Introduction
Pancreatic cancer, particularly pancreatic ductal
adenocarcinoma (PDAC) is extremely malignant, with a 5-year overall
survival rate of only ~9% (1). The
symptoms of pancreatic cancer are not clear in the early stages,
and most patients present with local invasion or distant metastasis
at the time of diagnosis. Therefore, the surgical resection rate is
low, with a previous study reporting a resection rate of 10–20%
(2). Patients with pancreatic cancer
often experience metastasis and recurrence following surgery, and
the prognosis is poor (3). Clinical
data have demonstrated that pancreatic cancer remains incurable and
that traditional chemoradiotherapy cannot prolong the survival of
patients (3–5). In the past, tumor cells were the main
subject of tumor research, particularly in studies investigating
the role of genes and signaling pathways in tumorigenesis and tumor
progression; these studies were used to find potential therapeutic
targets for pancreatic cancer, but no major targets have been
identified (6). Recent studies have
reported that the tumor microenvironment serves an important role
in tumorigenesis and development (7,8). These
studies have suggested that the malignant biological behavior of
tumors may be regulated by the tumor microenvironment (9–11).
Immunotherapy based on the tumor microenvironment is a feasible
treatment method; therefore, it is necessary to investigate the
immune microenvironment of pancreatic cancer and its influencing
factors.
The components of the tumor microenvironment include
tumor cells, interstitial cells, immune cells and cell regulatory
factors secreted by cells in the microenvironment or secreted by
other cells and transported to the microenvironment; the most
important of these components may be tumor immune cells, including
B lymphocytes, T lymphocytes, neutrophils, macrophages, dendritic
cells, and other immune cells that infiltrate tumor tissues
(7,8). Previous studies have shown that the
tumor microenvironment of pancreatic cancer is extremely complex
(12,13). Tang et al (14) have reported that the level of
CD8+ T lymphocyte infiltration in pancreatic tumor
tissues is lower compared with that in adjacent normal tissues.
Treg cells, which highly infiltrate tumor tissues, inhibit the
antitumor immunity of CD8+ T lymphocytes, which may be
associated with tumor immune escape (14). In another study, Liu et al
(15) reported that in 92 patients
with pancreatic cancer, the number of infiltrating CD4+
and CD8+ T lymphocytes in the adjacent normal tissues of
pancreatic cancer was higher compared with that in the tumor
tissues, but CD4+ FOXP3+ Treg cells were more
abundant in the tumor tissues. The survival analysis showed that
high infiltration of CD8+ T lymphocytes in adjacent
normal tissues and low infiltration of CD4+ T cells in
tumor tissues were good independent prognostic factors (15). Therefore, we hypothesized that immune
cell infiltration is an important part of the tumor
microenvironment.
Tumors are considered to be ‘organs’ composed of
different types of cells (7).
Compared with normal tissues, the information interactions between
cells in the tumor microenvironment are more complicated (8). A previous study has demonstrated that
these interactions are associated with the occurrence and
progression of tumors (16).
Chemokines are important mediators of interactions between
different types of cells in the tumor microenvironment (17). Chemokines not only act as chemotactic
molecules for targeted migration, but also serve an important
regulatory role in various physiological and pathological processes
of the cell (including infection, wound repair, inflammatory
response, tumorigenesis and invasion) (18). Chemokines in the tumor
microenvironment may act on tumor cells and other stromal cell
components, including neutrophils, macrophages, lymphocytes and
fibroblasts, to promote or inhibit tumor progression (19,20).
Furthermore, there may also be negative chemokines that support a
tumor microenvironment with no T lymphocyte infiltration (21). A lack of T lymphocyte infiltration in
the microenvironment may lead to a poor prognosis (22).
In the present study, chemokines or chemokine
receptors were screened to identify those differentially expressed
in pancreatic cancer compared with normal controls and associated
with patient prognosis.
Materials and methods
Public datasets from the gene
expression omnibus (GEO) database
The gene expression microarray and corresponding
clinical data of patients with pancreatic cancer were obtained from
the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The dataset
numbers were GSE56560 (23,24) and GSE28735 (25,26).
Screening of differentially expressed
(DE) chemokines and chemokine receptors
Initially, analysis of DE genes between pancreatic
tumor tissues and normal tissues was performed using the ‘limma’
package (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
in R (27,28), according to the screening criteria
(adjusted P≤0.10 and |log fold-change|≥1.5). DE chemokines and DE
chemokine receptors were determined by overlapping the DE genes
with a dataset containing all chemokines and their receptors; this
analysis was performed using the ‘VennDiagram’ package in R
(29).
Survival analysis of the TCGA
dataset
The survival analysis was performed on the Gene
Expression Profiling Interactive Analysis website (GEPIA;
http://gepia.cancer-pku.cn/). The data
for 178 patients with pancreatic cancer analyzed on the GEPIA
website were obtained from The Cancer Genome Atlas (TCGA,
http://cancergenome.nih.gov/).
Patients and tissue microarray
Two commercial tissue microarray (TMA) chips
consisting of 290 points were used. Of these, there were 128 pairs
of intra-tumoral tissues and adjacent peri-tumoral normal tissues
(Shanghai Outdo Biotech, Shanghai, China). All patients from the
TMA underwent surgical treatment. If the tumor was located in the
head of the pancreas, pancreaticoduodenectomy was performed. If the
tumor was located in the tail of the pancreas, pancreatic body tail
resection was performed. All patients had a surgical pathological
diagnosis. Clinicopathological data included pathological type,
operation time, survival status, follow-up time, survival time,
tissue code, tumor organ, sex, age, patient's main complaint, tumor
history, family tumor history, distant metastasis site, whether it
was a primary organ or metastatic, pathological classification,
pathological grade, tumor size, tumor site, description of the
tumor, texture of the tumor, tumor capsular invasion, pathological
morphology, tumor boundary, vascular invasion, lymph node invasion,
number of total and invaded lymph nodes, T stage, N stage, M stage
and American Joint Committee on Cancer stage (30). All patients received conventional
chemotherapy following surgery and were followed up regularly. The
follow-up information came from outpatient follow-up review or
telephone follow-up. Overall survival (OS) time refers to the time
between surgery and death, with OS as a prognostic indicator of
survival.
