Introduction
Nasopharyngeal carcinoma (NPC) is a type of head and
neck squamous cell carcinoma which had an incidence of 3.26/100,000
in China in 2013 (1). Previous
studies have revealed that multiple factors synergistically affect
the origin and progression of NPC, including viruses (specifically
Epstein-Barr virus), genetic changes and environmental factors
(2–4). In addition, an increased understanding
of the underlying molecular mechanisms may lead to the development
of more effective clinical therapies.
Long non-coding RNAs (lncRNAs) lack a protein-coding
capacity, but serve crucial roles in controlling gene expression
during cell development and differentiation (5). HOXA cluster antisense RNA 2 (HOXA-AS2)
has been implicated in a variety of cancer types, such as gastric
cancer, colorectal cancer and pancreatic cancer (6–8).
However, to the best of our knowledge, there is currently no
research on HOXA-AS2-mediated NPC progression.
MicroRNAs (miRNAs/miRs) are short non-coding RNAs
with a length of ~22 nucleotides, which may act as tumor
suppressors to target important oncogenes or act as oncogenes to
target crucial tumor suppressors (9). Numerous miRNAs, such as miR-101,
miR-19, miR-127 and miR-98, have been identified to be involved in
the progression of various cancer types, such as hepatocellular
carcinoma and non-small cell lung cancer miR-519 has been
extensively investigated, particularly its interaction with ELAV
like RNA binding protein 1 expression in regulating cancer cell
progression (10–12). Moreover, Yu et al (13) reported that miR-519 suppressed NPC
cell proliferation by targeting URG4/URGCP. However, studies on
miR-519-related mechanisms in cancer, particularly NPC, are
currently lacking.
Hypoxia-inducible factor 1 (HIF-1) is a
heterodimeric protein that consists of two proteins, HIF-1α and
HIF-1β (14). HIF-1α activates the
transcription of genes that are involved in crucial aspects of
cancer biology, including angiogenesis, cell survival, glucose
metabolism and invasion (15). Xie
et al (16) found that HIF-1α
was associated with poor overall survival, poor progression-free
survival, a higher rate of lymph node metastasis and a more
advanced tumor stage in NPC. Furthermore, Cha et al
(17) observed that miR-519 could
suppress HIF-1α expression and tumor angiogenesis, which suggested
that may be a potential direct interaction between miR-519 and
HIF-1α in NPC cells.
Programmed death-ligand 1 (PD-L1) serves an
immune-suppressive role by binding to its receptor PD-1 and
blocking T-cell activation (18,19). It
was recently revealed that PD-L1 expression is closely associated
with NPC prognosis and metastasis (20,21). For
example, Zhou et al (22)
demonstrated that PD-L1 predicted poor prognosis of NPC. In
addition, Fei et al (23)
observed that PD-L1 activated epithelial-to-mesenchymal transition
in NPC cells via PI3K/AKT signaling pathway.
The aim of the present study was to investigate the
biological role of HOXA-AS2 in NPC. To the best of our knowledge,
the present study was the first to investigate the role of the
HOXA-AS2/miR-519/HIF-1α or PD-L1 axis in regulating NPC
progression, using bioinformatics analysis and relevant in
vitro experiments. The current findings may provide a basis for
the development of HOXA-AS2-targeted clinical therapy.
Materials and methods
Clinical specimens and cell lines
Under the approval of the Ethics Committee of the
Zhuji People's Hospital of Zhejiang Province, 15 paired tumor
tissues and adjacent healthy tissues were collected from 15
patients with NPC (9 males and 6 females) with a mean age of 43
years (age range, 31–59 years) between March 2012 and August 2014
from Weifang Traditional Chinese Hospital (Weifang, China) and
written informed consent was obtained from the participants. Tumor
tissues were obtained using fiberoptic nasopharyngoscopy at the
tumor growth site, and the adjacent side (2-cm away from the tumor)
with an observed normal mucosal morphology was used for the
adjacent healthy tissues. None of the patients with NPC had
received chemotherapy, immunotherapy or radiotherapy prior to
surgery.
NPC cancer cell lines (SUNE1 and SUNE2) and a normal
nasopharyngeal epithelial cell line (NP69) were purchased from BeNa
Culture Collection, as was the 293T cell line, for in vitro
experiments. The cell lines were cultured in DMEM (Invitrogen;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Thermo
Fisher Scientific, Inc.), and placed at 37°C in a humidified
incubator containing 5% CO2.
Stable cell line generation and
transfection
The short hairpin (sh)RNA specific to HOXA-AS2
(shHOXA-AS2; 5′-UCAGCUGAUGGCGUAUCCAUGAU-3′) and its negative
control (shNC; 5′-GAUUCCCCGGACUUCUCACAG-3′), miR-519-mimics
(5′-GUGAUGAAACAACCUGUACUU-3′) and its negative control (NC mimics;
5′-UACCACUGACAAUCGCUACUG-3′), and miR-519-inhibitor
(5′-ACCCUUAAUCGACGUCGGGAG-3′) and its negative control (NC
inhibitor; 5′-GAGUAGAAGUUGUAAUCUGUC-3′) were synthesized by TsingKe
Biotechnology Co., Ltd. The full length of HIF-1α or PD-L1 was
subcloned into pcDNA3.1 (Shanghai GenePharma Co., Ltd.) to
overexpress HIF-1α or PD-L1, with empty pcDNA3.1 serving as the
control. Transfection of the cells with shHOXA-AS2 (10 nM) or shNC
(10 nM) and the miR-519-mimics (10 nM) or NC mimics (10 nM) and
miR-519-inhibitor (10 nM) or NC inhibitor (10 nM) was conducted
with Lipofectamine 2000 transfection reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). All functional experiments were performed
48 h post-transfection.
Bioinformatic analysis and dual
luciferase reporter assays
StarBase version 2.0 (http://starbase.sysu.edu.cn) was used to predict the
downstream target of HOXA-AS2 or miR-519. A dual luciferase
reporter assay was used to assess the direct binding site between
HOXA-AS2 and miR-519, as well as between miR-519 and HIF-1α.
