Introduction
In terms of incidence and mortality rate, lung
cancer ranks first among all types of cancer globally, with <20%
of patients surviving <5 years after diagnosis, in 2017
(1). There are two forms of lung
cancer: Non-small cell lung cancer (NSCLC) and small cell lung
cancer (2). NSCLC is further
subdivided into lung adenocarcinoma (LUAD), squamous cell carcinoma
and large cell carcinoma (3).
Adenocarcinoma accounts for the largest proportion of cases, and
its incidence has been increasing over the last 10 years, worldwide
(4).
The primary treatments available for patients with
lung cancer include surgery, chemo- and radiotherapy, molecular
targeted therapy and immunotherapy. In the past few decades,
researchers have improved our understanding of the role of the
immune system in cancer development, and thus, immunotherapy has
improved the field of tumor treatment. Of late, checkpoint
inhibitors have been developed for the treatment of lung cancer
(5,6); blockade of immune checkpoint proteins,
including programmed death-1 (PD-1)/programmed death-ligand1
(PD-L1) and cytotoxic T-lymphocyte-associated protein 4, has shown
promise in the treatment of several types of cancer, reducing tumor
burden and prolonging the survival time of patients (7).
By comprehensively exploring the prognostic value of
immune-related genes (IRGS), a recent study assessed individualized
immune characteristics to improve the prognoses of patients with
NSCLC (8). Previous studies have
reported that tumor-infiltrating B cells are closely associated
with a more favorable prognosis in NSCLC, cervical cancer and
breast cancer (9–11). Nielsen et al (11) reported that CD20+
tumor-infiltrating lymphocytes (TILs) colocalized with activated
CD8+ TILs expressed markers of antigen presentation. The
group proposed that the association between CD20+ TILs
and patient survival may reflect a supportive role in cytolytic
immune responses.
By investigating survival associated immune-related
genes, the present study aimed to elucidate the underlying
molecular mechanisms of immune genes in LUAD, with a view to
establish therapeutic targets and provide a basis for personalized
treatment.
Materials and methods
Patients
In total, 10 pairs of LUAD and adjacent normal
tissues were obtained from patients with LUAD (4 men and 6 women;
median age, 55 years; age range, 33–69 years), undergoing surgery
at the Jining Cancer Hospital (Jining, China) from November 2018 to
March 2019. None of the patients had received chemo- or
radiotherapy prior to surgery. The present study was approved by
the Medical Ethics Committees of Jining Cancer Hospital, and
written informed consent was provided by all patients prior to
surgery. A total of 497 patients were assessed from The Cancer
Genome Atlas (TCGA) database (cancer.gov/tcga), including 229 men and 268 women;
median age, 66 years; age range, 33–88 years.
Data acquisition and processing
The LUAD dataset (12) containing transcriptome RNA-sequencing
and clinical data of patients with LUAD was downloaded from TCGA
database. A total of 497 LUAD tissues and 54 normal lung tissues
were included in the present study. The list of IRGs was downloaded
from the Immunology Database and Analysis Portal (ImmPort) database
(13). The inclusion criteria were
as follows: Patients with histologically or cytologically confirmed
lung adenocarcinoma and patients with complete clinical
information. The exclusion criteria were as follows: Patients with
histologically or cytologically confirmed cancer other than lung
adenocarcinoma and patients with OS time <10 days.
Identification of differentially
expressed genes (DEGs), differentially expressed immune-related
genes (DEIRGs) and survival-associated immune related genes
(IRGs)
DEGs were identified using the edgeR package
(version 3.53) in Rand further analyzed. A |log2 fold
change| >2.0 and false discovery rate adjusted to P<0.01 were
set as the thresholds (14). In
addition, volcano and heat maps of the DEGs were constructed using
the gplots and heat map components of the
edgeR package, respectively. DEIRGs were obtained by
comparison with the immune gene lists. Survival-associated IRGs
were selected using univariate Cox regression analysis, which was
performed using the survival package in R.
Functional enrichment analysis
To understand the underlying biological mechanisms
of the IRGs, Gene Ontology (GO) annotation and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway analyses were performed using
The Database for Annotation, Visualization and Integrated Discovery
(david.ncifcrf.gov/) online tool
(15) and cluster profiler, an R
package for functional classification and enrichment of gene
clusters using the hypergeometric distribution (16,17). The
results of the GO and KEGG analyses were displayed using the GOplot
package in R, and analyses were based on a threshold of
P<0.01.
Development of the immune-related gene
prognostic model (IRGPM)
Overall survival time was measured from the date of
diagnosis to mortality or the last clinical evaluation.
Survival-associated IRGs were selected via univariate Cox
regression analysis using the R survival package. Using
multivariate Cox regression analysis via the Akaike Information
Criterion (18), patients with LUAD
were then divided into high-risk and low-risk groups according to
the median risks core value. The risk score was calculated using
the following formula:
Survival Risk
Score(SRS)=∑i=1k(Ci×Vi)
Where k represents the number of mRNAs,
Ci represents the coefficient of mRNA in
multivariate Cox regression analysis and Vi
represents the mRNA expression level. Kaplan-Meier plots were used
to divide patients into high and low risk score groups, according
to OS time.
Relationship between IRGPM and immune
cell infiltration
The Tumor Immune Estimation Resource (TIMER) online
database analyzes and creates a visualization of tumor infiltrating
immune cells (19). TIMER reanalyzes
gene expression data, which includes 10,897 samples across 32
cancer types from TCGA, to estimate the abundance of six subtypes
of tumor-infiltrating immune cells, including CD4 T cells, CD8 T
cells, B cells, macrophages, dendritic cells (DCs) and neutrophils.
Thus, TIMER can easily be used to determine the relationship
between immune cell infiltration and other parameters. Data
regarding immune infiltration levels among patients with LUAD were
obtained, and the association between IRGPM and immune cell
infiltration was assessed.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was obtained from the LUAD and
corresponding adjacent normal tissues of 10 patients using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and then reverse transcribed into cDNA using the First Strand
cDNA Synthesis kit (New England Biolabs, Inc.), according to the
manufacturer's protocol. PCR amplification was performed with a
SYBR Green PCR kit (ABM, Inc.), according to the manufacturer's
protocol, using the Applied Biosystems 7500Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer
sequences used for qPCR are presented in Table I. The following thermocycling
conditions were used for qPCR: Initial denaturation of 95°C for 10
min; 40 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30
sec; and a final extension at 75°C for 7 min. Relative mRNA
expression levels were measured using the 2−ΔΔCq method
(20) and normalized to the internal
reference gene GAPDH. All experiments were performed in
triplicate.
 | Table I.Primer sequences for reverse
transcription quantitative-PCR. |
Table I.
Primer sequences for reverse
transcription quantitative-PCR.
| Primer | Sequence,
5′→3′ |
|---|
| IL11 |
|
|
Forward |
GTGGCCAAGATACAGCTGTCGC |
|
Reverse |
GGTAGGACAGTAGGTCCGCTC |
| LGR4 |
|
|
Forward |
TCCACCTGGAAAGTCTGA |
|
Reverse |
GGTTAGATTTGATTACGCTGT |
| CRABP1 |
|
|
Forward |
ATTCTCGAGCCACCATGCCCAACTTC |
|
Reverse | ACAGGATCCC
TGCCTTCACTCTCGG |
| GAPDH |
|
|
Forward |
CAACGAATTTGGCTACAGCA |
|
Reverse |
AGGGGTCTACATGGCAACTG |
Statistical analysis
Survival analysis of data from patients in the
prognostic model was performed using the R survival package.
