Introduction
Lung cancer has a high degree of malignancy and
ranks first as the cause of cancer-associated mortality in the
United States (1). Non-small cell
lung cancer (NSCLC) accounts for 85% of all lung cancer cases in
the United States (2). Although the
diagnosis and treatment methods of NSCLC are continuously
improving, metastasis via the lymph nodes and blood system occurs
at an early stage, and due to insidious onset and rapid
progression, the prognosis of NSCLC remains unsatisfactory.
Therefore, an in-depth study on the mechanism of NSCLC metastasis
is of great significance for its prevention, and to develop novel
molecular targeted therapeutic drugs.
The JAK/STAT signaling pathway was found to have
sustained abnormal activation in various malignant tumor cells,
such as prostate cancer, sarcomas and lymphomas, regulating tumor
proliferation, apoptosis and metastasis (3). As a member of STAT family, STAT3 is
closely associated with the prognosis of NSCLC (4). JAK2 activates STAT3 by phosphorylation,
allowing it to enter the nucleus and perform its biological
function (3). It has been reported
that the activation of the JAK2/STAT3 signaling pathway induced
metastatic NSCLC (5). Due to the
importance of JAK2/STAT3 signaling in tumor development, targeted
therapy for this pathway has also become a popular research topic
in the study of NSCLC. At present, although several types of
JAK2/STAT3-targeted drugs are undergoing clinical trial (and have
made progress), their therapeutic effects remain unsatisfactory.
Therefore, identifying new targets for JAK2/STAT3 sensitization may
enhance the clinical effect of JAK2/STAT3 target-based therapy and
improve the prognosis of patients with NSCLC.
Sumoylation is a ubiquitin-like post-translational
modification that enhances the stability of protein and regulates
their distribution and localization (6). The transcriptional activity of
transcription factors can be modulated through sumoylation
(7). To date, four members of the
small ubiquitin-like modifier (SUMO) family (SUMO1, 2, 3 and 4)
have been cloned and identified (7).
SUMO4 is a newly discovered member of the sumoylation family, which
is mainly expressed in the immune-associated organs and kidneys. It
has been confirmed to be closely associated with type I diabetes
(8), coronary heart disease
(9), psoriasis (10), Behcet's disease (11) and other diseases. However, research
into the potential regulatory effects of SUMO4 on tumorigenesis is
somewhat lacking.
A Study has shown that SUMO4 negatively regulates
NF-kB transcriptional activity in a diabetic model (12). Mo et al (11–13)
found that SUMO4 decreased oxidative stress by increasing
antioxidant enzymatic activity and DNA damage signaling-associated
protein activity, thus activating the cellular self-protection
mechanisms (13). SUMO4 also
directly decreased the DNA-binding activity of the STAT protein,
leading to the inhibition of JAK/STAT signaling (14). These findings suggest that SUMO4 may
be associated with tumor development and progression. It has been
reported that SUMO4 expression was increased in thyroid cancer
(15). However, the expression and
function of SUMO4 during the tumorigenesis of NSCLC remain unknown.
The present study investigated the expression of SUMO4 in NSCLC and
identified the mechanisms of SUMO4 in augmenting the proliferation,
invasion and migration of NSCLC cells.
Materials and methods
NSCLC patient samples
A total of 100 NSCLC 10 adjacent non-cancerous
tissues (defined as tissue which is at least 3 cm from cancerous
region; samples were collected from 100 patients (71 men and 29
women) during surgery at Zhejiang Cancer Hospital (Zhejiang, China)
between January 2009 and March 2011. All patients gave written
informed consent to participate in the study and to allow their
samples to be biologically analyzed. The age of the patients with
NSCLC enrolled in the present study ranged from 39 to 76 years
(mean, 61.3 years). The exclusion criteria included patients with
other types of cancer and those who had received preoperative
chemoradiotherapy. Control samples were obtained from the same
patient at a site at least 3 cm from the tumor and were approved as
control samples by a pathologist. Tissue sections were fixed with
10% formalin for at least 24 h at room temperature and subsequently
embedded in paraffin for immunohistochemistry. The tumors were
staged according to the pathological tumor/node/metastasis (pTNM)
classification (7th edition) of the International Union against
Cancer (16). All procedures for
sample collection and processing were ratified by the International
Review Board of Zhejiang Cancer Hospital (Hangzhou, China; approval
no. IRB-2016-134).
Reagents
Tyrphostin AG490 was purchased from Sigma-Aldrich
(Merck KGaA). The Cell Counting Kit-8 (CCK-8) was purchased from
Dojindo Molecular Technologies, Inc. The Biotin-Streptavidin HRP
Detection kit was purchased from OriGene Technologies, Inc. (cat.
no. SP-9000). SUMO4-specific small interfering (si)RNA (forward,
5′-GGAUGGUUCUGUGGUGCAGTT-3′ and reverse,
5′-CUGCACCACAGAACCAUCCTT-3′) and negative control siRNA (forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′) were designed and synthesized by
Shanghai GenePharma Co., Ltd. Cisplatin was purchased from Jiangsu
Hanson Pharmaceutical Co. (http://www.hansoh.cn). The SUMO4 antibody was
purchased from Abcam (cat. no. ab126606), phosphorylated (p-)JAK2
and p-STAT3 antibodies were from Cell Signaling Technology Inc.
(cat. nos. 3771 and 9145), antibodies against vimentin, E-cadherin,
N-cadherin, JAK2 and STAT3 were purchased from ProteinTech Group,
Inc. (cat. nos. 10377-1-AP, 20874-1-AP, 22018-1-AP, 17670-1-AP and
10253-1-AP).
Cell lines
Human NSCLC cell lines A549, NCI-H1650 and SK-MES-1
were purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. All cell lines were cultured in DMEM
(Cytvia) supplemented with 10% fetal bovine serum and 1%
penicillin-streptomycin (Transgen Biotech Co., Ltd.) in humidified
air at 37°C (5% CO2).
Transfection
Cells were seeded into 6-well plates. At 60–70%
confluence (5×105 cells/well), SUMO4 siRNA (100 nM) and
scrambled control siRNA (100 nM) were transiently transfected into
the cells using Lipofectamine® 2000 Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to manufacturer's
instructions. The transfection reagent was removed after 5 h, and
the cells were harvested after 48 h.