Briefly, 293T cells were transfected with the pmirGLO reporter
vectors (Promega Corporation) containing wild-type or mutant
HOXA-AS2 and 3′-untranslated region (UTR) of HIF-1α, using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
Subsequently, 293T cells were co-transfected with the pmirGLO
reporter vectors and miR-519 mimic or miR-519 inhibitor. After 48 h
incubation at 37°C with 5% CO2, luciferase activity was
evaluated using the Dual-Luciferase Reporter Analysis system
(Promega Corporation). Firefly luciferase activity was normalized
to Renilla luciferase (Promega Corporation) gene
activity.
RNA isolation, reverse
transcription-quantitative PCR (RT-qPCR) and quantification
Total RNA from tissues and cell lines was extracted
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. RT
was conducted using the Takara PrimeScript kit (Takara
Biotechnology Co., Ltd.) at 37°C for 15 min. Subsequently, the
RT-qPCR assay was performed using the ViiATM 7 Real-Time PCR system
(Thermo Fisher Scientific, Inc.) with SYBR-Green Master Mix to
detect gene expression levels. The following amplification
conditions were used: Pre-denaturation at 95°C for 15 sec, followed
by 40 cycles of denaturation at 94°C for 30 sec, annealing at 60°C
for 20 sec and extension at 72°C for 40 sec. Relative gene
expression was calculated using the 2−ΔΔCq method
(24). GAPDH and U6 were set as an
internal control. The primer sequences were as follows: HOXA-AS2
forward, 5′-CCCGTAGGAAGAACCGATGA-3′ and reverse,
5′-TTTAGGCCTTCGCAGACAGC-3′; miR-519 forward,
5′-CATGCTGTGACCCTCCAAAG-3′ and reverse,
5′-GAGAAAACAAACAGAAAGCGCT-3′; PD-L1 forward,
5′-TGCGGACTACAAGCGAATCA-3′ and reverse,
5′-GATCCACGGAAATTCTCTGGTT-3′; HIF-1α forward,
5′-ACTTGGACGCTCTGCCTATG-3′ and reverse, 5′-TTGCGGGGGTTGTAGA-3′;
GAPDH forward, 5′-GAAGAGAGAGACCCTCACGCTG-3′ and reverse,
5′-ACTGTGAGGAGGGGAGATTCAGT-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACATATACTA-3′ and reverse,
5′-ACGAATTTGCGTGTCATCCTTGCG-3′.
Western blotting
Proteins were extracted using RIPA buffer (Beyotime
Institute of Biotechnology). Protein concentration was measured
with the bicinchoninic acid assay (Beyotime Institute of
Biotechnology). Following denaturation, 10 µg protein/lane was
separated by 10% SDS-PAGE. Proteins were transferred onto PVDF
membranes and blocked in 5% non-fat milk for 2 h at room
temperature. The membranes were incubated with primary antibodies
against HIF-1α (1:1,000; cat. no. sc-13515; Santa Cruz
Biotechnology, Inc.), PD-L1 (1:1,000; cat. no. ab205921; Abcam) and
GAPDH (1:1,000; cat. no. sc-47724; Santa Cruz Biotechnology, Inc.)
overnight at 4°C. Following primary incubation, membranes were
incubated with horseradish peroxidase-conjugated secondary
antibodies (1:1,000; goat anti-mouse IgG; cat. no. ab205719 and
goat anti-rabbit IgG; cat. no. ab205718; both Abcam) for 2 h at
room temperature. Protein bands were visualized using the Pierce
ECL Western Blotting kit (Pierce; Thermo Fisher Scientific, Inc.).
Protein expression was quantified using Image-Pro® Plus
software (version 6.0; Media Cybernetics, Inc.). GAPDH was used as
an endogenous control for data normalization.
MTT assay
Transfected cells were seeded into 96-well plates at
3×103 cells/well and cultured at 37°C. Following
incubation for 0, 24, 48 and 72 h, 20 µl MTT (Sigma-Aldrich; Merck
KGaA) was added into each well and incubated for a further 4 h at
37°C according to the manufacturer's instructions. Subsequently,
the medium was removed and 150 µl dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) was added to dissolve the formazan
crystals. The absorbance was measured at 450 nm using a microplate
reader (Molecular Devices LLC).
Wound healing assay
Transfected SUNE1 and SUNE2 cells were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented
without serum at a density of 1×104 cells/ml in a
humidified atmosphere of 5% CO2 at 37°C, and grown to a
fully confluent monolayer. After 6 h, culture medium was replaced
with serum-free medium and a sterile tip was used to create a
single-line scratch. The plates were then washed twice with PBS to
remove detached cells. After 24 h, the medium was replaced with
PBS, and the wound gap was observed. Images of the migrating cells
were acquired at 0 and 24 h using a light microscope (Nikon
Corporation; magnification, ×200) and measured using ImageJ
software version 1.8 (National Institutes of Health).
Cell invasion assay
Cell invasion was determined using Transwell
chambers (8 µm pore size; EMD Millipore) precoated with 100 µl
Matrigel (BD Biosciences) for 1 h at room temperature. SUNE1 and
SUNE2 cells (1×104 cells) were added to the top chamber
containing 150 µl RPMI-1640 without FBS. An extra 550 µl RPMI-1640
medium with 10% FBS was added to the bottom chamber. After 24 h, 4%
paraformaldehyde was added to fix the cells at room temperature for
20 min, followed by staining with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) for 20 min at room temperature. The
invading cells were placed under a light microscope (Nikon
Corporation; magnification, ×200) and photographed.
Statistical analysis
The software package SPSS 22.0 (IBM Corp.) was used
for subsequent statistical analysis. Each experiment was repeated
≥3 times and the data are presented as the mean ± SD. Comparisons
between NPC tissues and adjacent healthy tissues were performed
using a paired Student's t-test, while comparisons between the
experimental and control groups were performed using an unpaired
Student's t-test. Comparisons among multiple groups were performed
using one-way ANOVA followed by Tukey's test. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-519 inhibitor rescues HOXA-AS2
knockdown-attenuated progression of NPC
The expression of HOXA-AS2 was first examined in
both tissues and cell lines. Consistent with previous studies, the
RT-qPCR results demonstrated a significantly higher expression of
HOXA-AS2 in NPC tissues compared with the adjacent healthy tissues
(Fig. 1A). The same result was also
observed in NPC cell lines (SUNE1 and SUNE2) compared with the
normal nasopharyngeal epithelial cell line (Fig. 1B). Thus, the aberrant expression
pattern between tumor and healthy specimens indicated the important
role of HOXA-AS2 in NPC progression.