Survival curves were generated using the Kaplan-Meier method and a
log-rank test was used to compare the differences between the two
groups. To validate the performance of the prognostic signature,
the area under the survival receiver operating characteristic (ROC)
curve was calculated using the R survival ROC package (21). The expression levels of genes between
different groups were evaluated using the unpaired Student's t
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of DEGs
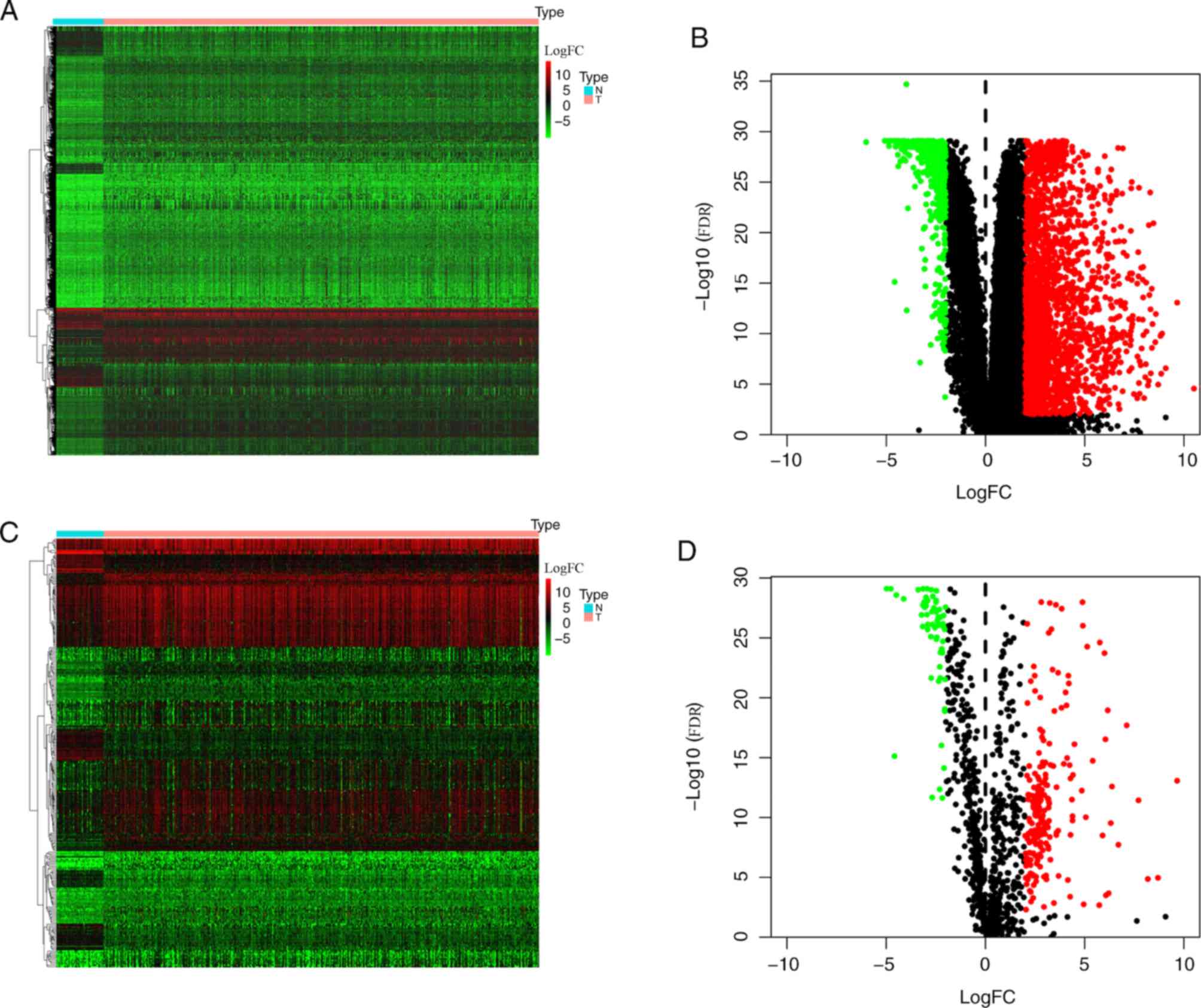
Using the edgeR package, 2,672 DEGs were identified
in patients with LUAD, 2,191 and 481 of which were upregulated and
downregulated, respectively (Fig. 1A and
B). Upon further comparison with immune gene lists from the
ImmPort database, 273 DEIRGs were identified, 210 and 63 of which
were up-and downregulated, respectively (Fig. 1C and D).
Construction of the prognostic
model
The median gene expression was set as the cut-off
value to divide all genes into two groups, high expression group
and low expression group. Univariate analysis was used to identify
survival-associated IRGS. The results demonstrated that high
expression levels of: S100P [hazard ratio (HR), 1.218; 95%
confidence interval (CI), 1.578–2.018; P=0.005), CPABP1 (HR, 1.343;
95% CI, 1.659–2.106; P=0.005), BIRC5 (HR, 1.645; 95% CI,
1.388–1.951; P=0.002), IGKV4-1 (HR, 1.665; 95% CI, 1.322–2.098;
P=0.009), IL11 (HR, 1.728; 95% CI, 1.349–2.636; P<0.001), INHA
(HR, 1.226; 95% CI, 0.972–2.265; P=0.004), INSL4 (HR, 1.978; 95%
CI, 1.493–2.872; P=0.007) and LGR4 (HR, 1.678; 95% CI, 1.433–2.172;
P=0.001) were associated with worse OS compared with the low
expression group. Conversely, low expression levels of: ADRB2 (HR,
0.711; 95% CI, 0.553–0.712; P=0.004) and VIPR1 (HR, 0.651; 95% CI,
0.413–0.732; P<0.001) were associated with worse OS compared
with the high expression group (Table
II). Based on multivariate Cox regression analysis of
survival-associated IRGs, a prognostic model was constructed which