Immunohistochemical staining and
interpretation
Immunohistochemical staining was performed on tissue
sections deparaffinized in xylene with an UltraSensitive SP
Mouse/Rabbit IHC kit (cat. no. KIT-9720; Maxim Biotech, Inc.)
according to the manufacturer's instructions and an IHC Biotin
Block kit (cat. no. BLK-0002; Maxim Biotech, Inc.) by an automated
immunostainer (cat. no. ST5010; Leica Microsystems, Inc.). The
protocol for the IHC Biotin Block kit is as follows: After the
antigen retrieval, the PBS was removed, Avidin solution (reagent A,
50 µl) was added dropwise to the TMA and incubated at room
temperature for 10 min. Subsequently, the TMA was rinsed twice with
PBS (3 min per rinse); the PBS was removed, and d-Biotin Solution
(reagent B, 50 µl) was added dropwise to the TMA and incubate for
10 min at room temperature, followed by further two rinses with PBS
(3 min per rinse). An anti-human CXCL5 monoclonal antibody (1:50;
cat. no. MAB 254-100; R&D Systems) was used for analysis. A
fully automatic digital pathology slice scanner (Aperio; Leica
Microsystems, Inc.) was used to scan the immunohistochemically
stained TMA chips to obtain a digital image. The microscopic images
were imported as digital photo files analyzed using QuantCenter in
Pannoramic Viewer software 1.15.4 (3DHISTECH, Ltd.). Following
preliminary observation, the unsatisfactory points of TMA in the
sectioning and staining images were eliminated. TMA images were
analyzed by two independent pathologists who were blinded to the
patient's clinical data. Histochemistry score (H-SCORE) was used to
evaluate the expression of CXCL5. H-SCORE is a scoring method for
tissue immunohistochemical results that reflects the positive ratio
and the positive intensity. The following formula was used:
H-SCORE=∑ (PI × I)=(percentage of cells of weak intensity ×1) +
(percentage of cells of moderate intensity ×2) + (percentage of
cells of strong intensity ×3). In the formula, PI represents the
percentage of positive cells to the total number of cells in this
position and I represents the intensity of staining. The H-SCORE is
0–300, with a higher score representing stronger positive staining.
The H-SCORE for each patient was calculated, and all patients were
divided into two groups (low and high CXCL5) according to the
cut-off value. The H-SCORE cut-off value was determined using
X-tile software version 3.6.1 (Yale University), which determines
the cut-off value by calculating the P-value corresponding to each
possible cut-off value; the cut-off corresponding to the lowest
P-value is considered the best. The calculations revealed that the
best cut-off value for the H-SCORE was 76.83.
Statistical analysis
In the present study, Student's t-test was used for
comparing quantitative data, and the χ2 test, Fisher's
exact test or likelihood ratio test were used for qualitative data.
The Kaplan-Meier method was used to generate survival curves, and
the log-rank method was used to compare the differences in survival
rates between the groups. Multivariate analysis was performed using
the Cox multivariate regression analysis model. The
clinicopathological factors with significant associations
(P<0.1) in the aforementioned Kaplan-Meier survival analysis
were subjected to Cox multivariate analysis. The statistical and
mapping software used in the present study were SPSS 22.0 (IBM
Corp.), GraphPad Prism 6.02 (GraphPad Software, Inc.), Adobe
Photoshop CS5, and Adobe Illustrator CS6 (both from Adobe Systems,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
CXCL5 is differentially expressed in
two datasets from the GEO database
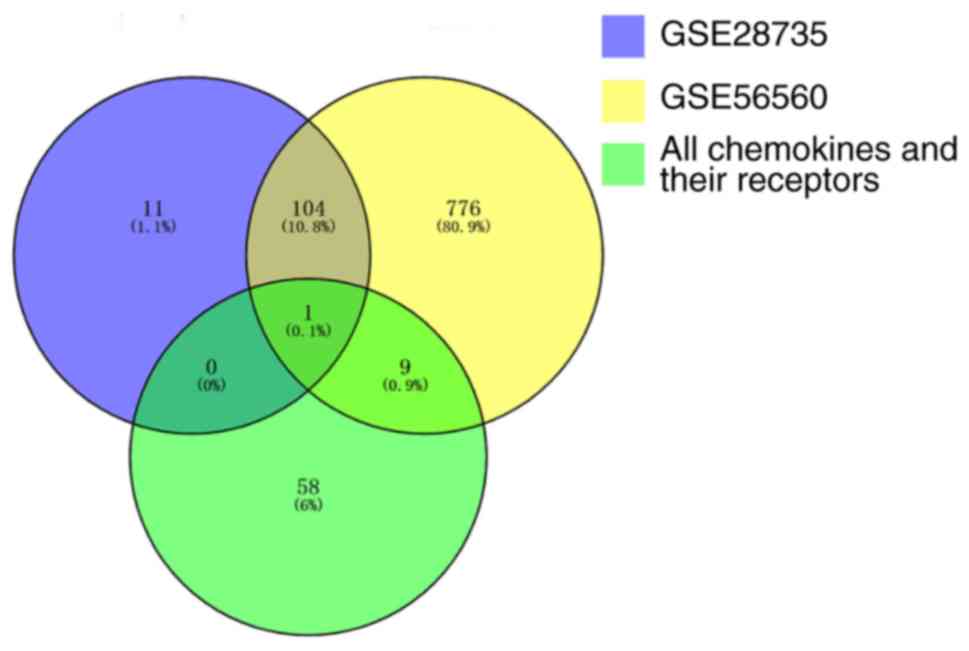
A total of 890 DE genes were identified between the
pancreatic tumor tissues and the normal tissues in the GSE56560
dataset, and 116 DE genes were identified in the GSE28735 dataset.
A total of 10 DE chemokines or DE chemokine receptors were
identified in the GSE56560 dataset, and only one (CXCL5) was
identified in the GSE28735 dataset. Therefore, CXCL5 was selected
for further analysis (Fig. 1).