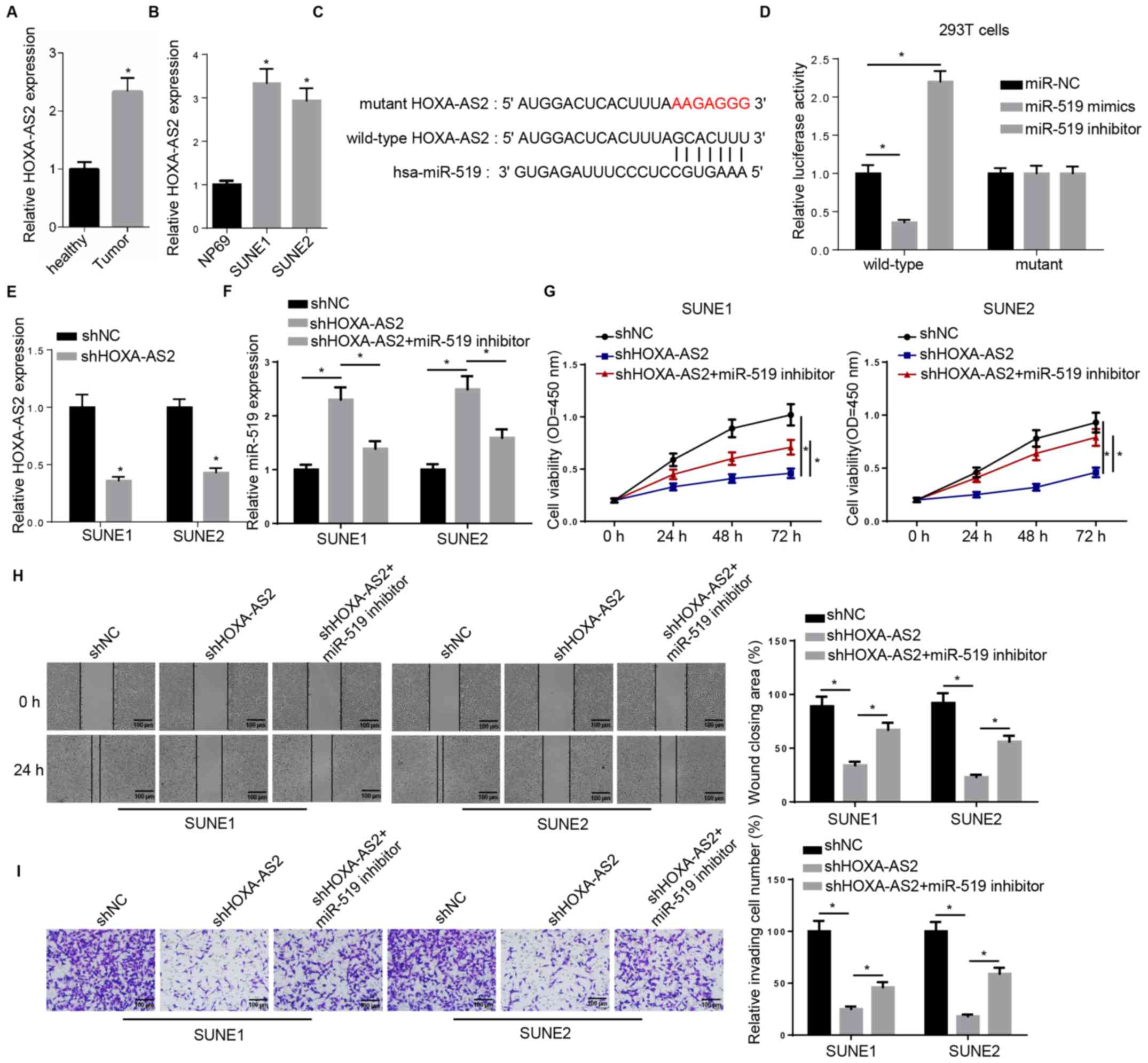 | Figure 1.miR-519 inhibitor rescues HOXA-AS2
knockdown-attenuated progression of NPC. (A) RT-qPCR analysis of
HOXA-AS2 expression in NPC tumor tissues (n=15) and the adjacent
healthy tissues (n=15). (B) RT-qPCR analysis of HOXA-AS2 expression
in the normal nasopharyngeal epithelial cell line (NP69) and the
NPC cancer cell lines (SUNE1 and SUNE2). (C) Bioinformatics
analysis identified the binding site between miR-519 and HOXA-AS2.
(D) Dual luciferase reporter assay demonstrated the direct
interaction between miR-519 and HOXA-AS2. (E) RT-qPCR analysis of
the expression of HOXA-AS2 in SUNE1 and SUNE2 cells transfected
with shNC and shHOXA-AS2. (F) RT-qPCR analysis of miR-519
expression in cells transfected with shNC, shHOXA-AS2 and
shHOXA-AS2 + miR-519 inhibitor. (G) MTT assay results of viability
of cells transfected with shNC, shHOXA-AS2 and shHOXA-AS2 + miR-519
inhibitor at 0, 24, 48 and 72 h. (H) Wound healing assay and
quantification demonstrated the migratory abilities of cells
transfected with shNC, shHOXA-AS2 and shHOXA-AS2 + miR-519
inhibitor at 0 and 24 h. (I) Transwell invasion assay and
quantification of the invasive abilities of cells transfected with
shNC, shHOXA-AS2 and shHOXA-AS2 + miR-519 inhibitor. Data are
presented as the mean ± SD. *P<0.05. miR, microRNA; NPC,
nasopharyngeal carcinoma; HOXA-AS2, HOXA cluster antisense RNA 2;
sh, short hairpin RNA; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; OD, optical density. |
starBase was used to identify miRNA-lncRNA
interactions, and it was found that there was a complementary
binding site between HOXA-AS2 and miR-519, which suggested that
miR-519 could bind to the 3′-untranslated region (UTR) of HOXA-AS2
(Fig. 1C). The wild-type and mutant
type of HOXA-AS2 were constructed and transfected into 293T cells,
and then miR-519 mimics or inhibitor were introduced into 293T
cells. Subsequently, a dual luciferase reporter assay was performed
to assess these findings. It was demonstrated that the miR-519
failed to bind to the 3′-UTR mutant type of HOXA-AS2, which did not
affect luciferase activity. However, in wild-type
HOXA-AS2-transfected 293T cells, the luciferase activity was
significantly decreased by adding exogenous miR-519 mimics to 293T
cells, whereas the luciferase activity was significantly increased
by the miR-519 inhibitor (Fig. 1D).