Immunohistochemical (IHC)
staining
IHC staining of tumor tissues was performed on 5-µm
sections. The general procedure for IHC staining was performed as
previously described (17). The
sections were blocked with blocking serum from the ABC Vectastain
kit (cat. no. PK-6100; Vector Laboratories, Inc.) at room
temperature for 30 min, followed by incubation with primary
antibody against SUMO4 (1:50; cat. no. ab126606; Abcam) overnight
at 4°C. Then the sections were incubated with a horseradish
peroxidase-conjugated mouse anti-rabbit Ig antibody at room
temperature for 1 h, followed by staining with the chromogen
diaminobenzidine (Zhongshan, Beijing, People's Republic of China)
until a brown color was shown. The slides were counterstained with
Mayer's hematoxylin at room temperature for 10 min. Random fields
from each slide were viewed under a light microscope (Olympus DP73;
Olympus Corporation) at ×20 magnification.
Western blotting
Proteins extracted from cells were denatured in RIPA
buffer (150 mM NaCl, 0.1% SDS, 25 mM Tris-HCl pH 7.6, 1% sodium
deoxycholate and 1% NP-40) with protease inhibitors. The
concentrations of protein were detected using a BCA protein assay
kit (cat. no. P0010S; Beyotime Institute of Biotechnology). Equal
amounts of total protein (50 µg) were analyzed by SDS-PAGE.
Proteins were separated on a 12% polyacrylamide gel and transferred
to a nitrocellulose membrane. The membrane was blocked for 1 h at
room temperature using blocking buffer (0.5% fat-free milk). After
blocking, the blot was probed with primary antibodies (dilution,
1:1,000) at 4°C overnight. Subsequently, the blot was incubated
with HRP-labeled secondary antibodies diluted in PBS buffer
(1:2,000; cat. no. SA00001-2; ProteinTech Group, Inc.). ECL reagent
was used for visualization. The images were obtained using the
Bio-Rad Imaging System. Signal quantification was obtained using
Quantity One software (version 4.6.6; Bio-Rad Laboratories, Inc.)
and normalized to GAPDH.
Wound-healing assay
A total of 5×105 H1650, A549 and sk-mes-1
cells were seeded independently into each well of 6-well plates
overnight. A scratch was made on the monolayers with a 200-µl
pipette tip, and washed with PBS to remove the detached cells.
Fresh DMEM/F12 medium without serum was added into each well of the
6-well plate. The wounded areas were observed and imaged using a
light microscope (magnification ×100) at 0 and 24 h. The migration
results were quantified using ImageJ software (version 1.52;
National Institutes of Health).
Transwell invasion assay
The trypsinized A549, H1650 and sk-mes-1 cells were
washed with PBS and resuspended in serum-free DMEM medium. Then,
200 µl cell suspension (1×105/well) was added to the
upper chamber with a 50 µl solidified Matrigel-coated membrane. The
lower chamber was filled with 800 µl DMEM supplemented with 10%
FBS. After 24 h incubation, the chambers were fixed with 100%
methanol for 20 min at room temperature, followed by staining with
0.1% crystal violet for 20 min at room temperature. Images were
captured with an Olympus fluorescence microscope (magnification
×100).
CCK-8 assay
Transfected H1650, A549 and sk-mes-1 cells were
collected (100 µl) at 24 h post-transfection and seeded into
96-well plates at a density of 3×103/well independently.
Following incubation for 0, 12, 24, 36 and 48 h at 37°C, 10 µl
CCK-8 reagent was added into each of the wells, and the cells were
incubated at 37°C for 2 h. The absorbance was measured at a
wavelength of 450 nm using a microplate reader (Bio-Rad
Laboratories, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS 23.0
software (IBM Corp.). All experiments were performed in triplicate
and data are presented as the mean ± standard error of the mean.
The association between the expression of SUMO4 and
clinicopathological parameters was examined using the χ2
test. The overall survival (OS) and disease-free survival (DFS)
curves were produced using the Kaplan-Meier method, the difference
in survival was determined using the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Unpaired Student's t-test was performed for western blotting and
wound-healing data analysis. One-way ANOVA with Tukey's test was
performed for invasion assay and phosphorylation analysis.
P<0.05 was considered to indicate a statistically significant
difference. Numerical data are presented as the mean ± standard
deviation.
