Introduction
Multiple myeloma (MM) is an incurable cancer of
plasma cells caused by the abnormal accumulation of monoclonal
plasma cells in the bone marrow (BM) (1), which accounts for ~10% of all
hematological malignancies (2). The
natural course of MM varies greatly. For example, some patients
live for only a few months, while other patients survive for
decades (3). The identification
patients who are at high risk may help optimize the choice of
personalized treatment and improve clinical outcomes (3). Currently, the Revised MM International
Staging System (R-ISS) is widely used in clinics for stratification
and can successfully classifies patients (3). The R-ISS staging system consists of
determining β2-microglobulin (β2-M), albumin
(ALB), lactate dehydrogenase (LDH) and cytogenetic heterogeneity
(4). In addition to stratification,
dynamically monitoring of the effect of therapy, termed ‘minimal
residual disease’ (MRD), is important for predicting relapse and
the prognosis in order to intervene in a timely manner and prolong
survival (5). However, the R-ISS is
not suitable for assessing MRD. For example, the cytogenetic
heterogeneity does not dynamically reflect the tumor load at
different stages of treatment. Moreover, diverse genetic
abnormalities exist in different patients, and numerous factors
influence β2-M, ALB and LDH levels (5).
Currently, multiparameter flow cytometry (MFC) of
the BM is routinely used for monitoring MRD (6). However, MM has a unique characteristic,
namely, myeloma cells are not evenly distributed in BM (foci), and
this characteristic often causes false negative results for MRD
(7). Therefore, identifying a novel
marker that is universal for the majority of patients, that
reflects the dynamic changes in tumor load and is able to be
conveniently assessed is urgently needed. For these purposes, the
present study aimed to develop a non-invasive approach.
IL-6 is a pivotal growth factor that regulates the
proliferation of MM via an autocrine mechanism from MM cells or a
paracrine mechanism from stromal cells (8). IL-6 exerts its biological effects by
binding to its receptor (IL-6R), and IL-6R leads to the activation
of the JAK/STAT signaling pathway (9). The activation of JAK family tyrosine
kinases recruits the STAT3 protein, which dimerizes and
translocates to the nucleus to transduce signals and initiate the
expression of genes involved in the proliferation and apoptosis of
myeloma cells (10).
MicroRNAs (miRNAs/miRs) can identify the 3′
untranslated region (3′UTR) of their specific target mRNAs and
subsequently inhibit the expression of the target gene at the
post-transcriptional level (11).
miR-451a was discovered in 2005 and is located on chromosome
17qll.2 (11). Furthermore,
miR-451a, which functions as a tumor suppressor, has been reported
to be associated with several types of cancer, including lung
cancer (12), hepatocellular
carcinoma (13), gliomas (14) and osteosarcoma (15), wherein it inhibits the proliferation,
migration and invasion of tumors (16). miR-451a has also been shown to target
IL-6R in several solid tumor types, and inhibits tumor
proliferation, migration and angiogenesis via the IL-6R signaling
pathway (17). However, whether
miR-451a inhibits tumor growth in MM remains unknown, and whether
there a targeted relationship between miR-451a and IL-6R in MM is
yet to be elucidated. Moreover, further investigations are required
to determine whether circulating miR-451a levels reflect the tumor
load in real time. The current study aimed to evaluate these
questions, and the present findings may provide a potential
prognostic strategy for MM.
Materials and methods
Ethics statement
This study was performed with the approval of the
Institutional Review Board of Sichuan Provincial People's Hospital
of China. Informed written consent was collected from each eligible
patient, and the whole study was performed in accordance with the
Declaration of Helsinki.
Study subjects
Between January 2017 and December 2018, 66 patients
with MM (age range, 38–88 years; 37 men; 29 women) and ten healthy
controls (age range, 31–83 years; six men; four women), who were
treated at the Sichuan Academy of Medical Sciences and Sichuan
Provincial People's Hospital were enrolled. The diagnostic criteria
for active MM were based on the International Myeloma Working Group
guidelines (18). The criteria for
classification were according to the subtype of abnormal
proliferative immunoglobulin. The criteria for staging were
assessed using the R-ISS (3). The
healthy control group consisted of transplant donors undergoing BM
transplantation, who were recruited to provide BM (~1 ml) and
peripheral blood samples (~5 ml).