Collectively, bioinformatics analysis and dual luciferase reporter
assay results suggested that HOXA-AS2 could directly interact with
miR-519.
The possible impact of the interaction between
HOXA-AS2 and miR-519 on NPC progression was further investigated.
The RT-qPCR results demonstrated that HOXA-AS2 expression was
significantly downregulated in shHOXA-AS2-transfected SUNE1 and
SUNE2 cells compared with sh negative control (shNC; Fig. 1E). In addition, the co-transfection
with the miR-519 inhibitor abolished HOXA-AS2 knockdown-mediated
promoting effects on the expression of miR-519 in SUNE1 and SUNE2
cells (Fig. 1F). The MTT assay
identified that the miR-519 inhibitor partly recovered the cell
viability attenuated by shHOXA-AS2 (Fig.
1G). Moreover, co-transfection with miR-519 inhibitor
significantly enhanced the migratory and invasive abilities of
shHOXA-AS2-transfected SUNE1 and SUNE2 cells, as detected using
wound healing and Transwell assays (Fig.
1H and I).
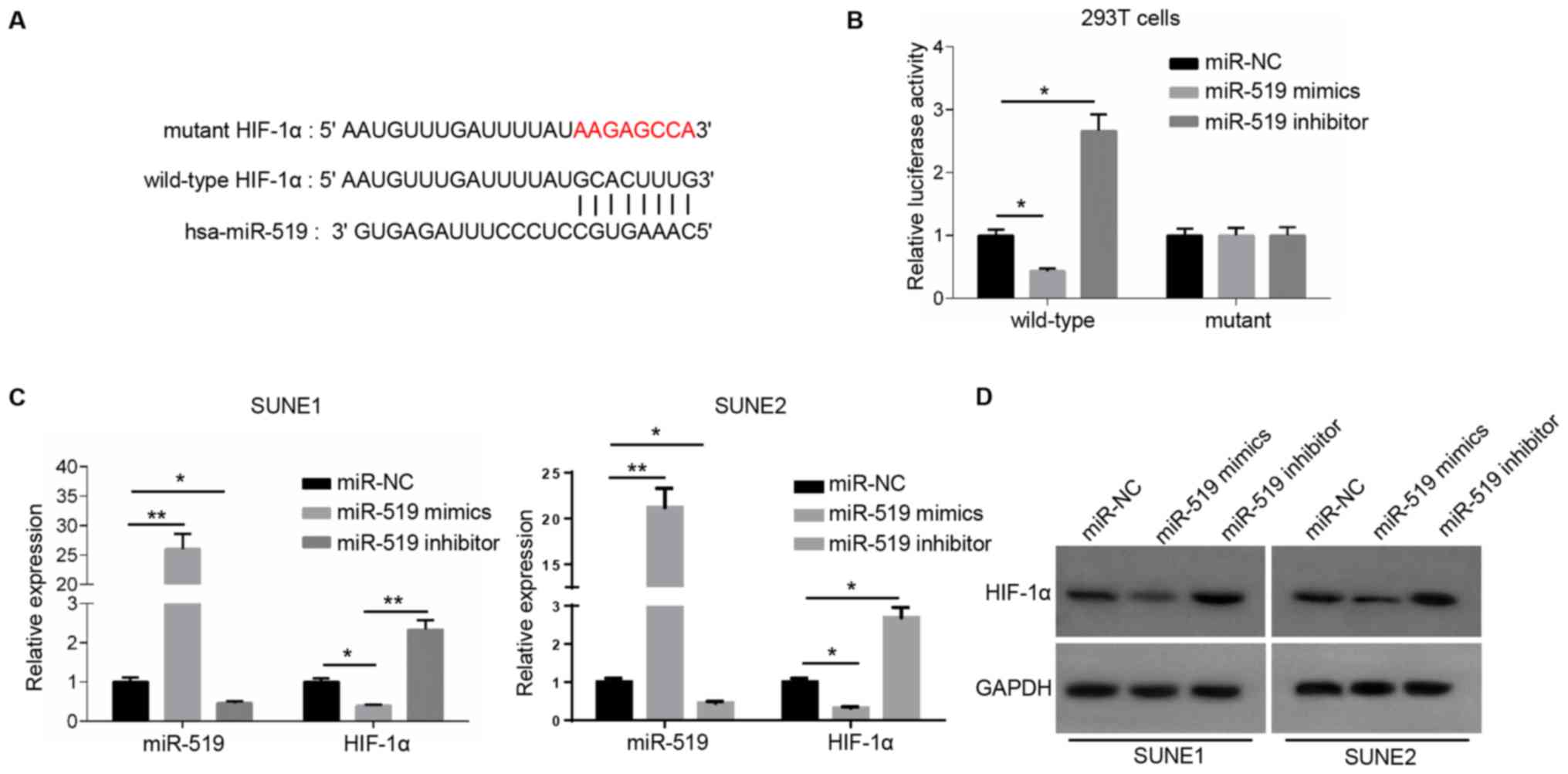
miR-519 directly binds to HIF-1α
It was demonstrated that HOXA-AS2 could directly
bind with miR-519 to regulate NPC cancer cell progression. However,
further research was required to elucidate the role of their
interaction in NPC. Therefore, the potential targets of miR-519
were also detected using starBase and dual luciferase reporter
assay to confirm their direct associations. There were a number of
potential targets (such as forkhead box Q1, PD-L1 and RCC1
domain-containing protein 1) of miR-519, and it was identified that
an important oncogene, HIF-1α, had a complementary binding site
with miR-519 (Fig. 2A). miR-519
could not bind to the mutant type of HIF-1α and did not affect the
luciferase activity, while the luciferase activity decreased
significantly by introducing miR-519 mimics into the wild-type
3′-UTR of HIF-1α-transfected 293T cells compared with miR-NC
(Fig. 2B).