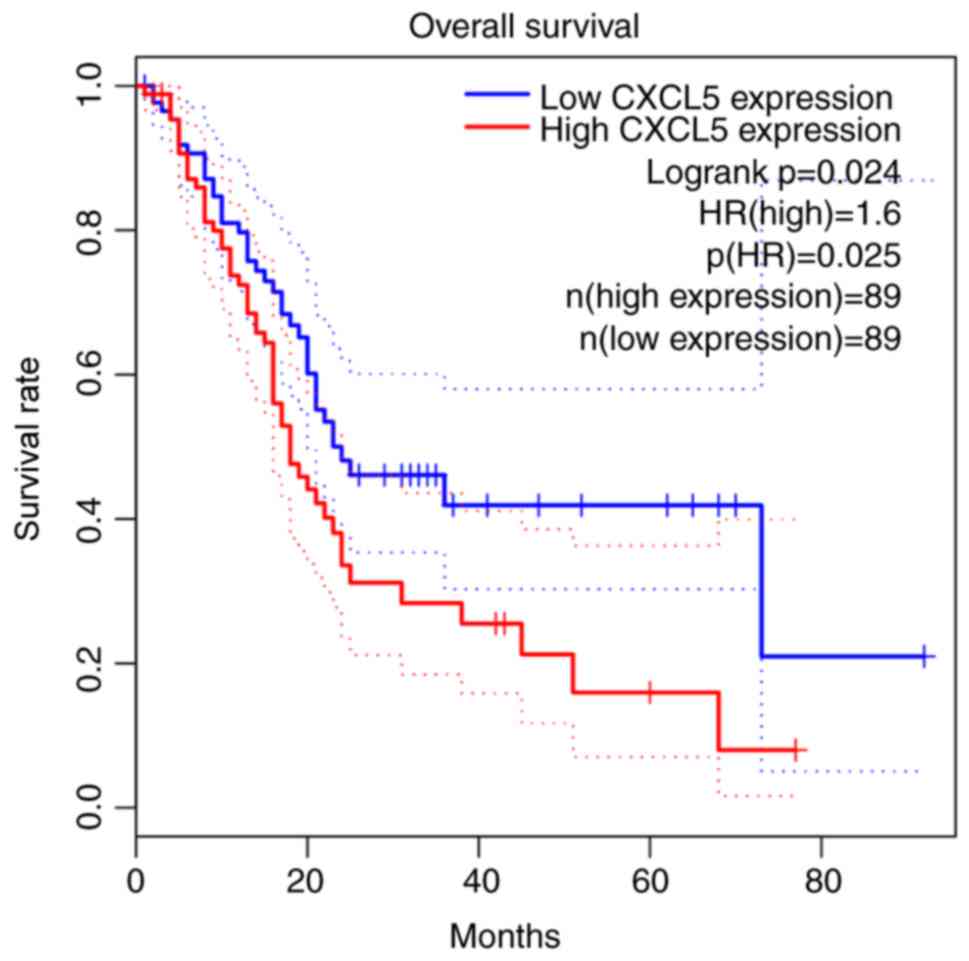
Patients with pancreatic cancer with
high CXCL5 expression in TCGA dataset have a poor prognosis
A total of 198 patients from TCGA dataset were
included in the present study; they were divided into groups of 89
patients with low CXCL5 expression and 89 patients with high CXCL5
expression according to the median value of CXCL5 expression. The
prognosis of the patients in the low CXCL5 group was significantly
improved compared with that of the patients in the high CXCL5 group
(P=0.024; Fig. 2).
Characteristics of 119 patients from
TMA dataset
Following removal of unsatisfactory points of TMA
chips in the sectioning and staining images, a total of 119
patients with pancreatic cancer were included in the study. Samples
of tumor and corresponding peritumoral healthy tissue were
collected from each patient. Table I
presents the clinicopathological characteristics of the patients
with pancreatic cancer.
 | Table I.Patient demographics and
clinicopathological factors. |
Table I.
Patient demographics and
clinicopathological factors.
| Factors | No. of
patients |
|---|
| Age, years |
|
|
<70 | 81 |
|
≥70 | 38 |
| Sex |
|
|
Male | 71 |
|
Female | 48 |
| Grade |
|
| I | 15 |
| II | 59 |
|
III | 45 |
| IV | 0 |
| Tumor size, cm |
|
| ≤4 | 76 |
|
>4 | 43 |
| Tumor site |
|
|
Head | 72 |
|
Other | 47 |
| Vascular
invasion |
|
| No | 64 |
|
Yes | 55 |
| T |
|
| T1 | 4 |
| T2 | 72 |
| T3 | 43 |
| T4 | 0 |
| N |
|
| N0 | 63 |
| N1 | 45 |
| N2 | 11 |
| M |
|
| M0 | 87 |
| M1 | 32 |
| TNM
stagea |
|
| I | 39 |
| II | 63 |
|
III | 11 |
| IV | 6 |
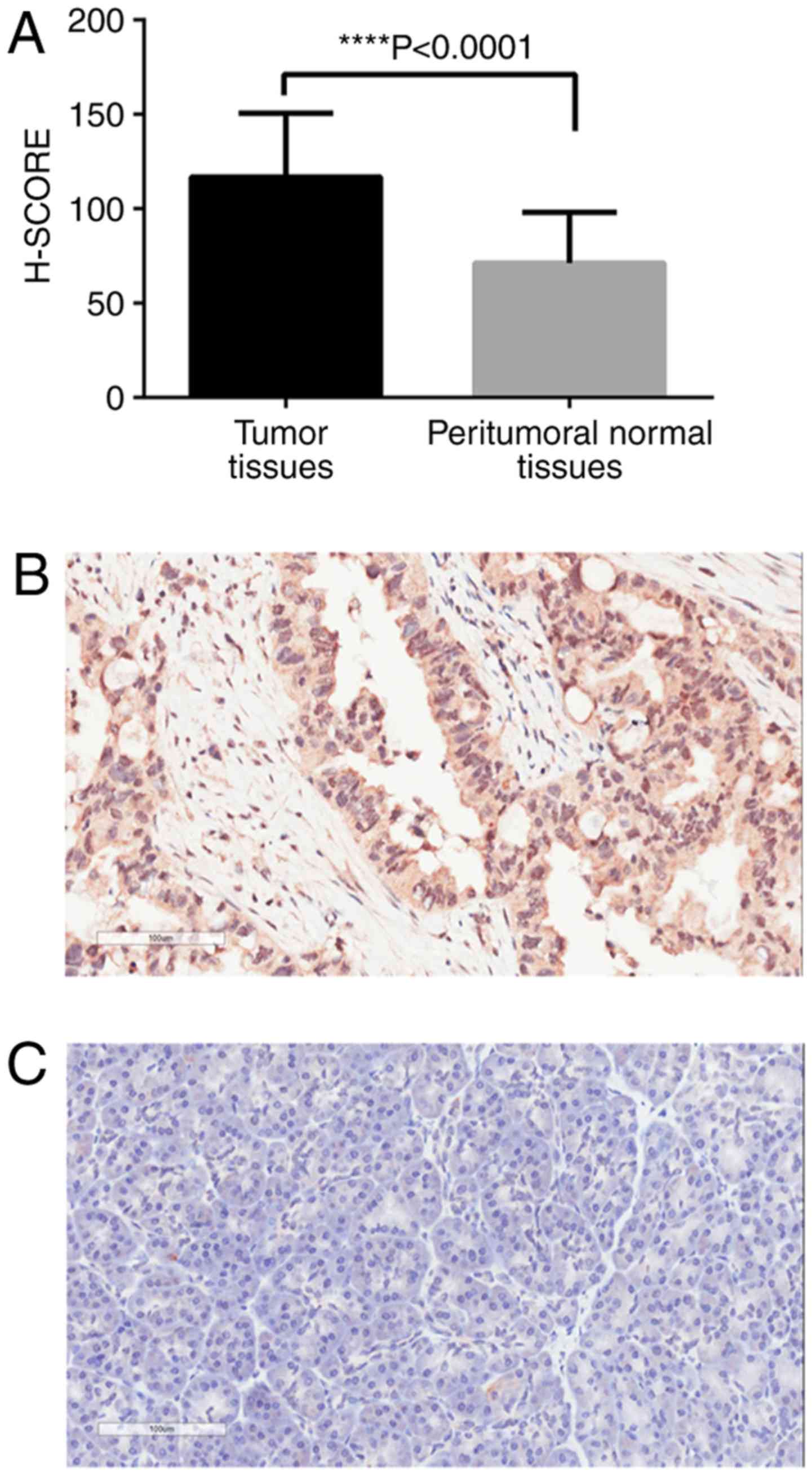
Expression of CXCL5 in the TMA
dataset
The mean H-SCORE of CXCL5 expression in pancreatic
cancer tissues in the TMA was 94.58, and the 95% confidence
interval (CI) was 89.35–99.82. The H-SCORE in adjacent peri-tumoral
normal tissues was 57.14, and the 95% CI was 54.58–59.69. The
expression level of CXCL5 in tumor tissues was significantly higher
than that in adjacent peritumoral healthy tissues, and the
difference was statistically significant (P<0.0001; Fig. 3A). Typical staining is presented in
Fig. 3B (pancreatic cancer tissues)
and C (adjacent normal tissues).
Association between CXCL5 expression
and clinicopathological factors
There was no significant association between the
expression of CXCL5 in pancreatic cancer tumor tissues and sex,
age, tumor site, tumor site, vascular invasion, histological grade,
T stage, N stage, or M stage (P>0.05; Table II).
 | Table II.Associations between CXCL5 and
clinicopathological factors. |
Table II.
Associations between CXCL5 and
clinicopathological factors.
|
|
| Expression level of
CXCL5 |
|
|---|
|
|
|
|
|
|---|
| Factors | No. of