Results
Overexpression of SUMO4 is associated
with a poor prognosis in human NSCLC
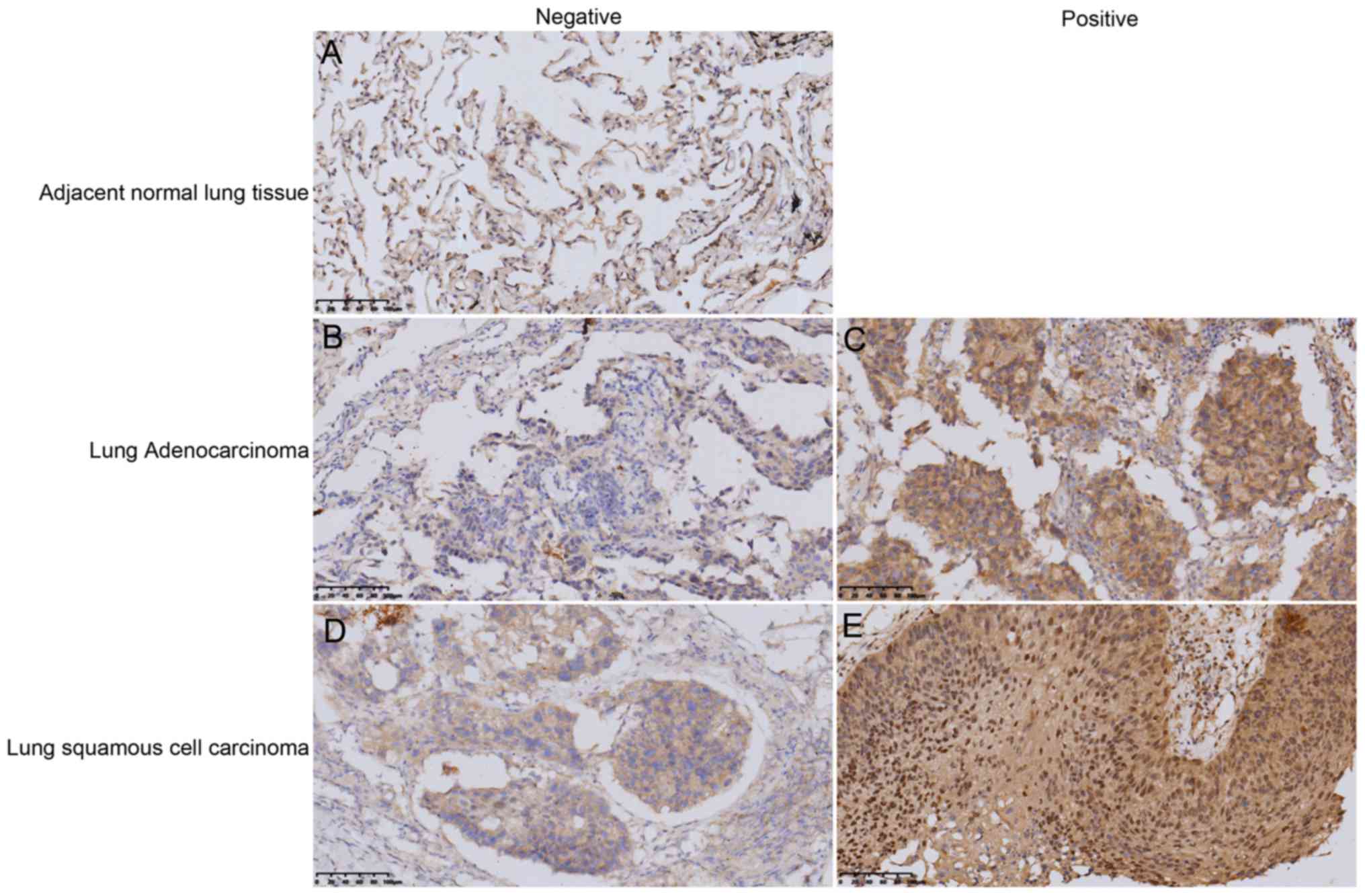
In the present study, 10 adjacent normal lung
tissues and 100 NSCLC tissues were collected for IHC staining, in
order to explore the association between SUMO4 expression and NSCLC
prognosis. The results revealed that SUMO4 was expressed
differently in the cytoplasm of these tissues. The tissues were
divided into ‘positive’ and ‘negative’ groups, according to SUMO4
expression (Fig. 1). The expression
of SUMO4 in NSCLC tissues were significantly higher compared with
the adjacent normal lung tissue (Fig.
1). Quantitative analysis of the IHC staining results indicated
that 69% of NSCLC samples were positive for SUMO4, while all
adjacent normal lung tissues were negative via IHC analysis
(Table I;
P=1.6862×10−5).
 | Table I.Summary of SUMO4 expression status in
NSCLC and adjacent normal tissues analyzed in the present
study. |
Table I.
Summary of SUMO4 expression status in
NSCLC and adjacent normal tissues analyzed in the present
study.
|
| SUMO4 expression |
|
|---|
|
|
|
|
|---|
| Group | Negative, n | Positive, n | Total, n |
|---|
| Adjacent normal
tissue | 10 | 0 | 10 |
| NSCLC tissue | 31 | 69 | 100 |
In order to demonstrate the association between
SUMO4 expression and NSCLC prognosis, the expression of SUMO4 was
systematically analyzed relative to the clinicopathological
characteristics of NSCLC from 100 patients. Based on the IHC
staining results of all samples, the following associations were
identified: Sex, tumor type, history of smoking and T stage were
significantly associated with the expression of SUMO4 (P<0.05),
however, age, body mass index (BMI), hypertension, diabetes,
history of drinking, tumor location, tumor size, tumor
differentiation, N stage, tumor stage and recurrence were not
associated with the expression of SUMO4 (P>0.05) (Table II).
 | Table II.Statistical analysis of the
association between the expression of SUMO4 and the
clinicopathological parameters of NSCLC. |
Table II.
Statistical analysis of the
association between the expression of SUMO4 and the
clinicopathological parameters of NSCLC.