divided the patients into two groups (high risk score group and low
risk score), according to OS time and the median risk score, using
the following formula: (Expression levels of CRABP1*0.00326)
+ (expression levels of IGKV4-1*-0.00036) + (expression
levels of IL-11*0.14555) + (expression levels of
INHA*0.00475) + (expression levels of LGR4*0.01757) +
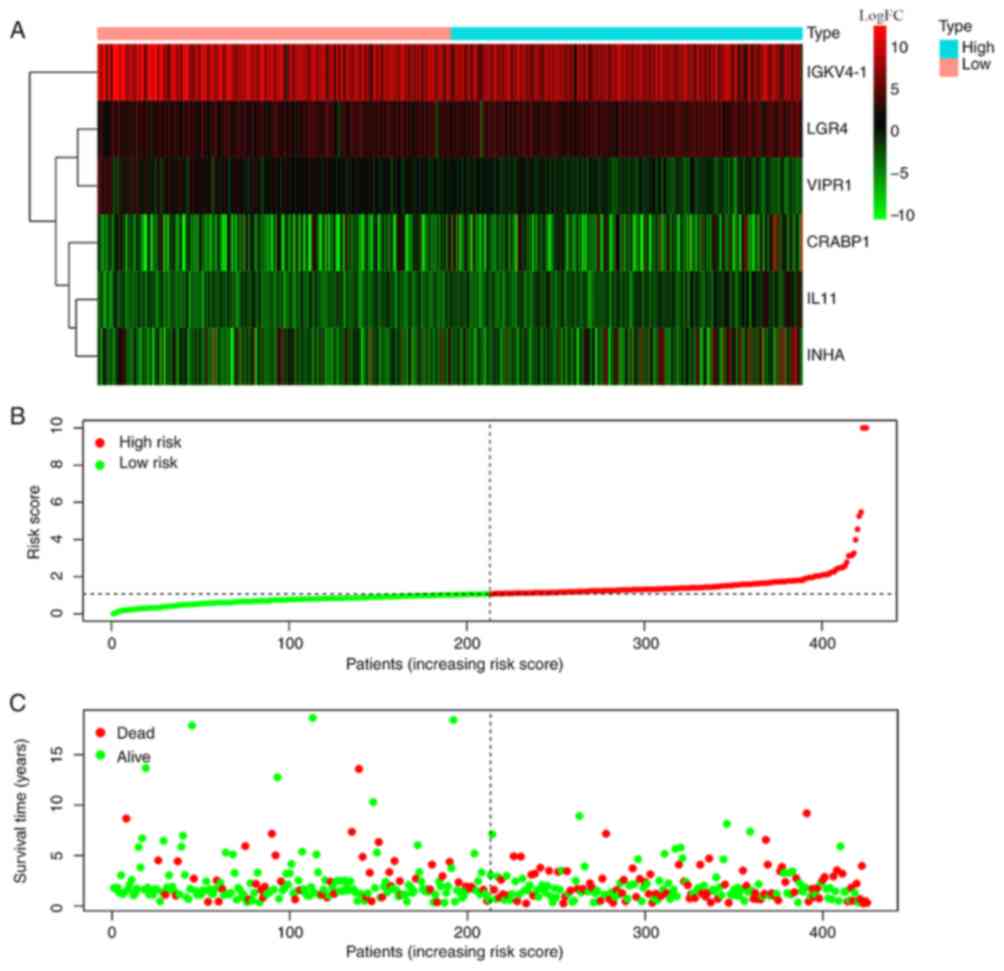
(expression levels of VIPR1*-0.17506). The results
demonstrated that the expression levels of IGKV4-1 and
VIPR1 in high risk score group were significantly higher
than low risk score group, while the expression levels of
CRABP1, IL11, INHA and LGR4 in high risk score group
were significantly lower than low risk score group (Fig. 2A). The risk coefficient (Fig. 2B) and mortality (Fig. 2C) were significantly higher in high
risk score group compared with the low risk score group,
respectively.
 | Table II.Univariate Cox regression analysis of
immune related genes of patients with lung adenocarcinoma. |
Table II.
Univariate Cox regression analysis of
immune related genes of patients with lung adenocarcinoma.
|
| Overall survival
Univariate analysis |
|---|
|
|
|
|---|
| Immune related
gene | HR (95% CI) | P-value |
|---|
| S100P | 1.218
(1.578–2.018) | 0.005 |
| CPABP1 | 1.343
(1.659–2.106) | 0.005 |
| BIRC5 | 1.645
(1.388–1.951) | 0.002 |
| IGKV4-1 | 1.665
(1.322–2.098) | 0.009 |
| IL11 | 1.728
(1.349–2.636) | <0.001 |
| INHA | 1.226
(0.972–2.265) | 0.004 |
| INSL4 | 1.978
(1.493–2.872) | 0.007 |
| ADRB2 | 0.711
(0.553–0.712) | 0.004 |
| LGR4 | 1.678
(1.433–2.172) | 0.001 |
| VIPR1 | 0.651
(0.413–0.732) | <0.001 |
Gene functional enrichment analysis of
differentially expressed IRGs
The biological functions of 273 IRGs were further
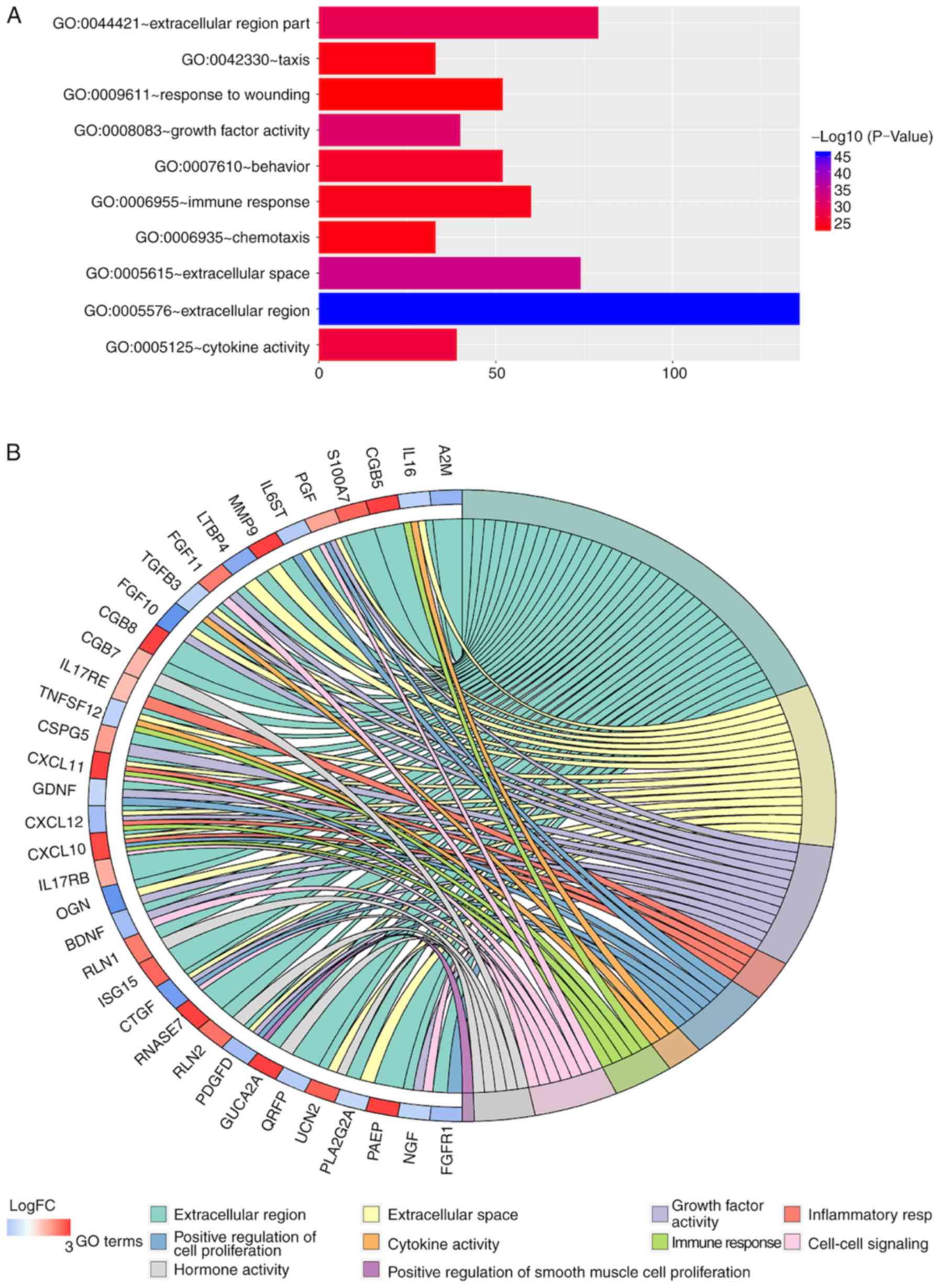
investigated using GO and KEGG analyses. The results showed that
the ‘extracellular region part’ was the most frequent GO biological
process category (P<0.05; Fig.