patients | Low (%) | High (%) | P-value |
|---|
| Sex |
|
|
| 0.969 |
|
Male | 71 | 19 (27) | 52 (73) |
|
|
Female | 48 | 13 (27) | 35 (73) |
|
| Age, years |
|
|
| 0.589 |
|
<70 | 81 | 23 (28) | 58 (72) |
|
|
≥70 | 38 | 9 (24) | 29 (76) |
|
| Tumor site |
|
|
| 0.879 |
|
Head | 72 | 19 (26) | 53 (74) |
|
|
Other | 47 | 13 (26) | 34 (74) |
|
| VI |
|
|
| 0.116 |
|
Yes | 64 | 21 (33) | 43 (67) |
|
| No | 55 | 11 (18) | 44 (82) |
|
| Grade |
|
|
| 0.421a |
| 1 | 15 | 6 (40) | 9 (60) |
|
| 2 | 59 | 16 (27) | 43 (73) |
|
| 3 | 45 | 10 (22) | 35 (78) |
|
| T |
|
|
| 0.139 |
|
T1+T2 | 76 | 17 (22) | 59 (78) |
|
|
T3+T4 | 43 | 15 (35) | 28 (65) |
|
| N |
|
|
| 0.981 |
| N0 | 63 | 17 (27) | 46 (73) |
|
|
N1+N2 | 56 | 15 (27) | 41 (73) |
|
| M |
|
|
| 0.854 |
| M0 | 87 | 23 (26) | 64 (74) |
|
| M1 | 32 | 9 (28) | 23 (72) |
|
| TNM stage |
|
|
| 0.800 |
|
I+II | 102 | 27 (26) | 75 (74) |
|
|
III+IV | 17 | 5 (29) | 12 (71) |
|
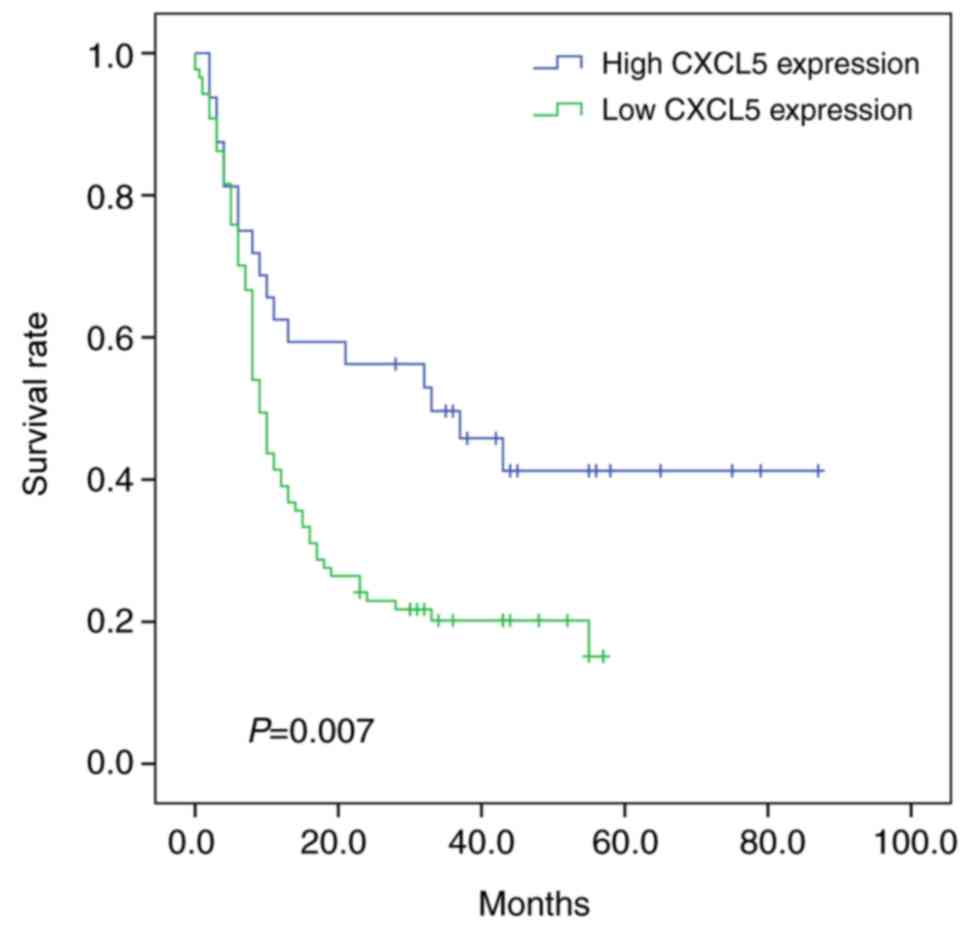
Patients with pancreatic cancer with
high CXCL5 expression had a poor prognosis: Analysis based on the
TMA dataset
OS was used as the prognostic indicator, and
Kaplan-Meier survival analysis showed that the prognosis of
patients in the CXCL5 high-expression group was significantly worse
than that of those in the low-expression group (P=0.007; Fig. 4). The median survival time of
patients in the high-expression group was 9 months with a 95% CI of
7.6–10.4 months, while the median survival time of patients in the
low-expression group was 33 months with a 95% CI of 7.4–58.6
months.
CXCL5 is an unfavorable independent
prognostic factor in patients with pancreatic cancer
Kaplan-Meier survival analysis showed that CXCL5 was
significantly associated with the OS of patients with pancreatic
cancer. It predicted a poor prognosis in patients with pancreatic
cancer (P=0.007). Among the clinicopathological factors, age
(P=0.031), histological grade (P=0.007), N stage (P=0.004) and
Tumor-Node-Metastasis (TNM) stage (P=0.002) were significantly
associated with poor overall survival (P<0.05). Since the N
stage and the M stage are factors related to TNM stage, only TNM
stage was included in the multifactor regression analysis. Cox
multivariate regression analysis showed that age (P=0.003),
histological grade (P=0.006), TNM stage (P=0.002) and CXCL5
(P=0.011) were independent prognostic factors for patients with
pancreatic cancer (Table III).
Therefore, CXCL5 is an independent factor for a poor prognosis in
patients with pancreatic cancer. The hazards ratio (95% CI) of
CXCL5 was 2.003 (1.176–3.412), indicating that in patients with
pancreatic cancer, the mortality risk of those with a high
expression of CXCL5 in tumor tissues is ~2-fold higher compared
with that of those with a low expression.