|
|
| SUMO4 expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameter | n | Negative, n | Positive, n | P-value |
|---|
| Age, years |
|
|
| 0.723 |
|
<60 | 33 | 11 | 22 |
|
| ≥60 | 67 | 20 | 47 |
|
| Sex |
|
|
| 0.017 |
| Male | 71 | 17 | 54 |
|
|
Female | 29 | 14 | 15 |
|
| Tumor type |
|
|
| 0.013 |
|
Adenocarcinoma | 46 | 20 | 26 |
|
|
Squamous cell carcinoma | 54 | 11 | 43 |
|
| BMI |
|
|
| 0.075 |
|
<22 | 52 | 12 | 40 |
|
|
≥22 | 48 | 19 | 29 |
|
| Hypertension |
|
|
| 0.108 |
|
With | 20 | 3 | 17 |
|
|
Without | 80 | 28 | 52 |
|
| Diabetes |
|
|
| 1.000 |
|
With | 7 | 2 | 5 |
|
|
Without | 93 | 29 | 64 |
|
| History of
drinking |
|
|
| 0.993 |
|
Moderate or heavy
drinking | 42 | 13 | 29 |
|
| Without
or light drinking | 58 | 18 | 40 |
|
| History of
smoking |
|
|
| 0.002 |
|
With | 64 | 13 | 51 |
|
|
Without | 36 | 18 | 18 |
|
| Tumor location |
|
|
| 0.510 |
| Left
lung | 37 | 10 | 27 |
|
| Right
lung | 63 | 21 | 42 |
|
| Tumor location |
|
|
| 0.327 |
| Upper
and middle lung | 54 | 19 | 35 |
|
| Lower
lung | 46 | 12 | 34 |
|
| Tumor size, cm |
|
|
| 0.262 |
| ≤3 | 34 | 13 | 21 |
|
|
>3 | 66 | 18 | 48 |
|
| Tumor
differentiation |
|
|
| 0.168 |
| Well
and moderate | 51 | 19 | 32 |
|
|
Poor | 49 | 12 | 37 |
|
| T stage |
|
|
| 0.025 |
| T1 and
T2 | 76 | 28 | 48 |
|
| T3 and
T4 | 24 | 3 | 21 |
|
| N stage |
|
|
| 0.137 |
| N0 | 47 | 18 | 29 |
|
| N1, N2
and N3 | 53 | 13 | 40 |
|
| M stage |
|
|
| 0.526 |
| M0 | 98 | 30 | 68 |
|
| M1 | 2 | 1 | 1 |
|
| Tumor stage |
|
|
| 0.186 |
| Stages
I and II | 58 | 21 | 37 |
|
| Stages
III and IV | 42 | 10 | 32 |
|
| Recurrence |
|
|
| 0.084 |
|
With | 64 | 16 | 48 |
|
|
Without | 36 | 15 | 21 |
|
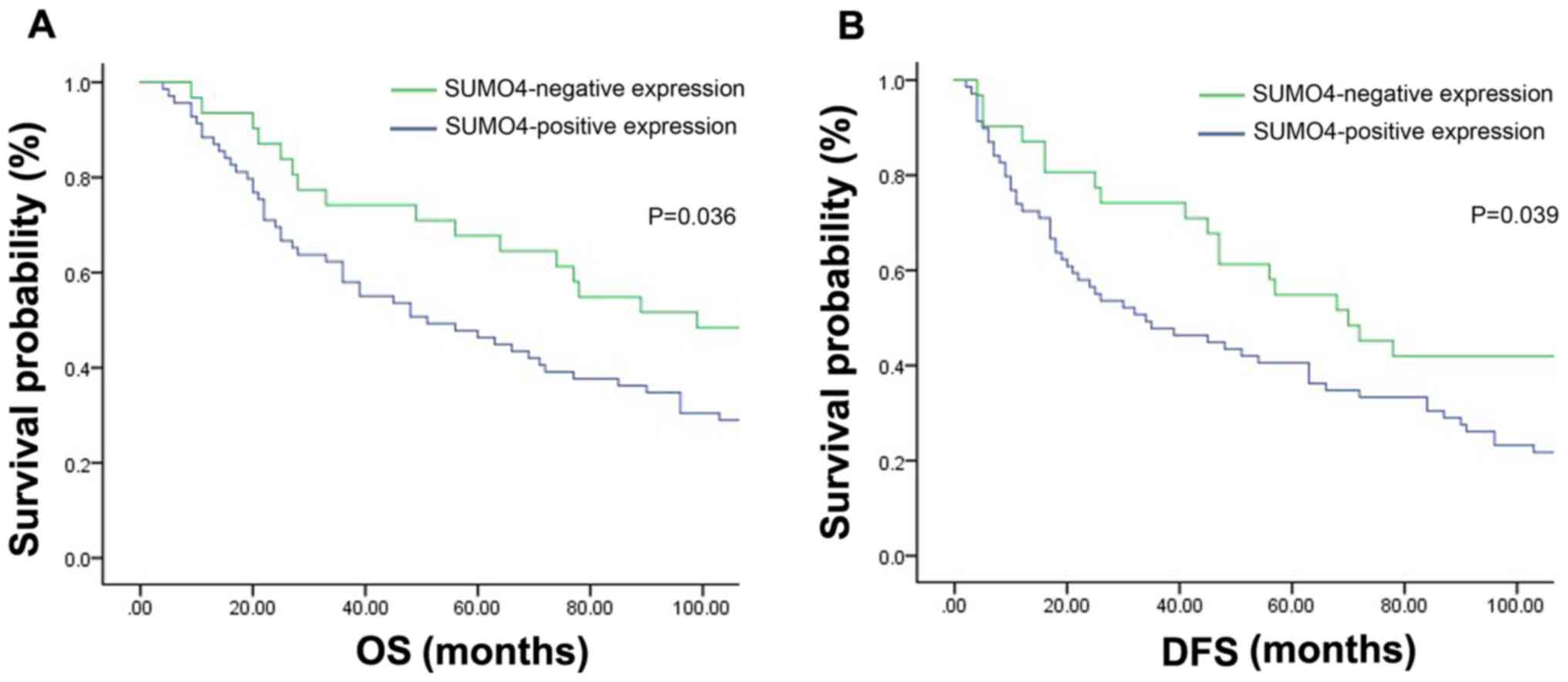
To clarify the associations between SUMO4 and NSCLC
prognosis, the OS and DFS curves of these patients were generated
using the Kaplan-Meier method. It was identified that positive
expression of SUMO4 was significantly associated with short DFS and
OS times (Fig. 2), indicating poor
prognosis.