3A). The top 10 enriched GO networks and top 40 genes involved
in GO networks are presented in Fig. 3A
and B, respectively.
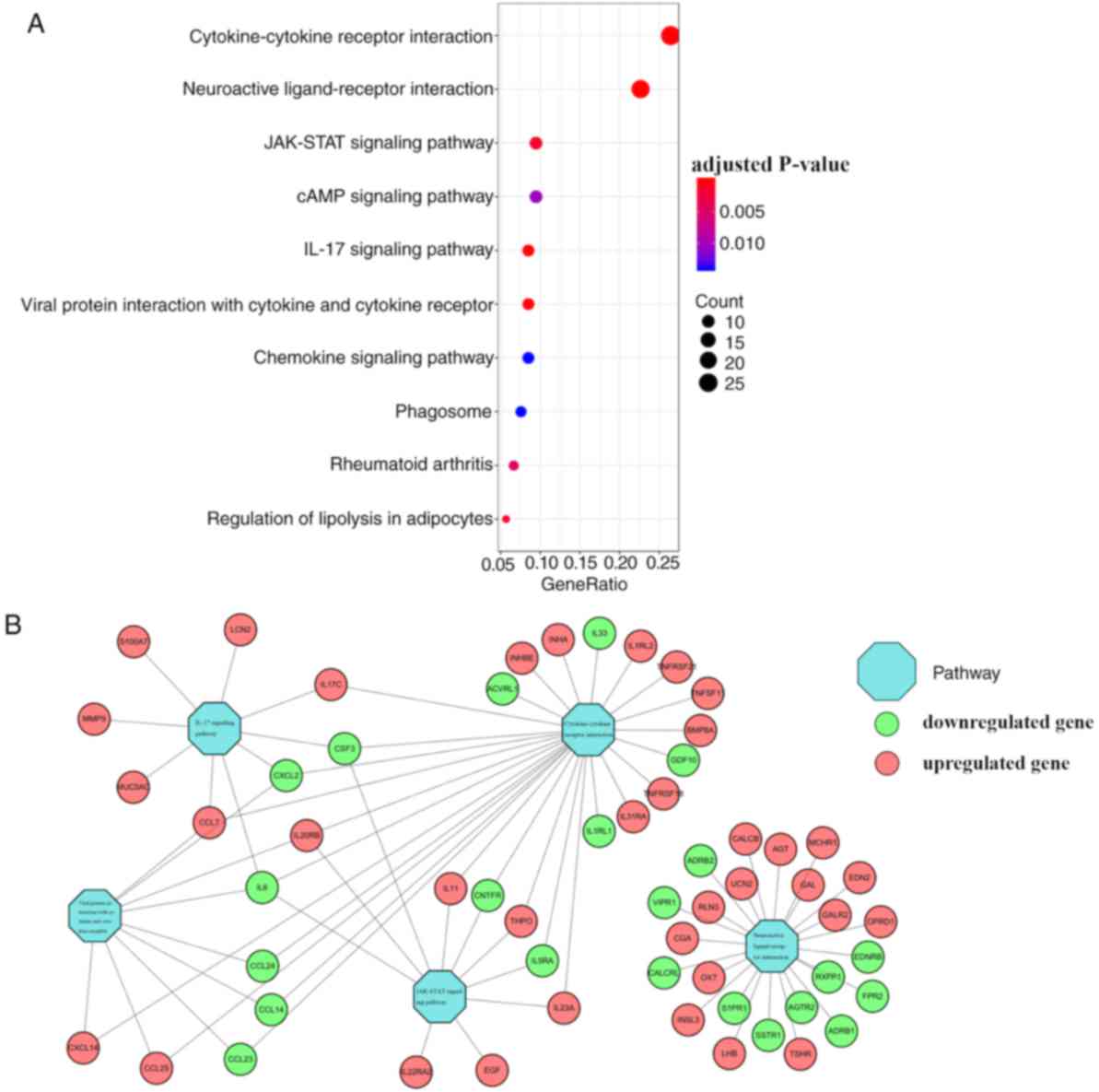
The top significantly enriched pathways were
obtained using KEGG pathway analysis (Fig. 4A); these were ‘cytokine-cytokine
receptor interaction’, ‘neuroactive ligand-receptor interaction’,
‘JAK-STAT signaling pathway’, ‘IL-17 signaling pathway’ and ‘viral
protein interaction with cytokine and cytokine receptor’. Based on
the relationship between IRGs and the top 5 KEGG pathways, a visual
network was constructed using Cytoscape version 3.6.1 (Fig. 4B).