Treatment
All patients were treated with bortezomib (1.3
mg/m2 subcutaneously on days 1, 4, 8 and 11 of every
21-day cycle or 1.3 mg/m2 subcutaneously on days 1, 8,
15 and 22 of every 35-day cycle; three manufacturers permitted by
medical insurance: QILU Pharmaceutical; Chia Tai Tianqing
Pharmaceutical; Hansoh Pharmaceutical) in combination with
dexamethasone (Tianjin Jinyao Pharmaceutical Co., Ltd.; 10 or 20 mg
on the day of, and the day after the administration of each dose of
bortezomib). In high-risk patients, either cyclophosphamide (300
mg/m2 on days 1 and 8; Baxter Oncology GmbH) or
lenalidomide (10 or 25 mg based on renal function measured from
days 1–21; Beijing Shuanglu Pharmaceutical Co., Ltd.) was added to
the combination. A total of 8–12 cycles were planned as induction
therapy for all patients, and after induction therapy, patients
entered maintenance therapy with either bortezomib (1.3
mg/m2 once every 2 weeks for 2 years) or lenalidomide
(25 mg once a day on days 1–21 and every 28 days until
relapse).
Sampling
Samples were initially collected from patients at
the first diagnosis and at the required follow-up time points (once
every 3 months); 5 ml blood was collected in serum collection
tubes. Whole blood was allowed to stand for ~1 h at room
temperature, before centrifugation at 1,200 × g for 10 min at 4°C,
followed by the separation of serum. Successive centrifugation was
performed for 10 min at 10,000 × g at 4°C to remove the cellular
debris. The resulting serum was aliquoted into Eppendorf tubes and
stored at −80°C to determine IL-6 and circulating miR-451a
levels.
BM samples collected from both patients and controls
were anticoagulated with EDTA-K2 (BD Biosciences) at
room temperature. Some BM samples were diluted 1:1 with RPMI-1640
basic medium (Gibco; Thermo Fisher Scientific, Inc.). Mononuclear
cells were isolated with Ficoll cell isolation medium at room
temperature (density 1.077 g/ml; Stemcell Technologies, Inc.).
Then, plasma cells were isolated using CD138 magnetic beads at room
temperature (Miltenyi Biotec, Inc.) for RNA extraction to analyze
miR-451a expression and proteins were extracted to measure IL-6R
levels. The remaining BM was used for MFC.
Analysis
Reverse transcription-quantitative PCR
(RT-qPCR)
To concentrate the circulating miRNA, the extraction
protocol was conducted by adding 10% PEG 8,000 solution (Beijing
Solarbio Science & Technology Co., Ltd.). Total RNA was
extracted from 500 µl serum using a mirVana miRNA Isolation kit
(cat. no. AM1561; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. Then, miR-451a was reverse transcribed
with a Bulge-LoopTM miRNA RT-qPCR Starter kit (Guangzhou RiboBio
Co., Ltd.). The reaction conditions were: 42°C for 60 min and 70°C
for 10 min. Relative quantification was performed via qPCR using
the SYBR Green PCR kit (Guangzhou RiboBio Co., Ltd.). External
references (cel-miR-39; Guangzhou RiboBio Co., Ltd.) were used to
compare miRNA levels among different groups. The fold change was
calculated based on the normalized mean differences
(2−ΔΔCq) (19). The PCR
cycling conditions were as follows: Initial denaturation at 95°C
for 10 min, followed by 40 cycles at 95°C for 2 sec, 60°C for 20
sec and 70°C for 10 sec. The following primers for miR-451a were
used: Sense, 5′-ACCGTTACCATTACT-3′ and antisense,
5′-CTCACACGACTCACGA-3′. All reactions, including no-template
controls, were performed in triplicate. Amplification and data
analysis were performed using an ABI Prism 7,000 thermocycler
(Applied Biosystems; Thermo Fisher Scientific, Inc.).