SUMO4 siRNA decreases cell migration,
invasiveness and epithelial-mesenchymal transition (EMT) in NSCLC
cells
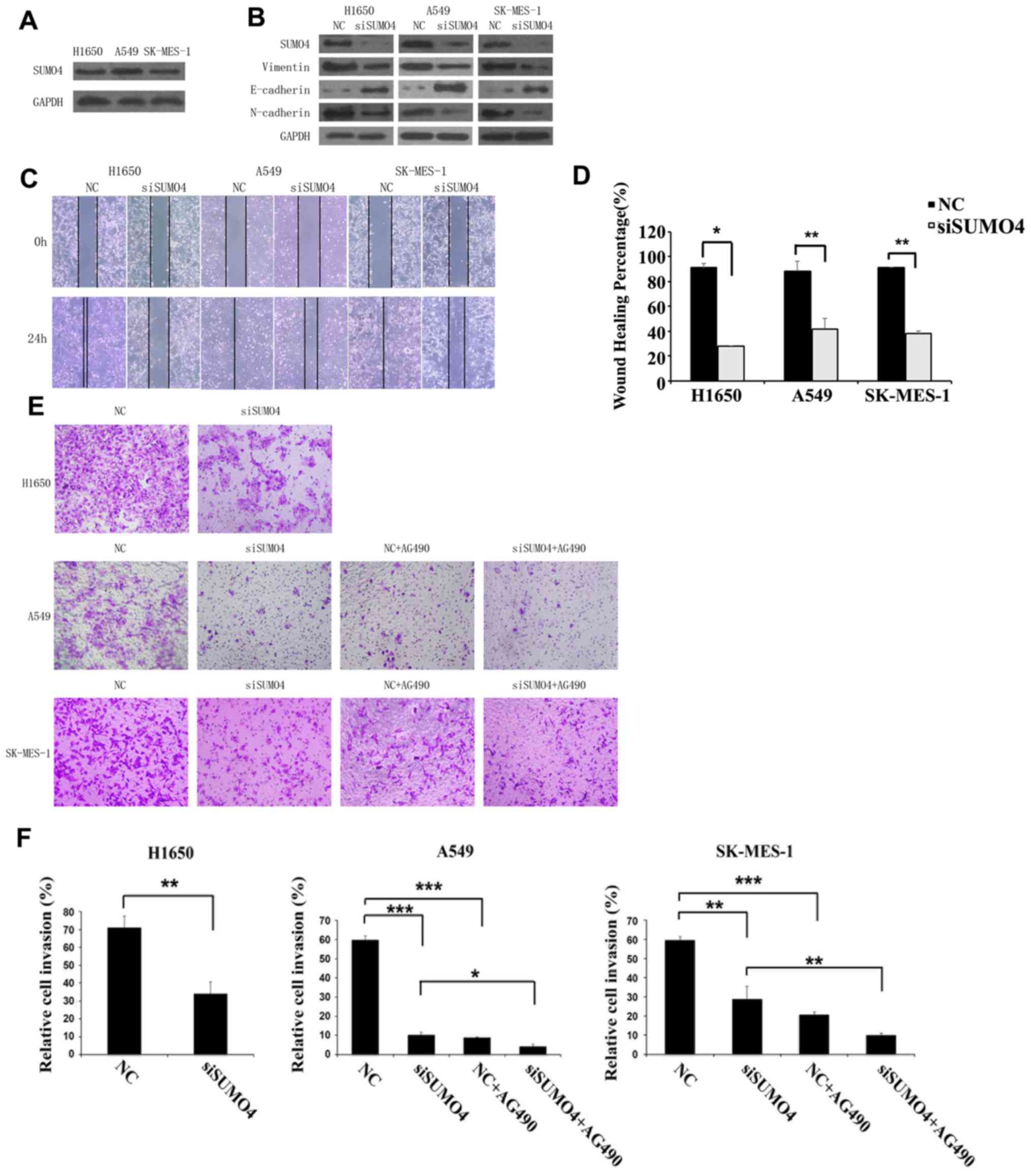
To explore whether SUMO4 regulates the migratory and
invasive abilities of NSCLC cells, SK-MES-1, NCI-H1650 and A549
cells which expressed SUMO4, were transfected with SUMO4 siRNA and
negative control siRNA (Fig. 3A).
EMT marker proteins including N-cadherin, E-cadherin and vimentin
were examined by western blotting as shown in Fig. 3B. A notable decrease in vimentin and
N-cadherin, as well as increase in E-cadherin was observed. Cell
migration and invasion were evaluated by wound healing and
Transwell assays (Fig. 3C-F). It was
found that SUMO4 siRNA significantly inhibited the wound closure
and invasion capacities of A549, H1975 and SK-MES-1 cells.
SUMO4 siRNA promotes the sensitivity
of NSCLC cells to chemotherapy
To identify whether SUMO4 expression affects NSCLC
chemosensitivity, control and SUMO4-siRNA transfected A549, H1975
and SK-MES-1 cells were treated with cisplatin at various
concentrations, and the CCK-8 assay was used to assess the
inhibition of cell proliferation rate. It was found that the
inhibitory effect on the proliferation of cells increased with
increasing concentrations of cisplatin. SUMO4 silencing
dose-dependently altered inhibition rate compared with the NC group