 | Table III.Univariate and multivariate analyses
of CXCL5 as a prognostic factor. |
Table III.
Univariate and multivariate analyses
of CXCL5 as a prognostic factor.
|
| Univariate
analysis | Cox analysis |
|---|
|
|
|
|
|---|
| Factors | MST | P-value | HR | 95% CI | P-value |
|---|
| Sex
(Male/female) | 10/10 | 0.785 |
|
|
|
| Age, years
(<70/≥70) | 14/8 | 0.031 | 1.993 | 1.260–3.152 | 0.003 |
| Grade
(I/II/III) | 13/8 | 0.007 | 1.844 | 1.191–2.856 | 0.006 |
| Tumor site
(Head/other) | 9/12 | 0.218 |
|
|
|
| Vascular invasion
(No/yes) | 10/10 | 0.729 |
|
|
|
| T
(T1+T2/T3+T4) | 10/11 | 0.220 |
|
|
|
| N (N0/N1/N2) | 15/10/8 | 0.004 |
|
|
|
| M (M0/M1) | 12/8 | 0.055 |
|
|
|
| TNM stage
(I+II/III+IV) | 13/8 | 0.002 | 2.401 | 1.372–4.200 | 0.002 |
| CXCL5
(Low/high) | 33/9 | 0.007 | 2.003 | 1.176–3.412 | 0.011 |
Discussion
There are a number of dysregulated chemokines and
chemokine receptors in tumor tissues, which serve a role in a
variety of cells and are the key regulatory molecules for
tumorigenesis, metastasis and immune escape (17). Chemokines and their receptors
interact in a variety of non-specific ways, which constitute the
multiple ligand and receptor axes (31). They may cause the cell to move
directionally and have a regulatory function in a variety of
cellular pathology processes and physiological processes (32). With regards to the role of chemokines
in the immune system, chemokines are usually divided into
inflammatory chemokines and endoenvironmental stability chemokines
(33). The latter type of chemokine
maintains the stability of the internal environment and is often
expressed in the lymphoid homing site. Inflammatory chemokines may
recruit immune effector cells to induce an inflammatory
microenvironment and are often expressed under pathological
conditions in which homeostasis is disrupted (33).
A recent study reported that certain chemokines
serve an important role in tumorigenesis; they serve an important
role in the migration, invasion, proliferation, chemotherapy
resistance, vascular and lymphangiogenesis, distant metastasis and
growth of stromal cells in pancreatic cancer (34). Previous studies have reported that
the axes of certain chemokines and their receptors not only
participate in the regulation of signaling pathways but also
regulate the differentiation and infiltration of T cells, serving a
key molecular role in tumor immune escape (19,20). The
CXCL12-CXCR4 axis has been identified to have immunosuppressive
effects in tumors. Studies have demonstrated that the CXCL12-CXCR4
action axis is associated with a poor prognosis in patients with
lung (35), esophageal (36), gastric (37), pancreatic (38), ovarian cancer (39) and other cancer types (40).