Primary plasma cells were purified from BM aspirates
obtained from patients with MM and healthy volunteers using CD138
magnetic beads (Miltenyi Biotec, Inc.). Total RNA of cells was
extracted using Tripure isolation reagent (Roche Diagnostics, Inc.)
and reverse transcribed into cDNAs with a first stand cDNA
synthesis kit using random primers (Roche Diagnostics). The
duration of RT was as following: 25°C for 10 min, 55°C for 30 min,
85°C for 5 min, 10°C for 15 min and end at 4°C. qPCR was performed
to analyze IL-6R expression using SYBR Premix Ex Taq (Takara Bio,
Inc.). The qPCR consisted of an initial denaturation step at 95°C
for 5 min, followed by 39 cycles at 95°C for 10 sec, 60°C for 30
sec, and a final elongation step at 95°C for 15 sec, 60°C for 1 min
and 95°C for 15 sec. The following primers for IL-6R were used:
Sense, 5′-CCTCTGCATTGCCATTGTTC-3′ and antisense,
5′-GAGATGAGAGGAACAAGCAC-3′.
ELISA
The serum level of IL-6 in serum was measured using
a human IL-6 ELISA kit (Abcam; cat. no. ab46027), according to the
manufacturer's instructions. After color development was stopped,
the absorbance was measured using a microtiter plate-computerized
reader set to a wavelength at 450 nm. The sensitivity for IL-6 was
2 pg/ml.
Western blotting
Western blotting was used to analyze the levels of
IL-6R in plasmocytes. Proteins were extracted from cells with a
lysis buffer containing PMSF (Beijing Solarbio Science &
Technology Co., Ltd.) and concentrations were measured using a BCA
protein assay kit (Tiangen Biotech Co., Ltd.). A total of 20–30 µg
protein extracts were subjected to electrophoresis on 10% SDS-PAGE
gels and transferred to nitrocellulose membranes. The membranes
were blocked with 10% skimmed milk for 2 h at room temperature and
then incubated with the following primary antibodies overnight at
4°C: Rabbit anti-IL-6R (1:400; Abcam; cat. no. ab27321) and rabbit
anti-GAPDH (1:1,000; Abcam; cat. no. ab8245). After incubation with
the secondary antibody conjugated with horseradish peroxidase
(1:2,000; Abcam; cat. no. ab6789) for 2 h at room temperature, the
proteins were visualized with the Molecular Imager Gel Doc XR
System (Bio-Rad Laboratories, Inc.) using Metal Enhanced DAB
Substrate kit (Beijing Solarbio Science & Technology Co.,
Ltd.).
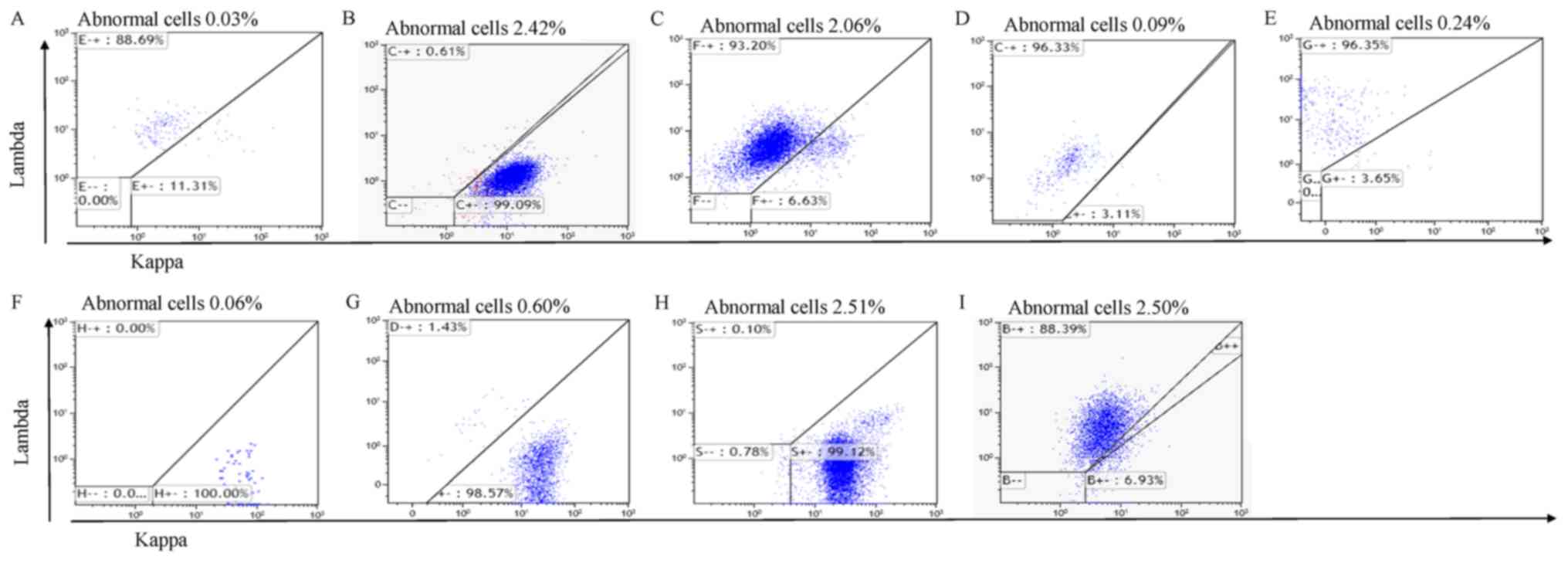
Flow cytometry
In follow-up experiments, MRD was assessed using
MFC. A fixative-free erythrocyte lysis buffer (Beckman Coulter,
Inc.) was used for the phenotypic characterization of the majority
of participants. Intraprep permeabilization reagent (cat. no.
A07803; Beckman Coulter, Inc.) was used to analyze intracellular
immunoglobulin levels at room temperature. The number and viability
of cells obtained were determined using staining cells with trypan