(Fig. 4).
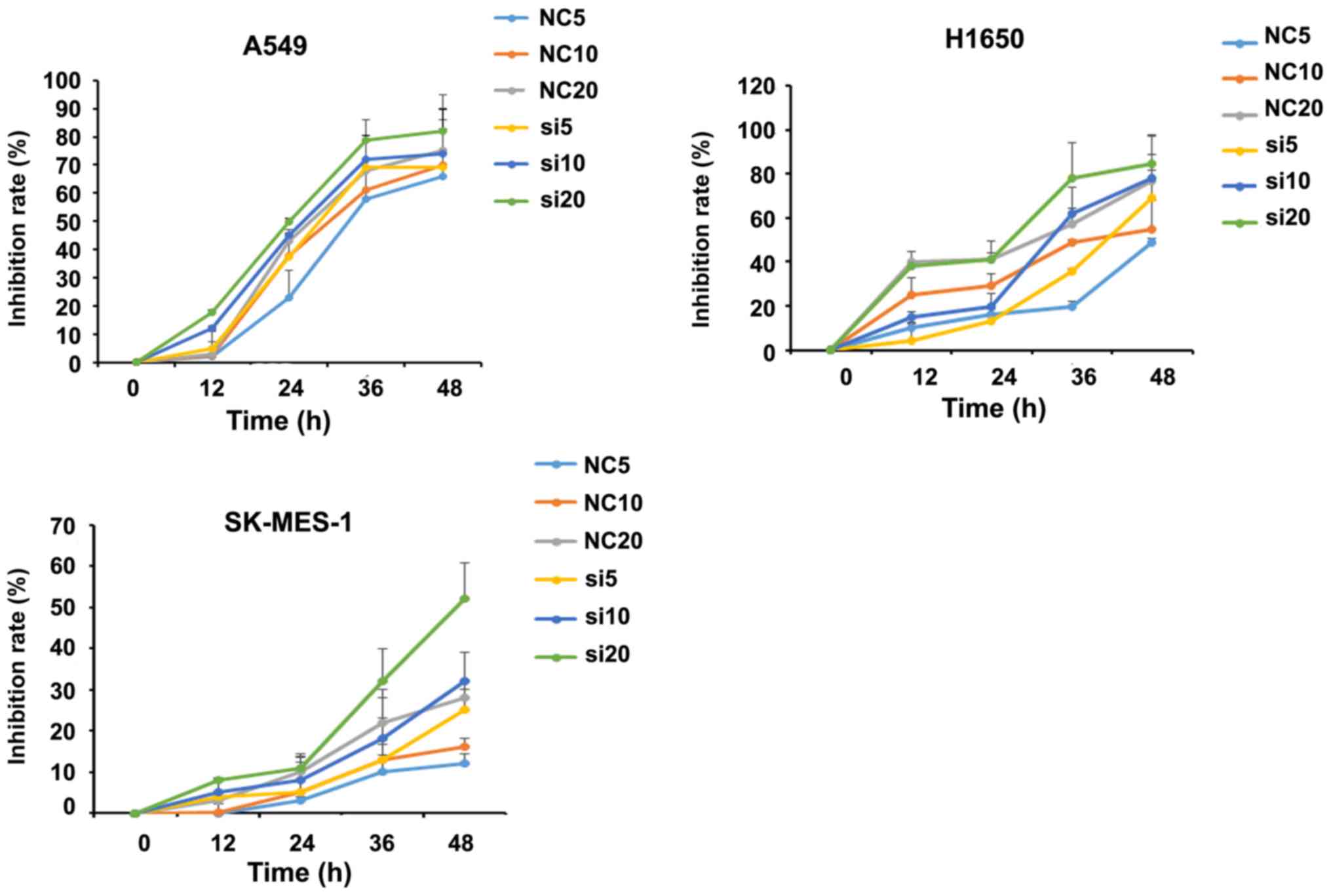 | Figure 4.Inhibitory effects of cisplatin on
three NSCLC cell lines tested by Cell Counting Kit-8 assay. NSCLC,
non-small cell lung cancer; SUMO4, small ubiquitin-like modifier 4;
siRNA, small interfering RNA; NC, negative control; NC5, NSCLC
cells with negative control siRNA plus 5 µM cisplatin; NC10, NSCLC
cells with negative control siRNA plus 10 µM cisplatin; NC20, NSCLC
cells with negative control siRNA plus 20 µM cisplatin; si5, NSCLC
cells with SUMO4 siRNA plus 5 µM cisplatin; si10, NSCLC cells with
SUMO4 siRNA plus 10 µM cisplatin; si20, NSCLC cells with SUMO4
siRNA plus 20 µM cisplatin. |
SUMO4 siRNA decreases cell
invasiveness via the JAK2/STAT3 pathway in NSCLC cells
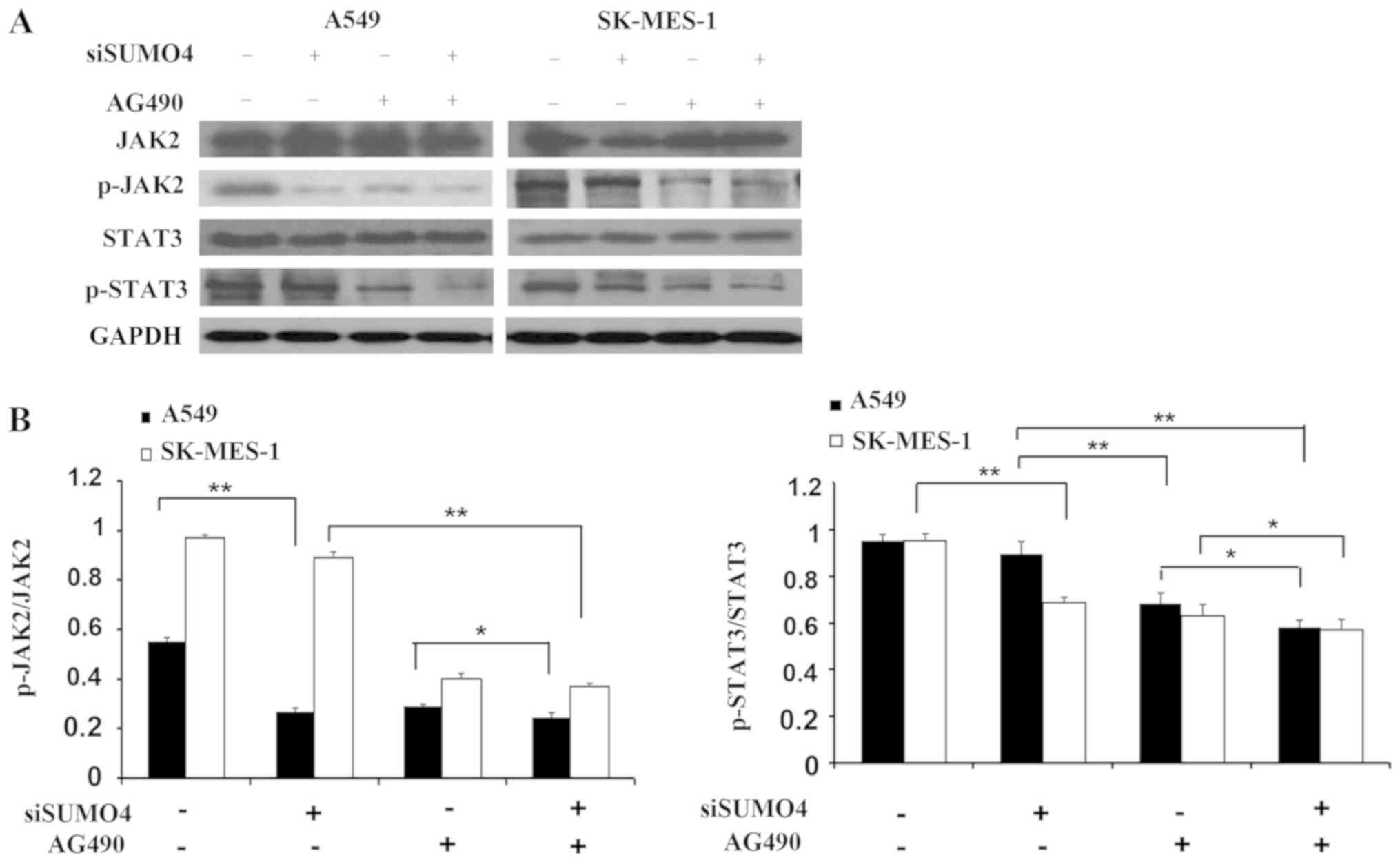
To explore the underlying molecular mechanism of the
augmented invasion by SUMO4 depletion, the JAK2/STAT3 pathway was
specifically analyzed in the present study. Using SUMO4 siRNA
combined with the application of JAK2 kinase inhibitor AG490, it
was found that knocking down SUMO4 slightly decreased JAK2 and
STAT3 activity, as reflected by the phosphorylated forms of JAK2
and STAT3 (Fig. 5). In addition,
inactivation of JAK2 by a specific inhibitor (AG490) resulted in
decreased invasive ability of cells, demonstrated by the transwell
invasion assay (Fig. 3D).