Subsequently, the expression of miR-519 was
significantly increased or decreased in NPC cells transfected with
miR-519 mimics or miR-519 inhibitor (Fig. 2C). RT-qPCR analysis and western
blotting were used to examine HIF-1α expression. The results
indicated that the mRNA and protein expression levels of HIF-1α
were significantly lower in miR-519 mimics-transfected NPC cells,
and significantly higher in miR-519 inhibitor-transfected NPC cells
compared with the miR-NC (Fig. 2C and
D). Collectively, these results suggested that miR-519 was able
to directly bind to HIF-1α and significantly inhibit its
expression.
HIF-1α overexpression partially
recovers HOXA-AS2-regulated NPC progression
Next, it was examined whether HOXA-AS2 can exert its
oncogenic effects on NPC progression via the miR-519/HIF-1α axis.
RT-qPCR results demonstrated that the expression of HIF-1α was
significantly increased in SUNE1 and SUNE2 cells transfected with
HIF-1α overexpression plasmid (Fig.
3A). HIF-1α overexpression plasmid was introduced into
shHOXA-AS2-expressing SUNE1 and SUNE2 cells and the results
indicated that HIF-1α overexpression abolished the inhibitory
effect of HOXA-AS2 knockdown on the expression of HIF-1α (Fig. 3B). The MTT assay demonstrated that
overexpression of HIF-1α (shHOXA-AS2 + HIF-1α) was able to
partially recover the cell proliferative ability attenuated by
shHOXA-AS2 alone (Fig. 3C). It was
further indicated that overexpression of HIF-1α in
shHOXA-AS2-transfected cells enhanced cell migratory and invasive
abilities, which were significantly decreased by shHOXA-AS2 alone
(Fig. 3D and E). Therefore, HIF-1α
may act as a potential downstream effector for HOXA-AS2/miR-519 in
NPC cells.
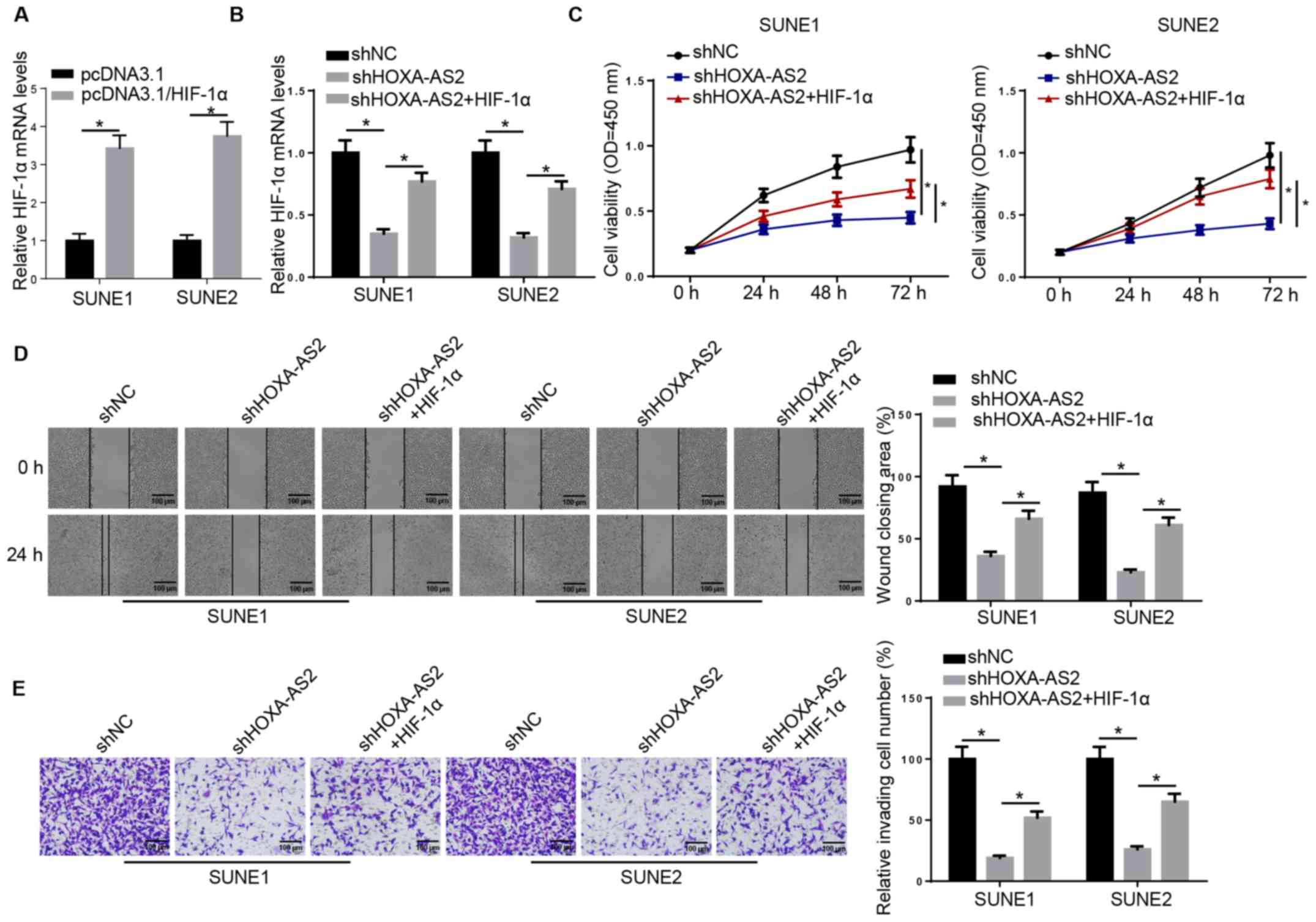 | Figure 3.HIF-1α overexpression partially
recovers HOXA-AS2-regulated nasopharyngeal carcinoma progression.