blue for 10 min at room temperature and counted under a microscope.
If the viability of recovered cells was ≥90%, 20 million cells were
stained with two independent 6-color panels (10 million each): One
tube contained CD19-FITC, CD20-PE, CD56-ECD, CD38-PECY5.5,
CD138-APC and CD45-PECy7, and the other tube contained CD19-FITC,
CD117-PE, CD56-ECD, CD38-PECY5.5, CD138-APC and CD45-PECy7. The
intracytoplasmic panel consisted of the markers Cyκ-APC,
Cyλ-APC750, CD19-PE, CD38-FITC and CD138-APC. In total, 10 million
cells suspended at a final volume of 200 µl/tube were labeled in
each tube. A minimum of 5 million events were recorded per tube in
the mode of middle speed in a flow cytometer (Navios' Beckman
Coulter, Inc.). The software used for analysis was Kaluza (version
no. 2.1; Beckman Coulter, Inc.). Cell populations are considered
abnormal if they have an atypical differentiation pattern, an
increased or decreased expression level of normal antigens, an
asynchronous maturational pattern or express aberrant antigens
(20).
miR-451a transfection
For miR-451a mimic transfections, the miR-451a mimic
(5′-AAACCGUUACCAUUACUGAGUU-3′; 50 nM; Guangzhou RiboBio Co., Ltd.)
and the corresponding negative control (50 nM; Guangzhou RiboBio
Co., Ltd.) were separately transfected into 293T cells (purchased
from China Center for Type Culture Collection) were performed
separately in the absence of any other treatments. The
transfections were performed using Lipofectamine 3000 (Thermo
Fisher Scientific, Inc.). Then, 48 h after transfection, the
expression of miR-451a was measured using RT-qPCR to confirm
successful transfections, and the subsequent effect was only
determined in cells expressing miR-451a.
Luciferase reporter assay
The online software TargetScan (TargetScan Human
7.0; http://www.targetscan.org/vert_70/) was used to
investigate the relationship between miR-451a and IL-6R. The
predicted binding site for miR-451a to human IL-6R 3′UTR was
5′-UUGCCAAA-3′ (17,21). The human IL-6R 3′UTR with a mutation
in the miR-451a seed sequence (mutant; mut) and 3′UTR (wild-type;
WT) were amplified and inserted between the restriction sites
XhoI and NotI of the firefly and Renilla
luciferase reporter vector pmiR-RB-REPORT (Guangzhou RiboBio Co.,
Ltd.), respectively. Cells were plated on 24-well plates at
1×105 cells/well and transfected with luciferase
reporter vectors (30 ng) using Lipofectamine 3,000 (Thermo Fisher
Scientific, Inc.). At 24 h post-transfection, firefly and
Renilla luciferase activities were measured using a Dual-Glo
Luciferase assay system (Promega Corporation), according to the
manufacturer's instructions. The Renilla luciferase signal
was normalized to that of the firefly luciferase signal for each
individual analysis.
