influenza viruses disrupt epithelial homeostasis as they directly
target epithelial cells in the lung mucosa (14). Severe infection causes excessive
epithelial injury, fluid buildup in the lung parenchyma and
increased susceptibility to secondary bacterial infections
(15). Endothelial cells are
considered to be the central regulators of cytokine storm in the
respiratory system during influenza virus infection (16). Previous studies have revealed that
infection with H1N1 triggers the release of proinflammatory
cytokines in human lung epithelial A549 cells (17,18).
Understanding how epithelial cells respond to influenza infection
is important for optimizing immune-based strategies to reduce the
burden of infection.
Several lines of evidence suggest that some miRNAs
function as crucial regulators in the pathological process of
influenza pneumonia. For example, Zhang et al (19) reported that miR-29c promotes H1N1
infection-induced inflammatory and antiviral signaling in A549
cells. Podsiad et al (20)
revealed that miR-155 increases mortality and impairs bacterial
clearance in post-influenza pneumonia via negative regulation of
the IL-23/IL-17 signaling pathway. Liu et al (21) demonstrated that inhibition of
miR-200c-3p ameliorates lung injury induced by H5N1 virus infection
by targeting angiotensin-converting enzyme 2. To the best of our
knowledge, the present study was the first to demonstrate the
downregulation of miR-4485 expression in patients with severe H1N1
pneumonia. Results from in vitro experiments demonstrated
that H1N1 infection markedly induced cell injury by inhibiting cell
viability, inducing cell apoptosis and enhancing the production of
cytokines. Additionally, H1N1 inoculation resulted in decreased
expression levels of miR-4485 in A549 cells. In addition,
overexpression of miR-4485 alleviated H1N1-induced cell injury.
There is emerging evidence that miRNAs serve crucial
roles in several biological processes by regulating their target
genes. In the present study, STAT3, an inflammation-related
transcription factor, was identified as a direct target gene of
miR-4485. Increasing evidence has suggested an association between
STAT3 and the development of pneumonia. For instance, STAT3
inhibition rescues mice from pulmonary Staphylococcus aureus
infections by targeting the antimicrobial protein regenerating
islet-derived 3γ (22). Traber et
al (23) demonstrated that STAT3
is required for neutrophil and macrophage recruitment during
pneumonia by inducing chemokine C-X-C motif ligand 5 expression.
Additionally, STAT3 inhibition could suppress
lipopolysaccharide-induced macrophage and inflammatory cell
infiltration in lung and bronchoalveolar lavage fluid (24). In the present study, STAT3 was
demonstrated to be a direct target gene of miR-4485. In addition,
knockdown of STAT3 could reverse the effects of miR-4485
downregulation on H1N1-induced cell injury. Overall, these findings
suggest that miR-4485 may serve a crucial role in H1N1-induced
severe pneumonia by inhibiting STAT3 expression. However, the
functional regulatory mechanisms of STAT3 and miR-4485 remain to be
elucidated.
Increasing evidence has revealed the critical roles
of the PI3K/AKT/mTOR signaling pathway in the host response against
pneumovirus infections. For instance, Klebsiella pneumoniae
induces autophagy and oxidative stress in Caenorhabditis
elegans through inhibition of the PI3K/AKT/mTOR signaling
pathway (25). Streptococcus
pneumonia induces autophagy in A549 cells via a
PI3K/AKT/mTOR-dependent mechanism (26). Yang et al (27) demonstrated that inhibition of
PI3K/AKT/mTOR decreases the reinfection of Streptococcus
pneumonia following influenza A virus infection in
community-acquired pneumonia. Therefore, the present study aimed to
explore whether the effects of miR-4485 on H1N1-induced severe
pneumonia could be mediated via the PI3K/AKT/mTOR signaling
pathway. In the present study, the PI3K/AKT/mTOR signaling pathway
was activated following inhibition of miR-4485, and was markedly
inhibited by STAT3 silencing, suggesting the regulatory functions
of miR-4485 on the STAT3-mediated PI3K/AKT/mTOR pathway. Therefore,
it was speculated that miR-4485 may target STAT3 to regulate the
activation of the PI3K/AKT/mTOR signaling pathway, thus mediating
the development of severe H1N1 pneumonia.