(A) RT-qPCR analysis demonstrated the expression of HIF-1α in SUNE1
and SUNE2 cells transfected with pcDNA3.1 and pcDNA3.1/HIF-1α, or
(B) transfected with shNC, shHOXA-AS2 and shHOXA-AS2 + HIF-1α. (C)
MTT assay results demonstrated the cell viability of cells
transfected with shNC, shHOXA-AS2 and shHOXA-AS2 + HIF-1α at 0, 24,
48 and 72 h. (D) Wound healing assay and quantification of
migratory abilities of cells transfected with shNC, shHOXA-AS2 and
shHOXA-AS2 + HIF-1α at 0 and 24 h. (E) Transwell invasion assay and
quantification of invasive abilities of cells transfected with
shNC, shHOXA-AS2 and shHOXA-AS2 + HIF-1α. Data are presented as the
mean ± SD. *P<0.05. HOXA-AS2, HOXA cluster antisense RNA 2; sh,
short hairpin RNA; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; OD, optical density; HIF-1α,
hypoxia-inducible factor-1α. |
PD-L1 acts as a downstream effector
for HOXA-AS2/miR-519 in NPC
miR-519 was also predicted to interact with a key
immune checkpoint, PD-L1, using the online program starBase
(Fig. 4A). Luciferase activity was
significantly downregulated by miR-519 in wild-type 3′-UTR of
PD-L1-transfected 293T cells, whereas no significant differences
were observed in the relative luciferase activity in mutant PD-L1
(Fig. 4B). Moreover, RT-qPCR was
used to examine PD-L1 expression, and found that PD-L1 mRNA
expression was negatively regulated by miR-519 (Fig. 4C).
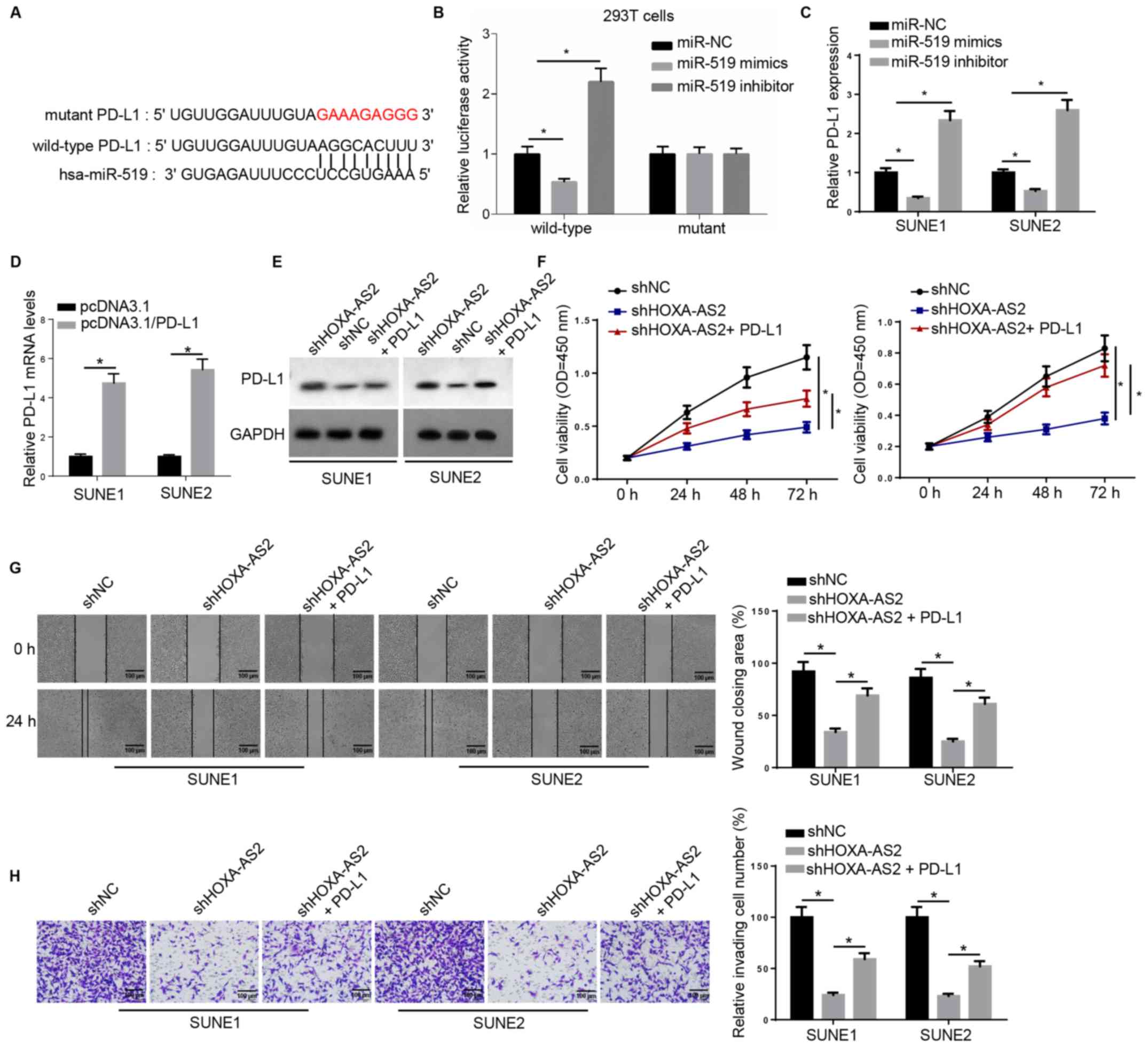 | Figure 4.PD-L1 acts as downstream effector for
HOXA-AS2/miR-519 in NPC. (A) Bioinformatics analysis identified the
binding site between miR-519 and PD-L1. (B) Dual luciferase
reporter assay of the luciferase activity of wild-type and mutant
3′-untranslated region of PD-L1 in 293T cells transfected with NC,
miR-519 mimics and miR-519 inhibitor. (C) Reverse
transcription-quantitative PCR results of PD-L1 expression in SUNE1
and SUNE2 cells transfected with NC, miR-519 mimics and miR-519
inhibitor, or (D) transfected with pcDNA3.1 and pcDNA3.1/PD-L1. (E)
Western blotting results of PD-L1 expression in cells transfected
with shNC, shHOXA-AS2 and shHOXA-AS2 + PD-L1. GAPDH was the loading
control. (F) MTT assay results of cell viability of cells
transfected with shNC, shHOXA-AS2 and shHOXA-AS2 + PD-L1 at 0, 24,
48 and 72 h. (G) Wound healing assay and quantification of
migratory abilities of cells transfected with shNC, shHOXA-AS2 and
shHOXA-AS2 + PD-L1 at 0 and 24 h. (H) Transwell invasion assay and
quantification of invasive abilities of cells transfected with
shNC, shHOXA-AS2 and shHOXA-AS2 + PD-L1. Data are presented as the
mean ± SD. *P<0.05. HOXA-AS2, HOXA cluster antisense RNA 2; sh,
short hairpin RNA; NC, negative control; OD, optical density;
PD-L1, programmed death-ligand 1. |
RT-qPCR results indicated that the expression of
PD-L1 was significantly increased in SUNE1 and SUNE2 cells
transfected with PD-L1 overexpression plasmid (Fig. 4D). Western blotting demonstrated that
PD-L1 protein expression was markedly decreased in cells with
shHOXA-AS2, while this effect was reversed by PD-L1 overexpression
(Fig. 4E). Moreover, PD-L1
overexpression significantly promoted the cell proliferative
ability that was attenuated by HOXA-AS2 knockdown (Fig. 4F). It was also identified that
overexpression of PD-L1 in shHOXA-AS2-transfected NPC cells
significantly enhanced cell migration and invasion compared with
NPC cells transfected with shHOXA-AS2 alone (Fig. 4G and H). Collectively, these findings
suggested that PD-L1 was also a critical downstream biomolecule
regulating HOXA-AS2-mediated NPC progression.