Statistical analysis
Data are presented as the medians and interquartile
ranges. The experiments were repeated three times. The
Kolmogorov-Smirnov test was used to analyze whether the data were
normally distributed. The Kruskal-Wallis rank sum test was used to
analyze differences in the data among the four groups. Dunn's test
was used as the post hoc test after Kruskal Wallis. Comparisons
between two groups were performed using the Mann-Whitney U test.
After the Mann-Whitney U test, a Bonferroni was conducted to
correct the P-value. Correlations were determined by calculating
Spearman's correlation coefficients. Statistical analyses were
performed using SPSS version 19.0 (IBM Corp.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Baseline characteristics and
treatments
In the present study, the median age of the patients
was 63.0 years (age range, 38–88 years), and 56.1% of patients were
male. The patients received immunomodulatory drugs or proteasome
inhibitors as induction therapy without autologous stem cell
transplantation. In total, 15 of these patients (22.7%) were R-ISS
stage I, 17 patients (25.8%) were R-ISS stage II and 34 patients
(51.5%) were R-ISS stage III (Table
I).
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
|
Characteristics | Patients |
|---|
| Sex |
|
|
Male | 37 (56.1%) |
|
Female | 29 (43.9%) |
| Age, years | 63 (38–88) |
| Isotype |
|
| IgG
type | 35 (53.0%) |
| IgA
type | 17 (25.8%) |
| IgM
type | 0 |
| IgE
type | 0 |
| IgD
type | 1 (1.5%) |
| Light
chain type | 13 (19.7%) |
| R-ISS Stage |
|
| I | 15 (22.7%) |
| II | 17 (25.8%) |
|
III | 34 (51.5%) |
| Cytogenetics |
|
| Gain
1q21 | 12 (13.6%) |
| 17p
deletion | 6 (9.0%) |
| t
(11;14) | 8 (12.1%) |
| t
(14;16) | 4 (6.0%) |
| t
(4;14) | 5 (7.6%) |
| t
(14;20) | 1 (1.5%) |
|
Trisomy | 27 (40.9%) |
|
Unknown | 3 (4.5%) |
| LDH level exceeding
the normal range | 11 (16.7%) |
Expression of miR-451a in the BM and
circulation of patients with MM