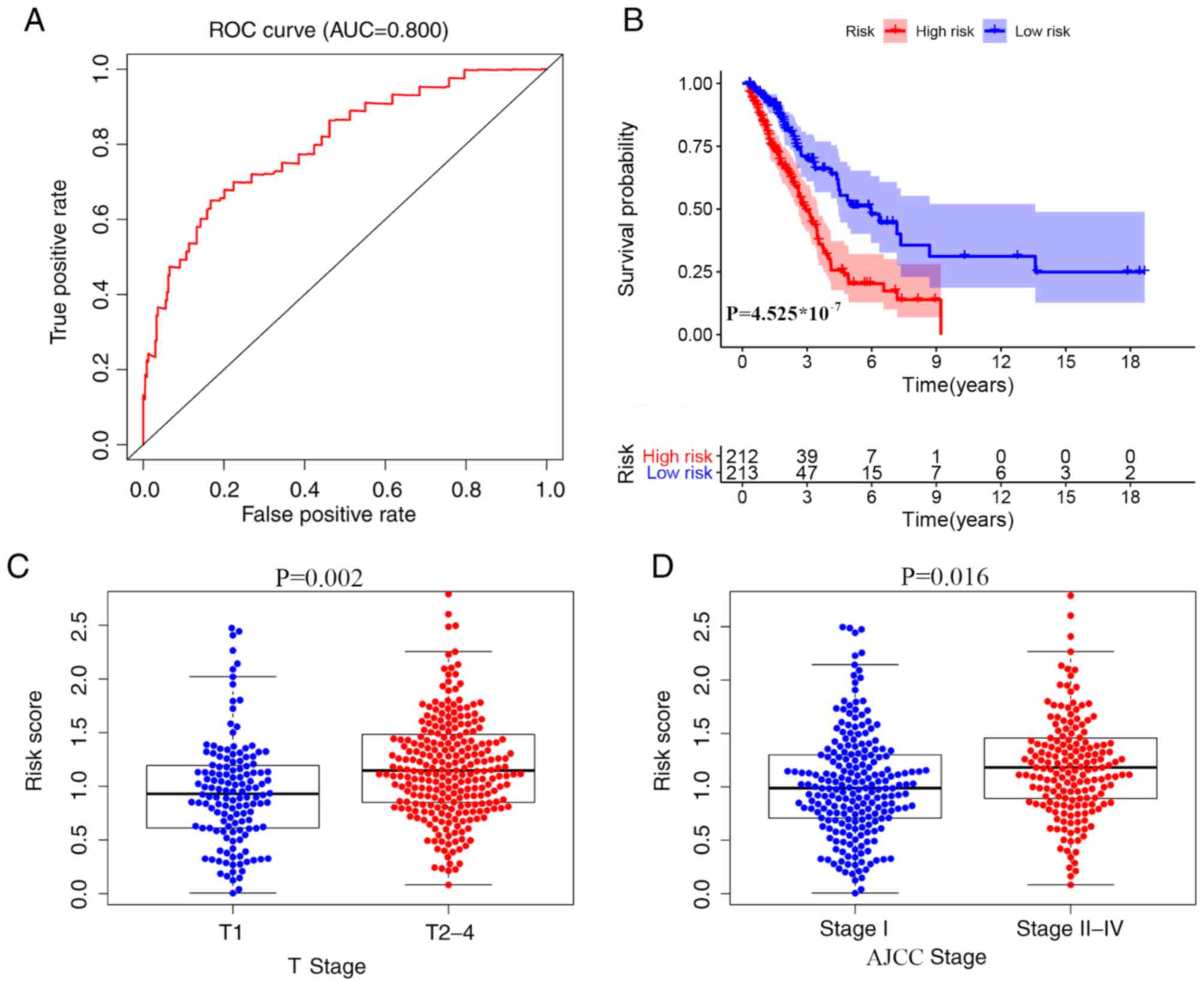
Clinical outcome of patients with LUAD
using the prognostic model
Kaplan-Meier plots were used to divide patients into
high and low risk score groups according to OS time. The area under
the ROC curve was 0.800, suggesting that a prognostic model based
on IRGs could be used to monitoring survival (Fig. 5A and B). Univariate analyses showed
that high American Joint Committee on Cancer (AJCC) stage (22) (HR, 1.645; 95% CI, 1.388–1.951;
P<0.001), high tumor stage (22)
(HR, 1.665; 95%; CI, 1.322–2.098; P<0.001), high node stage
(22) (HR, 1.928; 95% CI,
1.549–2.426; P<0.001) and high risk score (HR, 1.978; 95% CI,
1.493–2.872; P<0.001) were significant risk factors for a poor
prognosis (Table III). Using
multivariate analysis, a high risks core (HR, 2.071; 95% CI,
1.313–3.425; P<0.001) was found to be independently associated
with a less favorable OS time (Table
III). Collectively, these data indicate that the risk scores
are significantly higher among patients with advanced T (Fig. 5C) and high AJCC stages (Fig. 5D).
 | Table III.Univariate and Multivariate Cox
regression analysis of prognostic model (risk score) and clinical
features of patients with lung adenocarcinoma. |
Table III.
Univariate and Multivariate Cox
regression analysis of prognostic model (risk score) and clinical
features of patients with lung adenocarcinoma.
|
| Overall
survival |
|---|
|
|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Risk factors | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years |
| (>65
vs. ≤65) | 0.998
(0.978–1.018) | 0.842 | 1.002
(0.981–1.023) | 0.863 |
| Sex |
| (Male
vs. Female) | 0.843
(0.659–1.406) | 0.843 | 0.864
(0.581–1.286) | 0.472 |
| AJCC stage |
| (I–II
vs. III–IV) | 1.645
(1.388–1.951) | <0.001 | 1.547
(0.909–2.640) | 0.108 |
| T stage |
| (T1-2
vs. T3-4) | 1.665
(1.322–2.098) | <0.001 | 1.280
(0.985–1.662) | 0.065 |
| N stage |
| (N0 vs.
N1-3) | 1.928
(1.549–2.426) | <0.001 | 1.281
(0.795–2.062) | 0.309 |
| M stage |
| (M0 vs.
M1) | 1.226
(0.972–2.265) | 0.096 | 1.408
(0.541–1.966) | 0.277 |
| Risk score |
| (Low
vs. High) | 1.978
(1.493–2.872) | <0.001 | 2.071
(1.313–3.425) | <0.001 |
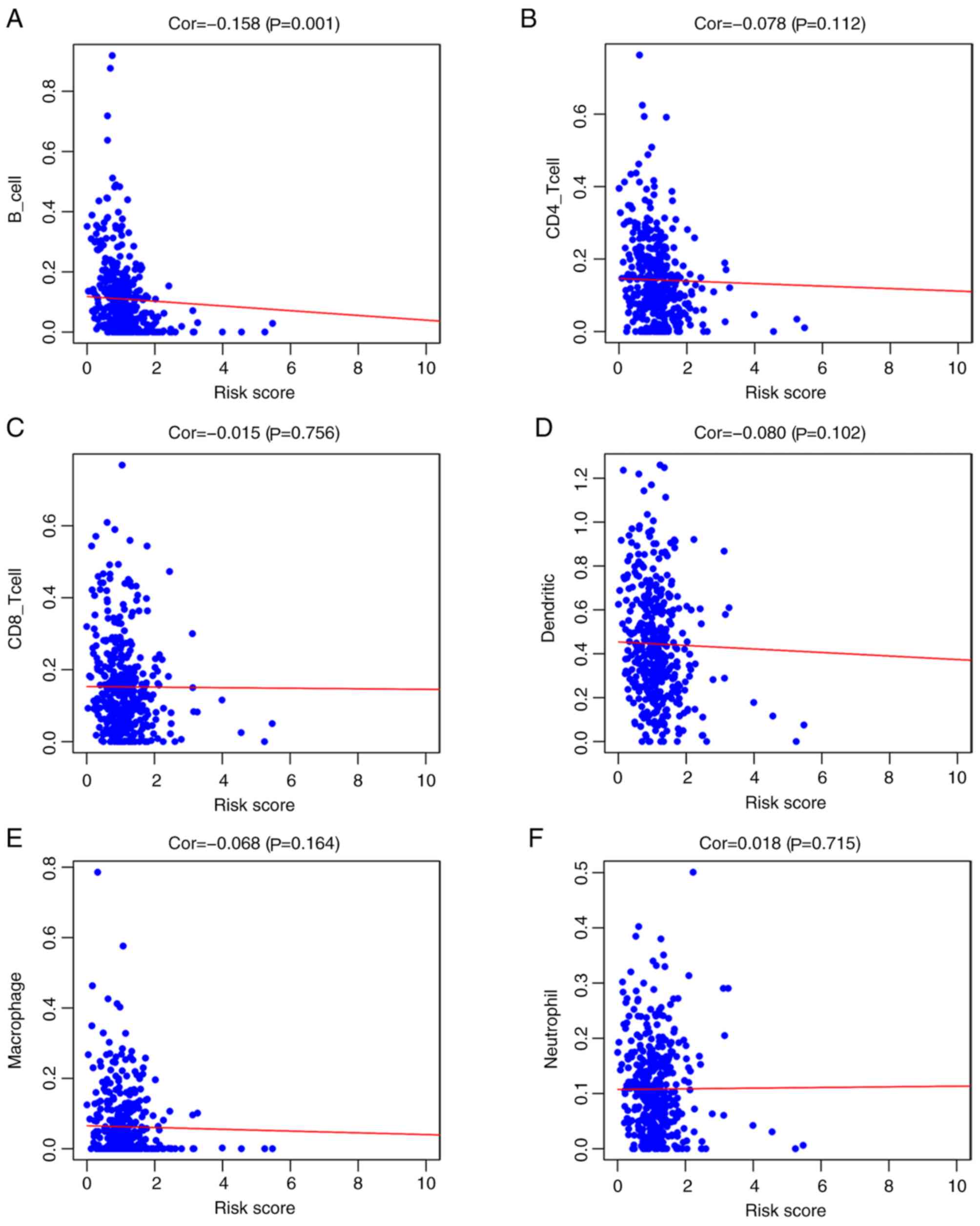
Correlation analysis of the prognostic
model and immune cell infiltration
Among the six immune cell types investigated (B
cells, CD4 T cells, CD8 T cells, DCs, macrophages and neutrophils),
the risk factors identified in the prognostic model were negatively
correlated with B cell infiltration (r=−0.158; P=0.001;
Fig. 6A); however, risk score was
not associated with CD4+ T cells (r=−0.078; P=0.112;
Fig. 6B), CD8+ T cells
(r=−0.015; P=0.756; Fig. 6C),
dendritic cells (r=−0.080; P=0.102; Fig.