Feig et al (41) reported that CXCL12, a chemokine
secreted by cancer-associated fibroblasts (CAFs) in pancreatic
cancer, may spread and coat the surface of tumor cells, and then
CXCL12 on the surface of tumor cells and mesenchymal CAFs binds to
CXCR4, a chemokine receptor on the surface of T lymphocytes,
leading to the clearance of T lymphocytes from tumor tissues. Karin
and Wildbaum (20) have proposed
that the CXCL12-CXCR4 axis inhibits the cellular immune response
and may induce Th0 cells to differentiate into regulatory T cells
(Tr1). At present, there are a variety of inhibitors that may
target the CXCL12-CXCR4 axis, including AMD3100 (42), TN14003 (43), and CTCE-9908 (44); therefore, cancer immunotherapy based
on cytokines is promising.
CXCL5 is also termed epithelial cell-derived
neutrophil-activating protein-78 (45). Studies have reported that CXCL5 has
strong effects on granulocyte chemotaxis and angiogenesis (46,47). The
target cells of CXCL5 include neutrophils and CXCR2+
monocytes of CXCR2+ myeloid-derived suppressor cells
(MDSCs) and tumor-associated receptors (48). Therefore, the role of the CXCL5-CXCR2
axis in tumors is also diverse. In colorectal cancer, the
preoperative serum CXCL5 level may be used as a novel prognostic
marker and is associated with the liver metastasis of colorectal
cancer (49). In bone metastatic
tumors of prostate cancer, CXCL5 mediates inflammatory cell
infiltration and accelerates the growth of metastatic tumors
(50). Gao et al (51) reported that the CXCL5-CXCR2 action
axis leads to bladder cancer invasion and metastasis by activating
the PI3K/AKT signaling pathway to upregulate MMP2/MMP9, suggesting
that CXCL5 may be a potential therapeutic target for bladder cancer
(51). In thyroid studies, CXCL5 was
found to promote the migration and invasion of thyroid cancer
cells, affecting tumor epithelial-mesenchymal transition, but not
proliferation, suggesting that CXCL5 may affect the
microenvironment of thyroid cancer (1). However, in another study, Cui et
al (52) suggested that the
CXCL5-CXCR2 axis promotes the proliferation of thyroid cancer cells
through the P38 and JNK signaling pathways and promotes the
progression of the cell cycle from the G1 phase to the S phase
(52). Soler-Cardona et al
(53) found that the lymph node
metastasis rate was significantly increased in melanoma with high
expression of CXCL5 compared with melanoma with low expression of
CXCL5; this may be associated with the fact that CXCL5 recruits
PD-1-expressing neutrophils into the tumor microenvironment and
interferes with the activation of antitumor immunity (53,54). In
other studies, CXCL5 was demonstrated to recruit MDSCs expressing
CXCR2 to promote the progression of colon cancer (48) and to promote tumor angiogenesis by
increasing the expression of FOXD1 (55). A previous study suggested that high
expression of CXCL5 in colorectal cancer is not sufficient to prove
it as a diagnostic or prognostic tumor marker (56), but it is considered a potential
biomarker and therapeutic target in lung cancer (57,58). A
meta-analysis involving 15 studies showed that CXCL5 was an adverse
prognostic biomarker in various tumors (59). CXC5 has also been shown to inhibit
tumor progression in colon cancer and kidney cancer, which
contradicts the aforementioned conclusions (60). At present, there are few studies on
CXCL5 in the field of pancreatic cancer. By analyzing three
pancreatic cancer data sets from the GEO database, Gu et al
(61) obtained 25 core genes,
including CXCL5, the functional enrichment of which was
demonstrated to be associated with the cell cycle of pancreatic
cancer cells, but no further study was conducted on CXCL5 (61).
The present study investigated the significance of
CXCL5 in pancreatic cancer. Initially, potential chemokines
associated with the prognosis of patients with pancreatic cancer
were searched for in the GEO and TCGA databases. CXCL5 was found to
be highly expressed in pancreatic cancer tissues and was associated
with a poor prognosis. Furthermore, the expression of CXCL5 in the
tumor tissues and adjacent peritumoral normal tissues of 119
patients with pancreatic cancer was analyzed by
immunohistochemistry. The present study reported that the
expression of CXCL5 in pancreatic cancer tumor tissues is higher
than that in adjacent peri-tumoral normal tissues, suggesting that
CXCL5 is involved in certain pancreatic cancer biological
processes. Finally, Kaplan-Meier survival analysis and Cox
multivariate analysis revealed that CXCL5 is an independent
prognostic factor in patients with pancreatic cancer. High
expression of CXCL5 in tumor tissues predicts a poor prognosis and
may be a potential biomarker and therapeutic target for pancreatic
cancer.
The chemokine, CXCL5, is highly expressed in
pancreatic cancer tissues, and high CXCL5 expression is associated
with a poor prognosis in patients with pancreatic cancer. The
association between CXCL5 and tumor-infiltrating lymphocytes
requires investigation in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Social
Development Research and Demonstration Application Project of the
Bureau of Science and Technology of Jiaxing City (grant no.
2019AY32025) and the Medical and Health Science and Technology
Project from the Health Committee of Provincial Zhejiang (grant no.
2020KY951).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available in The Cancer Genome Atlas (cancergenome.nih.gov), the Gene Expression Ominibus
database (ncbi.nlm.nih.gov/gds) and GEPIA (Gene
Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn). Tissue microarray and the
corresponding data were obtained from ShanghaiOutdo Biotech, China
and Shanghai National Engineering Research Centre for Biochips.
Authors' contributions
BW, ZZ and YS designed the present study. BW and MZ
performed the bioinformatics analysis. FC performed the
immunohistochemical staining. XW and JW performed the analysis of
the immunohistochemical staining and wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
1. Siegel RL, Miller KD and Jemal A:
Cancer statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saif MW: Pancreatic neoplasm in 2012: An
update. Tissue is an issue. JOP. 13:124–127. 2012.PubMed/NCBI
|
|
3
|
Ansari D, Tingstedt B, Andersson B,
Holmquist F, Sturesson C, Williamsson C, Sasor A, Borg D, Bauden M
and Andersson R: Pancreatic cancer: Yesterday, today and tomorrow.