Discussion
Continued advances in genomics and transcriptomics
have accelerate the discovery and emergence of potential biomarkers
of various diseases (25).
Furthermore, molecular therapies based on these key genes or
pathways are becoming increasingly important in clinical therapy.
Compared with other common cancer types, NPC is rare but often
difficult to diagnose early (26).
While the global incidence of NPC appears to be lower compared with
that of other types of cancer, it is more frequently encountered in
Asia (27). Considering the
limitations of the current research on NPC, it is important to
study the molecular mechanisms underlying NPC initiation,
progression and metastasis in greater depth.
lncRNAs and miRNAs often exert their oncogenic or
tumor-suppressive effects on cancer cells via multiple mechanisms.
For example, HOXA-AS2 can promote the progression of breast cancer,
osteosarcoma, papillary thyroid cancer and hepatocellular carcinoma
by interacting with miR-520c (28–31). In
the present study, it was demonstrated that a high expression level
of HOXA-AS2 was associated with an increased cell viability, as
well as significantly promoted NPC cell migration and invasion; as
HOXA-AS2 knockdown reduced these effects.
miRNAs generally function in RNA silencing and
post-transcriptional regulation of gene expression, and several
miRNAs, such as miR-273, miR-127 and miR-136, are evolutionarily
conserved (32). Jalali et al
(33) revealed that lncRNAs
potentially interact with other classes of non-coding RNAs,
including miRNAs, and modulate their regulatory role via
interactions. Therefore, in the present study starBase was used to
predict the potential targets of HOXA-AS2, and miR-519 was
selected. The results identified a direct interaction between
HOXA-AS2 and miR-519. Moreover, it was found that overexpression of
miR-519 inhibited HOXA-AS2 expression, thus inhibiting NPC
progression.
HIF-1α is a well-known functional gene and it has
been extensively investigated. HIF-1α was reported to participate
in inflammation (34) and is
associated with the response to hypoxia (35). It has been previously shown that
HIF-1α is important in tumor progression and cancer therapy
(36–38). The current results suggested that
miR-519 could directly bind to and significantly inhibit the
expression of HIF-1α. In addition, the in vitro experiments
demonstrated that overexpression of HIF-1α could recover NPC cancer
cell progression attenuated by miR-519.
As well as HIF-1α, the immune checkpoint pathway
PD-L1/PD-1 was identified to be involved in HOXA-AS2- and
miR-519-mediated NPC progression in the present study. Previous
studies indicated that abnormal expression of PD-L1 was associated
with NPC prognosis and metastasis (20,21). The
current findings were in agreement with the aforementioned previous
studies. However, considering the complex mechanism involved in
tumorigenesis, other potential downstream effectors should be
investigated and in vivo experiments must be performed in
future studies.
In conclusion, the results of the present study
suggested a novel HOXA-AS2/miR-519/HIF-1α axis that may be
underlying NPC progression, and provided additional insights into
the development of HOXA-AS2-based clinical therapy. However, some
limitations remain to be further addressed: Firstly, the lack of
microdissection and immunohistochemistry is a weakness of the
study. Secondly, whether other miRNAs or downstream effectors are
crucial to HOXA-AS2-regulated phenotypes of NPC must be elucidated
in future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
SW and SY designed the present study. SW and HY
performed all the experiments, analyzed the data and prepared the
figures. SW and SY drafted the initial manuscript. SY reviewed and
revised the manuscript. All authors have read and revised the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the Zhuji People's Hospital of Zhejiang Province (approval no.
2012A0122003) and written informed consent was obtained from the
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wei KR, Zheng RS, Zhang SW, Liang ZH, Li
ZM and Chen WQ: Nasopharyngeal carcinoma incidence and mortality in
China, 2013. Chin J Cancer. 36:902017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chang ET and Adami HO: The enigmatic
epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 15:1765–1777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jia WH and Qin HD: Non-viral environmental
risk factors for nasopharyngeal carcinoma: A systematic review.
Semin Cancer Biol. 22:117–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu MC: Diet and nasopharyngeal carcinoma.
FEMS Microbiol Immunol. 2:235–242. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xie M, Sun M, Zhu YN, Xia R, Liu YW, Ding
J, Ma HW, He XZ, Zhang ZH, Liu ZJ, et al: Long noncoding RNA
HOXA-AS2 promotes gastric cancer proliferation by epigenetically
silencing P21/PLK3/DDIT3 expression. Oncotarget. 6:33587–33601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tong G, Wu X, Cheng B, Li L, Li X, Li Z,
Nong Q, Chen X, Liu Y and Wang S: Knockdown of HOXA-AS2 suppresses
proliferation and induces apoptosis in colorectal cancer. Am J
Transl Res. 9:4545–4552. 2017.PubMed/NCBI
|
|
8
|
Lian Y, Li Z, Fan Y, Huang Q, Chen J, Liu
W, Xiao C and Xu H: The lncRNA-HOXA-AS2/EZH2/LSD1 oncogene complex
promotes cell proliferation in pancreatic cancer. Am J Transl Res.