6D), macrophages (r=−0.068; P=0.164; Fig. 6E) or neutrophils (r=0.018; P=0.715;
Fig. 6F).
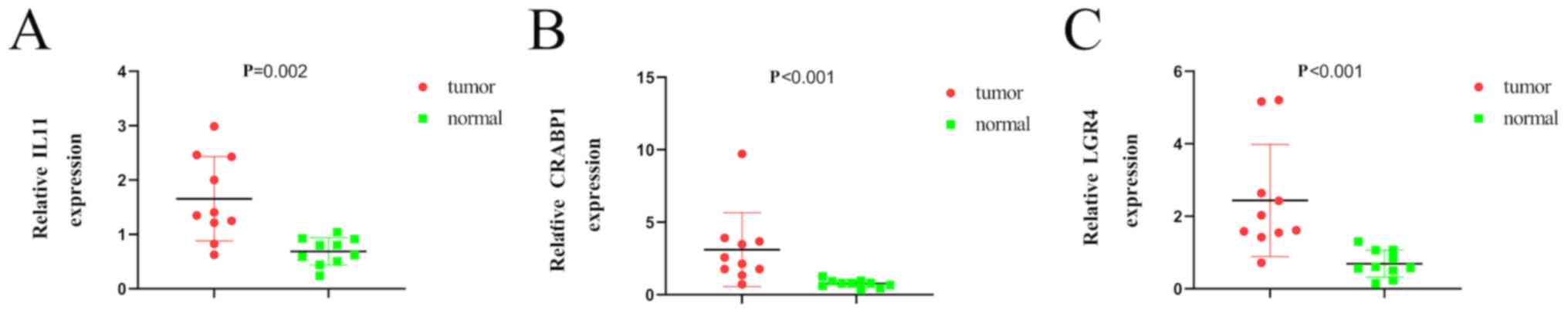
Analysis and validation of gene
expression
To further validate the expression of relevant key
genes in the prognostic model, three mRNAs (IL11, CARBP1 and LGR4)
were randomly selected and their expression levels were evaluated
in 10 pairs of LUAD and adjacent normal tissues. The expression
levels of IL11, CRABP1 and LGR4 were higher in tumor tissues
compared with adjacent normal tissues (Fig. 7A-C), which was consistent with the
findings observed in TCGA database.
Discussion
The incidence and mortality rates of lung cancer in
China are still increasing (23). A
previous study indicated that IRGs are promising prognostic
indicators of early stage lung cancer (8). A particular study screened for 40 genes
and classified patients into high-risk and low-risk groups
according to the immune signature; Patients with early non-squamous
lung cancer demonstrated independent prognostic factors (8). As is well known, the treatment of
advanced stage unresectable or metastatic lung cancer in China is
difficult (24). Immunotherapy, such
as immune checkpoint inhibitors, is primarily used for patients
with metastatic lung cancer (5,6);
therefore, the present study aimed to predict the prognoses of
patients with LUAD using IRGs.
A total of six IRGs associated with prognosis were
identified in patients with LUAD using TCGA database, and a
prognostic model was established based on these genes (CRABP1,
IGKV4-1, IL11, INHA, LGR4 and VIPR1). In this model, the
overall survival duration of patients with high-risk disease was
significantly shorter compared with patients with low-risk
disease.
CRABP1 is a member of the fatty acid binding
family of proteins, which binds to retinoic acid with high affinity
(25). There are few studies on
CRABP1 and lung cancer and the underlying molecular
mechanisms of CRABP1 function in lung cancer remain unclear.
Favorskaya et al demonstrated thatCRABP1 significantly
alters the expression levels of CRABP in NSCLC samples
(26). IGKV4-1 is inherently
autoreactive and has been implicated in B-cell mediated autoimmune
diseases and dysregulated B-cell tolerance (27–29).
However, the function of IGKV4-1 in lung cancer remains
unknown. VIPR1 is a G protein-coupled receptor that is widely
distributed in the normal tissues of humans, and that serves a role
in physiological functions (30).
Downregulation and deletion of VIPR1 have been detected in
patients with LUAD (31). Gong et
al (32) demonstrated that
LGR4 was expressed in LUAD tissues but not in normal lung
tissue. The group reported that LGR4 and IQGAP1
served a role in the regulation of tumor growth and metastasis in
lung cancer cells. IL-11 is a member of the IL-6
group and binds to its corresponding receptors. IL-11 is an
important inflammatory mediator that can affect the activity of a
variety of immune cells (33–35).
Increased IL-11 expression levels have been associated with
various types of cancer, including LUAD (36–38).
To further explain some of these potential
mechanisms, gene functional enrichment analysis was performed. It
was demonstrated that IRGs were primarily enriched in
‘cytokine-cytokine receptor interaction’, ‘neuroactive
ligand-receptor interaction’ and the ‘JAK-STAT signaling pathway’.
Among the above prognosis-related immune genes, IL-11 was
associated with ‘cytokine-cytokine receptor interaction’ and the
‘JAK-STAT signaling pathway’.
Cytokines are secreted glycoproteins that function
as intercellular mediators, promoting cellular proliferation,
differentiation and apoptosis (39).
On the other hand, cytokines secreted by tumors can promote the
recruitment of immunosuppressive cells, resulting in tumor
metastasis (40). Previous studies
have identified a variety of cytokines that can regulate
hematopoiesis, induce inflammatory responses and control immune
responses through the Janus kinase (JAK) signaling pathway
(41,42). The JAK family contains four members:
JAK1, JAK2, JAK3 and TYK2 (43). JAK
kinases are a potential target for the treatment of tumors due to
the oncogenic effects and the promotion of tumor inflammatory
responses via JAK signaling (41).