Future Oncol. 12:1929–1946. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin QJ, Yang F, Jin C and Fu DL: Current
status and progress of pancreatic cancer in China. World J
Gastroenterol. 21:7988–8003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aroldi F, Bertocchi P, Rosso E, Prochilo T
and Zaniboni A: Pancreatic cancer: Promises and failures of target
therapies. Rev Recent Clin Trials. 11:33–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu T and Dai Y: Tumor microenvironment and
therapeutic response. Cancer Lett. 387:61–68. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Frankel T, Lanfranca MP and Zou W: The
role of tumor microenvironment in cancer immunotherapy. Adv Exp Med
Biol. 1036:51–64. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki
M, Kosuge T, Kanai Y and Hiraoka N: Immune cell infiltration as an
indicator of the immune microenvironment of pancreatic cancer. Br J
Cancer. 108:914–923. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hinshaw DC and Shevde LA: The tumor
microenvironment innately modulates cancer progression. Cancer Res.
79:4557–4566. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu J, Chen J, Feng Y, Tian H and Chen X:
Tumor microenvironment as the ‘regulator’ and ‘target’ for gene
therapy. J Gene Med. 21:e30882019. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ho WJ, Jaffee EM and Zheng L: The tumour
microenvironment in pancreatic cancer-clinical challenges and
opportunities. Nat Rev Clin Oncol. 17:527–540. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dougan SK: The pancreatic cancer
microenvironment. Cancer J. 23:321–325. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Y, Xu X, Guo S, Zhang C, Tang Y, Tian
Y, Ni B, Lu B and Wang H: An increased abundance of
tumor-infiltrating regulatory T cells is correlated with the
progression and prognosis of pancreatic ductal adenocarcinoma. PLoS
One. 9:e915512014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu L, Zhao G, Wu W, Rong Y, Jin D, Wang
D, Lou W and Qin X: Low intratumoral regulatory T cells and high
peritumoral CD8(+) T cells relate to long-term survival in patients
with pancreatic ductal adenocarcinoma after pancreatectomy. Cancer
Immunol Immunother. 65:73–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lacalle RA, Blanco R, Carmona-Rodríguez L,
Martín-Leal A, Mira E and Mañes S: Chemokine receptor signaling and
the hallmarks of cancer. Int Rev Cell Mol Biol. 331:181–244. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bian X, Xiao YT, Wu T, Yao M, Du L, Ren S
and Wang J: Microvesicles and chemokines in tumor microenvironment:
Mediators of intercellular communications in tumor progression. Mol
Cancer. 18:502019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan KW, Evrard M, Tham M, Hong M, Huang C,
Kato M, Prevost-Blondel A, Donnadieu E, Ng LG and Abastado JP:
Tumor stroma and chemokines control T-cell migration into melanoma
following Temozolomide treatment. Oncoimmunology. 4:e9787092015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karin N and Wildbaum G: The role of
chemokines in shaping the balance between CD4(+) T Cell subsets and
its therapeutic implications in autoimmune and cancer diseases.
Front Immunol. 6:6092015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng W, Xue S and Chen Y: The role of
CXCL12 in tumor microenvironment. Gene. 641:105–110. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang J, Wang YF, Wu B, Zhong ZX, Wang KX,
Yang LQ, Wang YQ, Li YQ, Gao J and Li ZS: Intraepithelial attack
rather than intratumorally infiltration of CD8+T lymphocytes is a
favorable prognostic indicator in pancreatic ductal adenocarcinoma.
Curr Mol Med. 17:689–698. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Haider S, Wang J, Nagano A, Desai A,
Arumugam P, Dumartin L, Fitzgibbon J, Hagemann T, Marshall JF,
Kocher HM, et al: A multi-gene signature predicts outcome in
patients with pancreatic ductal adenocarcinoma. Genome Med.
6:1052014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Dumartin L, Mafficini A, Ulug P,
Sangaralingam A, Alamiry NA, Radon TP, Salvia R, Lawlor RT, Lemoine
NR, et al: Splice variants as novel targets in pancreatic ductal
adenocarcinoma. Sci Rep. 7:29802017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang G, He P, Tan H, Budhu A, Gaedcke J,
Ghadimi BM, Ried T, Yfantis HG, Lee DH, Maitra A, et al:
Integration of metabolomics and transcriptomics revealed a fatty
acid network exerting growth inhibitory effects in human pancreatic
cancer. Clin Cancer Res. 19:4983–4993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang G, Schetter A, He P, Funamizu N,
Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, et al:
DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity
and predicts clinical outcome in pancreatic ductal adenocarcinoma.
PLoS One. 7:e315072012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang J, Wang Y, Luo Y, Fu J, Zhang Y, Li
Y, Xiao Z, Lou Y, Qiu Y and Zhu F: Computational advances of tumor
marker selection and sample classification in cancer proteomics.
Comput Struct Biotechnol J. 18:2012–2025. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
R Core Team R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna, Austria: 2014, http://www.R-project.org/
|
|
29
|
Chen H and Boutros PC: VennDiagram: A
package for the generation of highly-customizable Venn and Euler
diagrams in R. BMC Bioinformatics. 12:352011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Allen PJ, Kuk D, Castillo CF, Basturk O,
Wolfgang CL, Cameron JL, Lillemoe KD, Ferrone CR, Morales-Oyarvide
V, He J, et al: Multi-institutional validation study of the
American joint commission on cancer (8th edition) changes for T and
N staging in patients with pancreatic adenocarcinoma. Ann Surg.
265:185–191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Legler DF and Thelen M: Chemokines:
Chemistry, biochemistry and biological function. Chimia (Aarau).
70:856–859. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luster AD: Chemokines-chemotactic
cytokines that mediate inflammation. N Engl J Med. 338:436–445.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zlotnik A and Yoshie O: The chemokine
superfamily revisited. Immunity. 36:705–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu PF, Lu ZP, Cai BB, Tian L, Zou C, Jiang
KR and Miao Y: Role of CXCL12/CXCR4 signaling axis in pancreatic
cancer. Chin Med J (Engl). 126:3371–3374. 2013.PubMed/NCBI
|
|
35
|
Wald O, Shapira OM and Izhar U:
CXCR4/CXCL12 axis in non small cell lung cancer (NSCLC) pathologic
roles and therapeutic potential. Theranostics. 3:26–33. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Goto M and Liu M: Chemokines and their
receptors as biomarkers in esophageal cancer. Esophagus.
17:113–121. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee HJ and Jo DY: The role of the
CXCR4/CXCL12 axis and its clinical implications in gastric cancer.