Discussion
Sumoylation has been shown to play important roles
in various biological processes such as signal transmission,
nuclear transportation, gene expression regulation and cell cycle
regulation (18). It also
participates in the regulation of mitochondrion division, as well
as the maintenance of genome integrity (18). Sumoylation is closely associated with
several human diseases, such as cancer, diabetes, Parkinson's
disease and Alzheimer's disease (19). For example, sumoylation of β-catenin
regulated the proliferation of myeloma (20), CDK6 sumoylation regulated the cell
cycle arrest of glioma cells (21),
and sumoylation of Forkhead box protein K2 regulated the
sensitivity of breast cancer cells to paclitaxel (22). These results identified that
sumoylation played an important role in the development of
tumors.
In 2004, Guo et al (23) and Bohren et al (24) simultaneously discovered the SUMO4
gene on chromosome 6q25, which contains 702 nucleotide residues and
encodes 95 amino acids. Guo et al (23) also found a close association between
SUMO4 and type I diabetes, which may be a novel sensitivity gene
for type I diabetes (20). In
addition, it was also confirmed that SUMO4 decreased the activity
of NF-kB by binding with IKB (23).
Mo et al (13) showed that
SUMO4 initiated cell self-protection mechanisms by increasing the
activity of antioxidant enzyme and DNA damage signaling protein,
thus decreasing oxidative stress. These results indicated that
SUMO4 may play an important role in the development of cancer. In
previous studies, it was found that SUMO4 expression was
significantly increased in thyroid cancer (13). The present findings confirmed that
SUMO4 was associated with tumor progression. However, the study of
SUMO4 in NSCLC has not been reported, and the mechanism of SUMO4 in
tumors is still unclear.
The present study first demonstrated the expression
of SUMO4 in NSCLC. The expression of SUMO4 in NSCLC tissues was
significantly higher than in the adjacent normal lung tissues. The
results showed that SUMO4 may play an important role in the
occurrence and development of NSCLC. In the clinicopathological
characteristics analysis, the SUMO4 positivity in men, squamous
cell carcinoma, patients with a smoking history, T3 and T4 stage
was found to be significantly higher compared with women, those
with adenocarcinoma and those without a smoking history, and T1 and
T2 stage tumors (P<0.05). However, older patients with a lower
BMI, hypertension, diabetes, moderate or heavy drinking history,
larger tumors, poor differentiation, lymph node metastasis, tumor
stage III or IV, and with recurrence were associated with SUMO4
positivity, although not significantly so. These results showed
that SUMO4 was closely associated with T stage, but there is no
significant difference between N stage and tumor stage, which may
be due to the limited number of specimens. Interestingly, it was
found that men with squamous cell carcinoma and a history of
smoking had high positivity for SUMO4. Since the smoking rate in
men is significantly higher compared with women, the association
between smoking and squamous cell carcinoma is higher compared with
between smoking and adenocarcinoma. Therefore, it is speculated
that smoking may increase the positivity for SUMO4 and result in
the occurrence and development of NSCLC. In the survival analysis,
patients positive for SUMO4 expression were found to have poor
prognosis. The results showed that SUMO4 may be a novel prognostic
factor for NSCLC.
The mechanism underlying the regulatory effect of
SUMO4 in the A549, H1650 and SK-MES-1 cell lines was further
explored. The expression of SUMO4 enhanced invasion and migratory
abilities, and increased EMT in all three cell lines. These results
imply that SUMO4 expression could promote the metastatic capability
of NSCLC. It was found that SUMO4 expression decreased cisplatin
chemosensitivity. Thus, SUMO4 expression may be involved in
chemotherapy resistance in NSCLC.
Wang et al (14) found that SUMO4 decreased the DNA
binding activity of STAT, thus inhibiting the JAK/STAT signaling
pathway. In the present study, the inhibition of SUMO4 was found to
inhibit invasion and migration by downregulating the activation of
the JAK2/STAT3 pathway in NSCLC cells. SUMO4 may regulate the
stability of molecules which activate JAK2-STAT3 signaling
pathways, which lead to tumor progression. However, the molecular
mechanism still requires further exploration.
There are limitations of the present study. Firstly,
due to the limited number of specimens, some subgroups had
insufficient samples. The sample size needs to be increased to
obtain more accurate results. Secondly, in vivo studies
should be performed to explore the effect of SUMO4 on NSCLC
metastasis. Furthermore, the mechanisms underlying the effect of
SUMO4 on JAK2/STAT3 signal pathway activation requires further
exploration, by constructing SUMO4 expression plasmids, in future
studies.
In conclusion, SUMO4 was found to be expressed in
NSCLC and is significantly associated with sex, tumor type, history
of smoking, T stage and poor prognosis in NSCLC. SUMO4 plays a
significant role in cell invasion and migration via JAK2/STAT3
pathway activation in NSCLC cell lines, which implies that SUMO4
may be a potential therapeutic target for NSCLC with positive
expression of SUMO4.