In summary, the present study revealed that miR-4485
expression was downregulated in patients with severe H1N1
pneumonia, and that downregulation of miR-4485 may promote H1N1
infection-induced A549 cell injury by targeting STAT3 via the
activation of the PI3K/AKT/mTOR signaling pathway. These findings
will provide a potential therapeutic target for the treatment of
severe H1N1 pneumonia.
Acknowledgements
Not applicable.
Funding
The study was funded by the Key Subject Project of
Gansu Provincial Hospital (grant. no. 20GSSY1-14).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LG and QW conducted the majority of the experiments,
wrote the manuscript and analyzed the data. DZ designed the study
and revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by the
Human Ethics Committee of Gansu Provincial People's Hospital
(approval no. GSYYR20180912).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gurav YK, Chadha MS, Tandale BV, Potdar
VA, Pawar SD, Shil P, Deoshatwar AR, Aarthy R and Bhushan A:
Influenza A(H1N1)pdm09 outbreak detected in inter-seasonal months
during the surveillance of influenza-like illness in Pune, India,
2012–2015. Epidemiol Infect. 145:1898–1909. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peteranderl C, Herold S and Schmoldt C:
Human influenza virus infections. Semin Respir Crit Care Med.
37:487–500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li H, Yang SG, Gu L, Zhang Y, Yan XX,
Liang ZA, Zhang W, Jia HY, Chen W, Liu M, et al: National influenza
apdm09 clinical investigation group of, effect of
low-to-moderate-dose corticosteroids on mortality of hospitalized
adolescents and adults with influenza A(H1N1)pdm09 viral pneumonia.
Influenza Other Respir Viruses. 11:345–354. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kalogianni DP, Kalligosfyri PM, Kyriakou
IK and Christopoulos TK: Advances in microRNA analysis. Anal
Bioanal Chem. 410:695–713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hoffmann J, Machado D, Terrier O, Pouzol
S, Messaoudi M, Basualdo W, Espínola EE, Guillen RM, Rosa-Calatrava
M, Picot V, et al: Paranhos-baccala, viral and bacterial
co-infection in severe pneumonia triggers innate immune responses
and specifically enhances IP-10: A translational study. Sci Rep.
6:385322016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin J, Wang Y, Zou YQ, Chen X, Huang B,
Liu J, Xu YM, Li J, Zhang J, Yang WM, et al: Differential miRNA
expression in pleural effusions derived from extracellular vesicles
of patients with lung cancer, pulmonary tuberculosis, or pneumonia.
Tumour Biol. 14:10072016.
|
|
7
|
Yang B, Jing C, Wang J, Guo X, Chen Y, Xu
R, Peng L, Liu J and Li L: Identification of microRNAs associated
with lymphangiogenesis in human gastric cancer. Clin Transl Oncol.
16:374–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen J, Wu X, Cai B, Su Z, Li L, An Y and
Wang L: Circulating microRNAs as potential biomarkers of HBV
infection persistence. Infect Genet Evol. 54:152–157. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sripada L, Singh K, Lipatova AV, Singh A,
Prajapati P, Tomar D, Bhatelia K, Roy M, Singh R, Godbole MM, et
al: Hsa-MiR-4485 regulates mitochondrial functions and inhibits the
tumorigenicity of breast cancer cells. J Mol Med (Berl).