9:5496–5506. 2017.PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ristimäki A: Tumor suppressor effect of
the microRNA miR-519 is mediated via the mRNA-binding protein HuR.
Cell Cycle. 9:12342010. View Article : Google Scholar
|
|
11
|
Abdelmohsen K, Kim MM, Srikantan S,
Mercken EM, Brennan SE, Wilson GM, de Cabo R and Gorospe M: miR-519
suppresses tumor growth by reducing HuR levels. Cell Cycle.
9:1354–1359. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abdelmohsen K, Srikantan S, Kuwano Y and
Gorospe M: miR-519 reduces cell proliferation by lowering
RNA-binding protein HuR levels. Proc Natl Acad Sci USA.
105:20297–20302. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu G, Zhang T, Jing Y, Bao Q, Tang Q and
Zhang Y: miR-519 suppresses nasopharyngeal carcinoma cell
proliferation by targeting oncogene URG4/URGCP. Life Sci.
175:47–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Semenza GL: Hypoxia-inducible factor 1
(HIF-1) pathway. Sci STKE. 2007:cm82007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie W, Liu L, He H and Yang K: Prognostic
value of hypoxia-inducible factor-1 alpha in nasopharyngeal
carcinoma: A meta-analysis. Int J Biol Markers. Jun 10–2018.(Epub
ahead of print). doi: doi.org/10.1177/1724600818778756. View Article : Google Scholar
|
|
17
|
Cha ST, Chen PS, Johansson G, Chu CY, Wang
MY, Jeng YM, Yu SL, Chen JS, Chang KJ, Jee SH, et al: MicroRNA-519c
suppresses hypoxia-inducible factor-1alpha expression and tumor
angiogenesis. Cancer Res. 70:2675–2678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Giatromanolaki A, Koukourakis IM, Balaska
K, Mitrakas AG, Harris AL and Koukourakis MI: Programmed death-1
receptor (PD-1) and PD-ligand-1 (PD-L1) expression in non-small
cell lung cancer and the immune-suppressive effect of anaerobic
glycolysis. Med Oncol. 36:762019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang Z, Tian Y, Cai W, Weng Z and Li Y,
Zhang H, Bao Y and Li Y: High-affinity human PD-L1 variants
attenuate the suppression of T cell activation. Oncotarget.
8:88360–88375. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang ZL, Liu S, Wang GN, Zheng SH, Ding
SR, Tao YL, Chen C, Liu SR, Yang X, Chang H, et al: The prognostic
significance of PD-L1 and PD-1 expression in patients with
nasopharyngeal carcinoma: A systematic review and meta-analysis.
Cancer Cell Int. 19:1412019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao Y, Chan KI, Xiao G, Chen Y, Qiu X, Hao
H, Mak SC and Lin T: Expression and clinical significance of PD-L1
and BRAF expression in nasopharyngeal carcinoma. BMC Cancer.
19:10222019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou Y, Shi D, Miao J, Wu H, Chen J, Zhou
X, Hu D, Zhao C, Deng W and Xie C: PD-L1 predicts poor prognosis
for nasopharyngeal carcinoma irrespective of PD-1 and EBV-DNA load.
Sci Rep. 7:436272017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fei Z, Deng Z, Zhou L, Li K, Xia X and Xie
R: PD-L1 induces epithelial-mesenchymal transition in
nasopharyngeal carcinoma cells through activation of the PI3K/AKT
pathway. Oncol Res. 27:801–807. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ilyas M: Next-generation sequencing in
diagnostic pathology. Pathobiology. 84:292–305. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luftig M: Heavy LIFting: Tumor promotion
and radioresistance in NPC. J Clin Invest. 123:4999–5001. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang Y, Wang J, Wu F, Song Y, Zhao S and
Zhang Q: Long non-coding RNA HOXA-AS2 promotes proliferation and
invasion of breast cancer by acting as a miR-520c-3p sponge.
Oncotarget. 8:46090–46103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Zhang R, Cheng G, Xu R and Han X:
Long non-coding RNA HOXA-AS2 promotes migration and invasion by
acting as a ceRNA of miR-520c-3p in osteosarcoma cells. Cell Cycle.
17:1637–1648. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xia F, Chen Y, Jiang B, Du X, Peng Y, Wang
W, Huang W, Feng T and Li X: Long noncoding RNA HOXA-AS2 promotes
papillary thyroid cancer progression by regulating
miR-520c-3p/S100A4 pathway. Cell Physiol Biochem. 50:1659–1672.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Xu J, Zhang S, An J, Zhang J,
Huang J and Jin Y: HOXA-AS2 promotes proliferation and induces
epithelial-mesenchymal transition via the miR-520c-3p/GPC3 axis in
hepatocellular carcinoma. Cell Physiol Biochem. 50:2124–2138. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jalali S, Bhartiya D, Lalwani MK,
Sivasubbu S and Scaria V: Systematic transcriptome wide analysis of
lncRNA-miRNA interactions. PLoS One. 8:e538232013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cramer T, Yamanishi Y, Clausen BE, Förster
I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V,
et al: HIF-1alpha is essential for myeloid cell-mediated
inflammation. Cell. 112:645–657. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rius J, Guma M, Schachtrup C, Akassoglou
K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG and Karin M:
NF-kappaB links innate immunity to the hypoxic response through
transcriptional regulation of HIF-1alpha. Nature. 453:807–811.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
LaGory EL and Giaccia AJ: The
ever-expanding role of HIF in tumour and stromal biology. Nat Cell
Biol. 18:356–365. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Unwith S, Zhao H, Hennah L and Ma D: The
potential role of HIF on tumour progression and dissemination. Int
J Cancer. 136:2491–2503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu T, Tang B and Sun X: Development of
inhibitors targeting hypoxia-inducible factor 1 and 2 for cancer
therapy. Yonsei Med J. 58:489–496. 2017. View Article : Google Scholar : PubMed/NCBI
|