Histol Histopathol. 27:1155–1161. 2012.PubMed/NCBI
|
|
38
|
Sleightholm RL, Neilsen BK, Li J, Steele
MM, Singh RK, Hollingsworth MA and Oupicky D: Emerging roles of the
CXCL12/CXCR4 axis in pancreatic cancer progression and therapy.
Pharmacol Ther. 179:158–170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Salomonnson E, Stacer AC, Ehrlich A, Luker
KE and Luker GD: Imaging CXCL12-CXCR4 signaling in ovarian cancer
therapy. PLoS One. 8:e515002013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Samarendra H, Jones K, Petrinic T, Silva
MA, Reddy S, Soonawalla Z and Gordon-Weeks A: A meta-analysis of
CXCL12 expression for cancer prognosis. Br J Cancer. 117:124–135.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Feig C, Jones JO, Kraman M, Wells RJ,
Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL,
et al: Targeting CXCL12 from FAP-expressing carcinoma-associated
fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic
cancer. Proc Natl Acad Sci USA. 110:20212–20217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 348:124–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Beider K, Begin M, Abraham M, Wald H,
Weiss ID, Wald O, Pikarsky E, Zeira E, Eizenberg O, Galun E, et al:
CXCR4 antagonist 4F-benzoyl-TN14003 inhibits leukemia and multiple
myeloma tumor growth. Exp Hematol. 39:282–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wong D, Kandagatla P, Korz W and Chinni
SR: Targeting CXCR4 with CTCE-9908 inhibits prostate tumor
metastasis. BMC Urol. 14:122014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Okabe H, Beppu T, Ueda M, Hayashi H,
Ishiko T, Masuda T, Otao R, Horlad H, Mima K, Miyake K, et al:
Identification of CXCL5/ENA-78 as a factor involved in the
interaction between cholangiocarcinoma cells and cancer-associated
fibroblasts. Int J Cancer. 131:2234–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Walz A, Burgener R, Car B, Baggiolini M,
Kunkel SL and Strieter RM: Structure and neutrophil-activating
properties of a novel inflammatory peptide (ENA-78) with homology
to interleukin 8. J Exp Med. 174:1355–1362. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Keane MP, Belperio JA, Xue YY, Burdick MD
and Strieter RM: Depletion of CXCR2 inhibits tumor growth and
angiogenesis in a murine model of lung cancer. J Immunol.
172:2853–2860. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Katoh H, Wang D, Daikoku T, Sun H, Dey SK
and Dubois RN: CXCR2-expressing myeloid-derived suppressor cells
are essential to promote colitis-associated tumorigenesis. Cancer
Cell. 24:631–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kawamura M, Toiyama Y, Tanaka K, Saigusa
S, Okugawa Y, Hiro J, Uchida K, Mohri Y, Inoue Y and Kusunoki M:
CXCL5, a promoter of cell proliferation, migration and invasion, is
a novel serum prognostic marker in patients with colorectal cancer.
Eur J Cancer. 48:2244–2251. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Roca H, Jones JD, Purica MC, Weidner S,
Koh AJ, Kuo R, Wilkinson JE, Wang Y, Daignault-Newton S, Pienta KJ,
et al: Apoptosis-induced CXCL5 accelerates inflammation and growth
of prostate tumor metastases in bone. J Clin Invest. 128:248–266.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao Y, Guan Z, Chen J, Xie H, Yang Z, Fan
J, Wang X and Li L: CXCL5/CXCR2 axis promotes bladder cancer cell
migration and invasion by activating PI3K/AKT-induced upregulation
of MMP2/MMP9. Int J Oncol. 47:690–700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cui D, Zhao Y and Xu J: Activation of
CXCL5-CXCR2 axis promotes proliferation and accelerates G1 to S
phase transition of papillary thyroid carcinoma cells and activates
JNK and p38 pathways. Cancer Biol Ther. 20:608–616. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Soler-Cardona A, Forsthuber A, Lipp K,
Ebersberger S, Heinz M, Schossleitner K, Buchberger E, Gröger M,
Petzelbauer P, Hoeller C, et al: CXCL5 facilitates melanoma
cell-neutrophil interaction and lymph node metastasis. J Invest
Dermatol. 138:1627–1635. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Forsthuber A, Lipp K, Andersen L,
Ebersberger S, Graña-Castro O, Ellmeier W, Petzelbauer P,
Lichtenberger BM and Loewe R: CXCL5 as regulator of neutrophil
function in cutaneous melanoma. J Invest Dermatol. 139:186–194.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen C, Xu ZQ, Zong YP, Ou BC, Shen XH,
Feng H, Zheng MH, Zhao JK and Lu AG: CXCL5 induces tumor
angiogenesis via enhancing the expression of FOXD1 mediated by the
AKT/NF-κB pathway in colorectal cancer. Cell Death Dis. 10:1782019.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yildirim K, Colak E, Aktimur R, Gun S,
Taskin MH, Nigdelioglu A, Aktimur SH, Karagöz F and Ozlem N:
Clinical value of CXCL5 for determining of colorectal cancer. Asian
Pac J Cancer Prev. 19:2481–2484. 2018.PubMed/NCBI
|
|
57
|
Wang L, Shi L, Gu J, Zhan C, Xi J, Ding J
and Ge D: CXCL5 regulation of proliferation and migration in human
non-small cell lung cancer cells. J Physiol Biochem. 74:313–324.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu K, Yu S, Liu Q, Bai X, Zheng X and Wu
K: The clinical significance of CXCL5 in non-small cell lung
cancer. Onco Targets Ther. 10:5561–5573. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hu B, Fan H, Lv X, Chen S and Shao Z:
Prognostic significance of CXCL5 expression in cancer patients: A
meta-analysis. Cancer Cell Int. 18:682018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Speetjens FM, Kuppen PJ, Sandel MH, Menon
AG, Burg D, van de Velde CJ, Tollenaar RA, de Bont HJ and
Nagelkerke JF: Disrupted expression of CXCL5 in colorectal cancer
is associated with rapid tumor formation in rats and poor prognosis
in patients. Clin Cancer Res. 14:2276–2284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gu Y, Feng Q, Liu H, Zhou Q, Hu A,
Yamaguchi T, Xia S and Kobayashi H: Bioinformatic evidences and
analysis of putative biomarkers in pancreatic ductal
adenocarcinoma. Heliyon. 5:e023782019. View Article : Google Scholar : PubMed/NCBI
|