When cytokines bind to their cognate receptors, JAK is activated
and phosphorylates downstream signaling and transcriptional
activator (STAT), ultimately leading to tumor invasion,
angiogenesis, apoptosis and metastasis (41). IL-11 activates downstream
JAK/STAT signaling proteins via a gp130 homodimer (42). The suppressor of cytokine signaling
proteins regulate JAK/STAT signaling pathways by serving as
feedback inhibitors of activated JAK (44). Currently, few studies have
investigated IL-11 and JAK signaling pathways in tumors, and
the underlying molecular mechanisms of action remain unknown. In
the present study, KEGG analysis revealed that IRGs are mainly
enriched in these two signaling pathways. Further network
construction revealed that IL-11 is closely associated with
these two pathways. The explanation of this relationship between
the two pathways in the present study may provide a basis for
determining the prognosis of lung cancer.
In the present study, the immune gene-related
prognostic index was not only associated with the prognosis of LUAD
but was also negatively correlated with immune B cell infiltration.
Tumor-infiltrating B lymphocytes have seemingly conflicting effects
in tumors. On the one hand, B cells function in the inhibition of
tumor cell proliferation via antigen restricted tumoricidal
responses; on the other hand, B cells also act by suppressing the
immune system, thus promoting tumor growth, proliferation and
metastasis (45). Tumor-infiltrating
B lymphocyte-derived lymphotoxin has been reported to promote the
progression of androgen-independent prostate cancer by activating
the Nuclear Factor κ-B and STAT3 signaling pathways (46). Previous studies have shown that
tumor-infiltrating CD20+ B cells reside in close
proximity to CD8+ T cells, and in patients with ovarian
cancer, infiltration of CD20+ B and CD8+ T
cells prolongs DSS (disease-specific survival) compared with
CD8+ T cell infiltration alone (11). Pinto et al (47) demonstrated that patients with LUAD,
with mutated K-RAS had associated B cell infiltration. B
cells also exert a number of anti-tumor effects. First, they can
stimulate other immune cells to produce cytokines, particularly
those that enhance the activity of CTL (cytotoxic T-lymphocytes)
(48). Secondly, B cells secrete
granzyme B to directly kill tumor cells (48). Furthermore, B cells can suppress
pancreatic cancer via antibody-dependent mechanisms (48,49).
However, the functions of a number of prognosis-related genes in
lung cancer remain unclear. For example, the IGKV4-1 gene
encodes a B cell receptor (50).
Previous studies have not described the relationship between
IGKV4-1 and lung cancer. It is unclear whether the
IGKV4-1 gene serves a role in B cell infiltration in lung
cancer, and further research is required to investigate the
possible underlying molecular mechanisms. Due to the negative
correlation between the prognostic model and B cell infiltration in
the present study, some patients may have had low risk scores due
to the anti-tumor effects exerted by infiltrating B
lymphocytes.
The present study was not without limitations. The
primary limitation was the small sample size, which will be
increased in future studies. Although the conclusions of the
present study were drawn based on evidence from TCGA database, only
gene expression was verified. Thus, it remains critical to further
verify the applicability of survival models.
In summary, the present study identified
prognosis-related immune genes using TCGA database and established
a prognostic model for patients with LUAD. Using multivariate
analysis with other clinicopathological features, such as age,
gender and TNM stage, risk score was revealed to be an independent
prognostic factor, hence, the present model can predict the
prognoses of patients with LUAD. In addition, the prognostic model
was associated with B cell infiltration and the present study may
provide novel evidence for the prognosis and immunotherapy of
patients with LUAD in the future.
Acknowledgements
The authors would like to thank Dr Wei Shan
(Department of Gastrointestinal Surgery, Renmin Hospital of Wuhan
University) for his assistance regarding statistics.
Funding
The present study was supported by The Science and
Education for Health Foundation of Suzhou for Youth (grant nos.
kjxw2018030 and kjxw2018032), The Science and Technology Project
Foundation of Suzhou (grant no. SS201651), The Education Research
Project Foundation of Nanjing Medical University (grant no.
FZS-ZD-201701), The Jiangsu Province Medical Key Discipline (grant
no. ZDXKC2016007) and Suzhou Oncology Clinical Center (grant no.
Szzx201506).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in The Cancer Genome Atlas repository
(https://portal.gdc.cancer.gov/).
Authors' contributions
ZLD and WJW conceived and designed the study. YQH
and JPS performed the statistical analysis. HW, MSW and YW were
involved in the writing of the manuscript and in the interpretation
of the results. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Medical Ethics
Committees of Jining Cancer Hospital (approval no. 20190067).
Written informed consent was provided by all patients prior to the
study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AJCC
|
American Joint Committee on Cancer
|
|
CI
|
confidence interval
|
|
DCs
|
dendritic cells
|
|
DEIRGs
|
differentially expressed
immune-related genes
|
|
GO
|
gene ontology
|
|
HR
|
hazard ratio
|
|
IRGPM
|
immune-related gene prognostic
model
|
|
IRGs
|
immune-related genes
|
|
JAK-STAT
|
Janus kinase-signaling and
transcriptional activator
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
LUAD
|
lung adenocarcinoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
OS
|
overall survival
|
|
PD-1
|
programmed death-1
|
|
PD-L1
|
programmed death-ligand 1
|
|
ROC
|
receiver operating characteristic
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TILs
|
tumor-infiltrating lymphocytes
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sher T, Dy GK and Adjei AA: Small cell
lung cancer. Mayo Clin Proc. 83:355–367. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Duffy MJ and O'Byrne K: Tissue and blood
biomarkers in lung cancer: A review. Adv Clin Chem. 86:1–21. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meza R, Meernik C, Jeon J and Cote ML:
Lung cancer incidence trends by gender, race and histology in the
United States, 1973–2010. PLoS One. 10:e01213232015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garon EB, Rizvi NA, Hui R, Leighl N,
Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L,
et al: Pembrolizumab for the treatment of non-small-cell lung
cancer. N Engl J Med. 372:2018–2028. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hellmann MD, Rizvi NA, Goldman JW,
Gettinger SN, Borghaei H, Brahmer JR, Ready NE, Gerber DE, Chow LQ,
Juergens RA, et al: Nivolumab plus ipilimumab as first-line
treatment for advanced non-small-cell lung cancer (CheckMate 012):
Results of an open-label, phase 1, multicohort study. Lancet Oncol.
18:31–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Postow MA, Callahan MK and Wolchok JD:
Immune checkpoint blockade in cancer therapy. J Clin Oncol.
33:1974–1982. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li B, Cui Y, Diehn M and Li R: Development
and validation of an individualized immune prognostic signature in
early-stage nonsquamous non-small cell lung cancer. JAMA Oncol.
3:1529–1537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Shibli KI, Donnem T, Al-Saad S, Persson
M, Bremnes RM and Busund LT: Prognostic effect of epithelial and
stromal lymphocyte infiltration in non-small cell lung cancer. Clin
Cancer Res. 14:5220–5227. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nedergaard BS, Ladekarl M, Nyengaard JR
and Nielsen K: A comparative study of the cellular immune response
in patients with stage IB cervical squamous cell carcinoma. Low
numbers of several immune cell subtypes are strongly associated
with relapse of disease within 5 years. Gynecol Oncol. 108:106–111.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nielsen JS, Sahota RA, Milne K, Kost SE,
Nesslinger NJ, Watson PH and Nelson BH: CD20+ tumor -infiltrating
lymphocytes have an atypical CD27- memory phenotype and together
with CD8+ T cells promote favorable prognosis in ovarian cancer.