Acknowledgements
The authors of the present study would like to thank
Dr Wei Gao from Zhejiang University City College (Hangzhou, China)
and Dr Guoping Cheng from Zhejiang Cancer Hospital (Hangzhou,
China) for their technical assistance, and Dr Lei Cai from Zhejiang
Cancer Hospital (Hangzhou, China) for providing technical
assistance on lung tumor immunohistochemical analyses. The authors
would also like to thank Dr Xiaowei Zeng for technical support on
the in vitro experiments, and Dr Kaiyi Tao, Dr Xun Yang and
Dr Jinxiao Liang [all from Zhejiang Cancer Hospital (Hangzhou,
China)] for collection, analysis and interpretation of data.
Funding
The present study was supported by grants from
Medical Health Science and Technology Project of Zhejiang
Provincial Health Commission (grant nos. 2017184728 and
2018241087).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JXL performed in vitro experiments and
drafted the initial manuscript. WG, GPC and LC collected lung
cancer tumor and non-tumor adjacent tissues, analyzed and
interpreted the patient data. XWZ performed the in vitro
experiments. Data analysis was performed by KYT. JXL and XY
conceived the study and finalized the manuscript. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Zhejiang Cancer Hospital and written informed consent
was provided by all patients. All procedures involving human
participants were performed in accordance with the ethical
standards of the institutional and national research committee
(Hangzhou, China; approval no. IRB-2016-134).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pencik J, Pham HT, Schmoellerl J, Javaheri
T, Schlederer M, Culig Z, Merkel O, Moriggl R, Grebien F and Kenner
L: JAK-STAT signaling in cancer: From cytokines to non-coding
genome. Cytokine. 87:26–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu Y, Zhao Q, Wang Z and Liu XY: Activated
STAT3 correlates with prognosis of non-small cell lung cancer and
indicates new anticancer strategies. Cancer Chemother Pharmacol.
75:917–922. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song Q, Liu B, Li X, Zhang Q, Cao L, Xu M,
Meng Z, Wu X and Xu K: MiR-26a-5p potentiates metastasis of human
lung cancer cells by regulating ITGβ8- JAK2/STAT3 axis. Biochem
Biophys Res Commun. 501:494–500. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilson VG: Introduction to sumoylation.
Adv Exp Med Biol. 963:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang Y, He Y, Wang X, Liang Z, He G, Zhang
P, Zhu H, Xu N and Liang S: Protein SUMOylation modification and
its associations with disease. Open Biol. 7:1701672017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang J, Chen Z, Zhou Z, Yang P and Wang
CY: Sumoylation modulates the susceptibility to type 1 diabetes.
Adv Exp Med Biol. 963:299–322. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li H, Yang Z, Pu LM, Li X, Ruan Y, Yang F,
Meng S, Yang D, Yao W, Fu H, et al: Adiponectin receptor 1 and
small ubiquitin-like modifier 4 polymorphisms are associated with
risk of coronary artery disease without diabetes. J Geriatr
Cardiol. 13:776–782. 2016.PubMed/NCBI
|
|
10
|
Alzolibani AA, Settin A, Ahmed AA, Ismail
H, Elhefni N and Robaee AA: Genetic polymorphisms of NFκB1 −94
del/ins ATTG, NFκB1A 2758 A>G and SUMO rs237025 G>A in
psoriasis. Int J Health Sci (Qassim). 9:25–33. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hou S, Kijlstra A and Yang P: The genetics
of behçet's disease in a Chinese population. Front Med. 6:354–359.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen S, Yang T, Liu F, Guo Y, Yang H, Xu
J, Song J, Zhu Z and Liu D: Inflammatory factor-specific
sumoylation regulates NF-κB signalling in glomerular cells from
diabetic rats. Inflamm Res. 63:23–31. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mo YY, Yu Y, Ee PL and Beck WT:
Overexpression of a dominant negative mutant Ubc9 is associated
with increased sensitivity to anticancer drugs. Cancer Res.
64:2793–2798. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang CY and She JX: SUMO4 and its role in
type I diabetes pathogenesis. Diabetes Metab Res Rev. 24:93–102.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen QY, Wu HH, Liang JX, XU JY and Liang
Y: Clinical significance of SUMO4 expression in papillary thyroid
carcinoma. Chin J Pathophysiology. 31:1422–1426. 2015.
|
|
16
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumours, International Union
Against Cancer. 7th. A John Wiley & Sons, Ltd., Publication;
2009, http://www.inen.sld.pe/portal/documentos/pdf/educacion/13072015_TNM%20Classification.pdf
|
|
17
|
Gillett CE: Immunohistochemistry. Methods
Mol Med. 120:191–200. 2006.PubMed/NCBI
|
|
18
|
Wilson VG and Rangasamy D: Incellular
targeting of proteins by sumoylation. Exp Cell Res. 271:57–65.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sarge KD and Park-Sarge OK: Sumo and its
role in human diseases. Int Rev Cell Mol Biol. 288:167–183. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang HJ, Zhou LL, Fu WJ, Zhang CY, Jiang
H, Du J and Hou J: β-Catenin SUMOylation is involved in the
dysregulated proliferation of myeloma cells. Am J Cancer Res.
5:309–320. 2014.PubMed/NCBI
|
|
21
|
Bellail AC, Olson JJ and Hao C: SUMO1
modification stabilizes CDK6 protein and drives the cell cycle and
glioblastoma progression. Nat Commun. 23:42342014. View Article : Google Scholar
|
|
22
|
Nestal de Moraes G, Ji Z, Fan LY, Yao S,
Zona S, Sharrocks AD and Lam EW: SUMOylation modulates
FOXK2-mediated paclitaxel sensitivity in breast cancer cells.
Oncogenesis. 7:292018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo D, Li M, Zhang Y, Yang P, Eckenrode S,
Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, et al: A
functional variant of sumo4, a new i kappa b alpha modifier, is
associated with type 1 diabetes. Nat Genet. 36:837–841. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bohren KM, Nadkarni V, Song JH, Gabbay KH
and Owerbach D: A M55V polymorphism in a novel SUMO gene (SUMO-4)
differentially activates heat shock transcription factors and is
associated with susceptibility to type I diabetes mellitus. J Biol
Chem. 279:27233–27238. 2004. View Article : Google Scholar : PubMed/NCBI
|