95:641–651. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang S, Feng C, Zhai YZ, Zhou X, Li B,
Wang LL, Chen W, Lv FQ and Li TS: Identification of miRNA
biomarkers of pneumonia using RNA-sequencing and bioinformatics
analysis. Exp Ther Med. 13:1235–1244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang SG, Cao B, Liang LR, Li XL, Xiao YH,
Cao ZX, Jia HY, Yu HJ, Xu Z, Gu L, et al: Antiviral therapy and
outcomes of patients with pneumonia caused by influenza a pandemic
(H1N1) virus. PLoS One. 7:e296522012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao YR, Wang D, Liu Y, Shan L and Zhou
JL: The PI3K/Akt, p38MAPK, and JAK2/STAT3 signaling pathways
mediate the protection of SO2 against acute lung injury induced by
limb ischemia/reperfusion in rats. J Physiol Sci. 66:229–239. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Traylor ZP, Aeffner F and Davis IC:
Influenza A H1N1 induces declines in alveolar gas exchange in mice
consistent with rapid post-infection progression from acute lung
injury to ARDS. Influenza Other Respir Viruses. 7:472–479. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Siemens N, Oehmcke-Hecht S, Mettenleiter
TC, Kreikemeyer B, Valentin-Weigand P and Hammerschmidt S: Port
d'Entree for respiratory infections-does the influenza a virus pave
the way for bacteria? Front Microbiol. 8:26022017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Jiang H, Shen SM, Wen CX, Xing Z
and Shi Y: Inhibition of autophagy and chemokine induction by
sphingosine 1-phosphate receptor 1 through NF-κB signaling in human
pulmonary endothelial cells infected with influenza a viruses. PLoS
One. 13:e0205344 View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Teijaro JR, Walsh KB, Cahalan S, Fremgen
DM, Roberts E, Scott F, Martinborough E, Peach R, Oldstone MB and
Rosen H: Endothelial cells are central orchestrators of cytokine
amplification during influenza virus infection. Cell. 146:980–991.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo X, Zhu Z, Zhang W, Meng X, Zhu Y, Han
P, Zhou X, Hu Y and Wang R: Nuclear translocation of HIF-1α induced
by influenza A (H1N1) infection is critical to the production of
proinflammatory cytokines. Emerg Microbes Infect. 6:e392017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Dong C, Sun X, Li Z, Zhang M,
Guan Z and Duan M: Induction of the cellular miR-29c by influenza
virus inhibits the innate immune response through protection of A20
mrna. Biochem Biophys Res Commun. 450:755–761. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Podsiad A, Standiford TJ, Ballinger MN,
Eakin R, Park P, Kunkel SL, Moore BB and Bhan U: MicroRNA-155
regulates host immune response to postviral bacterial pneumonia via
IL-23/IL-17 pathway. Am J Physiol Lung Cell Mol Physiol.
310:L465–L475. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Q, Du J, Yu X, Xu J, Huang F, Li X,
Zhang C, Li X, Chang J, Shang D, et al: MiRNA-200c-3p is crucial in
acute respiratory distress syndrome. Cell Discov. 3:170212017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choi SM, McAleer JP, Zheng M, Pociask DA,
Kaplan MH, Qin S, Reinhart TA and Kolls JK: Innate stat3-mediated
induction of the antimicrobial protein Reg3g is required for host
defense against MRSA pneumonia. J Exp Med. 210:551–561. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Traber KE, Hilliard KL, Allen E, Wasserman
GA, Yamamoto K, Jones MR, Mizgerd JP and Quinton LJ: Induction of
STAT3-dependent CXCL5 expression and neutrophil recruitment by
oncostatin-M during pneumonia. Am J Respir Cell Mol Biol.
53:479–488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao J, Yu H, Liu Y, Gibson SA, Yan Z, Xu
X, Gaggar A, Li PK, Li C, Wei S, et al: Protective effect of
suppressing STAT3 activity in LPS-induced acute lung injury. Am J
Physiol Lung Cell Mol Physiol. 311:L868–L880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kamaladevi A and Balamurugan K: Global
proteomics revealed klebsiella pneumoniae induced autophagy and
oxidative stress in caenorhabditis elegans by inhibiting
PI3K/AKT/mTOR pathway during infection. Front Cell Infect
Microbiol. 7:3932017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li P, Shi J, He Q, Hu Q, Wang YY, Zhang
LJ, Chan WT and Chen WX: Streptococcus pneumoniae induces autophagy
through the inhibition of the PI3K-I/Akt/mTOR pathway and ROS
hypergeneration in A549 cells. PLoS One. 10:e01227532015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang Z, Zou X, Feng P, Zhan H, Xiong D and
Lang J: Inhibition of the PI3K/AKT signaling pathway or
overexpression of beclin1 blocks reinfection of streptococcus
pneumoniae after infection of influenza a virus in severe
community-acquired pneumonia. Inflammation. 42:1741–1753. 2019.
View Article : Google Scholar : PubMed/NCBI
|