Clin Cancer Res. 18:3281–3292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bhattacharya S, Andorf S, Gomes L, Dunn P,
Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J,
et al: ImmPort: Disseminating data to the public for the future of
immunology. Immunol Res. 58:234–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wagenmakers EJ and Farrell S: AIC model
selection using Akaike weights. Psychon Bull Rev. 11:192–196. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heagerty PJ, Lumley T and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Detterbeck FC, Boffa DJ, Kim AW and Tanoue
LT: The eighth edition lung cancer stage classification. Chest.
151:193–203. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chinese Association for Clinical
Oncologists, . Clinical practice guideline for stage IV primary
lung cancer in China(2020 version). Zhonghua Zhong Liu Za Zhi.
42:1–16. 2020.(In Chinese). PubMed/NCBI
|
|
25
|
Dong D, Ruuska SE, Levinthal DJ and Noy N:
Distinct roles for cellular retinoic acid-binding proteins I and II
in regulating signaling by retinoic acid. J Biol Chem.
274:23695–23698. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Favorskaya I, Kainov Y, Chemeris G,
Komelkov A, Zborovskaya I and Tchevkina E: Expression and clinical
significance of CRABP1 and CRABP2 in non-small cell lung cancer.
Tumour Biol. 35:10295–10300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yurasov S, Wardemann H, Hammersen J,
Tsuiji M, Meffre E, Pascual V and Nussenzweig MC: Defective B cell
tolerance checkpoints in systemic lupus erythematosus. J Exp Med.
201:703–711. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pugh-Bernard AE, Silverman GJ, Cappione
AJ, Villano ME, Ryan DH, Insel RA and Sanz I: Regulation of
inherently autoreactive VH4-34 B cells in the maintenance of human
B cell tolerance. J Clin Invest. 108:1061–1070. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cappione A III, Anolik JH, Pugh-Bernard A,
Barnard J, Dutcher P, Silverman G and Sanz I: Germinal center
exclusion of autoreactive B cells is defective in human systemic
lupus erythematosus. J Clin Invest. 115:3205–3216. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Reubi JC, Laderach U, Waser B, Gebbers JO,
Robberecht P and Laissue JA: Vasoactive intestinal
peptide/pituitary adenylate cyclase-activating peptide receptor
subtypes in human tumors and their tissues of origin. Cancer Res.
60:3105–3112. 2000.PubMed/NCBI
|
|
31
|
Mlakar V, Strazisar M, Sok M and Glavac D:
Oligonucleotide DNA microarray profiling of lung adenocarcinoma
revealed significant downregulation and deletions of vasoactive
intestinal peptide receptor 1. Cancer Invest. 28:487–494. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gong X, Yi J, Carmon KS, Crumbley CA,
Xiong W, Thomas A, Fan X, Guo S, An Z, Chang JT and Liu QJ:
Aberrant RSPO3-LGR4 signaling in Keap1-deficient lung
adenocarcinomas promotes tumor aggressiveness. Oncogene.
34:4692–4701. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Zhang Y, Dutta DJ, Argaw AT,
Bonnamain V, Seto J, Braun DA, Zameer A, Hayot F, Lòpez CB, et al:
Proapoptotic and antiapoptotic actions of Stat1 versus Stat3
underlie neuroprotective and immunoregulatory functions of IL-11. J
Immunol. 187:1129–1141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Trepicchio WL, Wang L, Bozza M and Dorner
AJ: IL-11 regulates macrophage effector function through the
inhibition of nuclear factor-kappaB. J Immunol. 159:5661–5670.
1997.PubMed/NCBI
|
|
35
|
Curti A, Ratta M, Corinti S, Girolomoni G,
Ricci F, Tazzari P, Siena M, Grande A, Fogli M, Tura S and Lemoli
RM: Interleukin-11 induces Th2 polarization of human CD4(+) T
cells. Blood. 97:2758–2763. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pastor MD, Nogal A, Molina-Pinelo S,
Quintanal-Villalonga Á, Meléndez R, Ferrer I, Romero-Romero B, De
Miguel MJ, López-Campos JL, Corral J, et al: IL-11 and CCL-1: Novel
protein diagnostic biomarkers of lung adenocarcinoma in
bronchoalveolar lavage fluid (BALF). J Thorac Oncol. 11:2183–2192.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamazumi K, Nakayama T, Kusaba T, Wen CY,
Yoshizaki A, Yakata Y, Nagayasu T and Sekine I: Expression of
interleukin-11 and interleukin-11 receptor alpha in human
colorectal adenocarcinoma; immunohistochemical analyses and
correlation with clinicopathological factors. World J
Gastroenterol. 12:317–321. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hanavadi S, Martin TA, Watkins G, Mansel
RE and Jiang WG: Expression of interleukin 11 and its receptor and
their prognostic value in human breast cancer. Ann Surg Oncol.
13:802–808. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Morris R, Kershaw NJ and Babon JJ: The
molecular details of cytokine signaling via the JAK/STAT pathway.
Protein Sci. 27:1984–2009. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
O'Shea JJ, Schwartz DM, Villarino AV,
Gadina M, McInnes IB and Laurence A: The JAK-STAT pathway: Impact
on human disease and therapeutic intervention. Annu Rev Med.
66:311–328. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Buzzelli JN, O'Connor L, Scurr M, Chung
Nien Chin S, Catubig A, Ng GZ, Oshima M, Oshima H, Giraud AS,
Sutton P, et al: Overexpression of IL-11 promotes premalignant
gastric epithelial hyperplasia in isolation from germline
gp130-JAK-STAT driver mutations. Am J Physiol Gastrointest Liver
Physiol. 316:G251–G262. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Stark GR and Darnell JE Jr: The JAK-STAT
pathway at twenty. Immunity. 36:503–514. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Inagaki-Ohara K, Kondo T, Ito M and
Yoshimura A: SOCS, inflammation, and cancer. JAKSTAT.
2:e240532013.PubMed/NCBI
|
|
45
|
Guo S, Contratto M, Miller G, Leichman L
and Wu J: Immunotherapy in pancreatic cancer: Unleash its potential
through novel combinations. World J Clin Oncol. 8:230–240. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ammirante M, Luo JL, Grivennikov S,
Nedospasov S and Karin M: B-cell-derived lymphotoxin promotes
castration-resistant prostate cancer. Nature. 464:302–305. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pinto R, Petriella D, Lacalamita R,
Montrone M, Catino A, Pizzutilo P, Botticella MA, Zito FA, Del Bene
G, Zonno A, et al: KRAS-driven lung adenocarcinoma and B cell
infiltration: Novel insights for immunotherapy. Cancers (Basel).
11(pii): E11452019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tsou P, Katayama H, Ostrin EJ and Hanash
SM: The emerging role of B cells in tumor immunity. Cancer Res.
76:5597–5601. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lee KE, Spata M, Bayne LJ, Buza EL, Durham
AC, Allman D, Vonderheide RH and Simon MC: Hif1a deletion reveals
pro-neoplastic function of B cells in pancreatic neoplasia. Cancer
Discov. 6:256–269. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Asare A, Kanaparthi S, Lim N, Phippard D,
Vincenti F, Friedewald J, Pavlakis M, Poggio E, Heeger P, Mannon R,
et al: B cell receptor genes associated with tolerance identify a
cohort of immunosuppressed patients with improved renal allograft
graft function. Am J Transplant. 17:2627–2639. 2017. View Article : Google Scholar : PubMed/NCBI
|