Low circulating miR-451a levels were detected in
patients with MM, and were only 0.39 times the value of the control
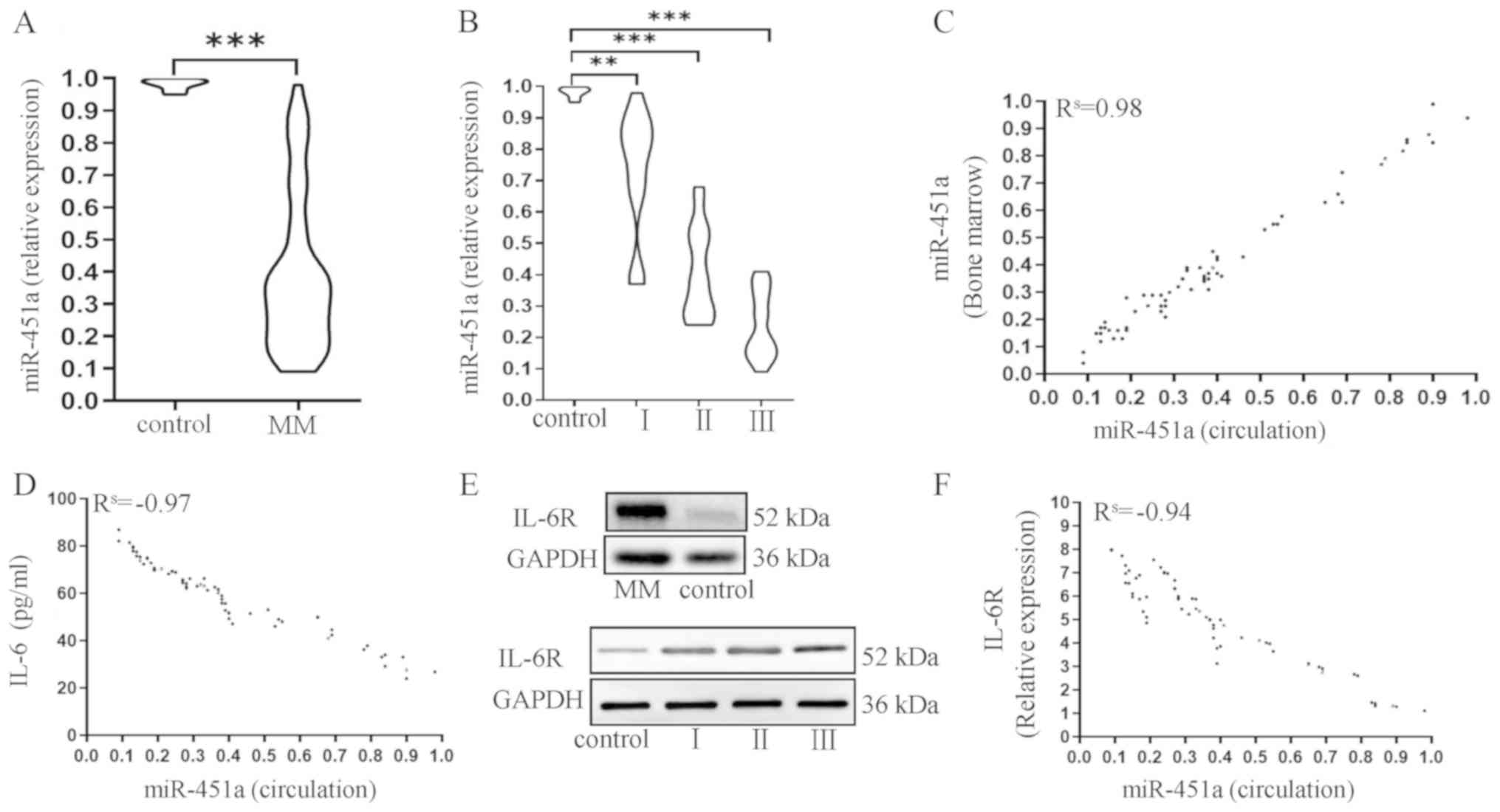
group (U=4.00; P<0.001; Fig. 1A).
Among the 66 patients with MM, the median expression of miR-451a
was 0.73 times the value of the control group in R-ISS stage I MM;
and that in R-ISS stage II MM was 0.41 times the value of the
control group (Fig. 1B). Moreover,
patients with R-ISS stage III MM presented the lowest level, at
0.24 times the value of the control group (Fig. 1B).
The expression of miR-451a was compared between
cells in the BM and circulation to evaluate the consistency. The
expression of miR-451a in the circulation exhibited a strong
positive correlation with miR-451a expression in BM
(Rs=0.98; P<0.001; Fig.
1C). The concentration of IL-6 was strongly, negatively
correlated with miR-451a expression (Rs=−0.97;
P<0.0001; Fig. 1D). The plasma
cells from patients with MM demonstrated an upregulated expression
of IL-6R compared with the control subjects via western blotting.
Furthermore, the expression levels increased gradually from stage I
to stage III (Fig. 1E). IL-6R levels
were strongly and negatively correlated with miR-451a expression
(Rs=−0.95; P<0.001; Fig.
1F).
After two courses of consolidation chemotherapy, 19
patients achieved complete remission (CR). In the 2-year follow-up
period, MRD was observed using MFC and changes in circulating
miR-451a expression were evaluated. For 10 patients, the expression
of circulating miR-451a remained steady in the follow-up period
(seven patients with R-ISS stage I, two with R-ISS stage II and one
with R-ISS stage III). The change in miR-451a expression was
<50%. The 10 patients were in continuous CR (Fig. 2A). However, the other nine patients
(two patients with R-ISS stage I, three with R-ISS stage II and
four with R-ISS stage III) demonstrated an abrupt decrease in
circulating miR-451a expression during the follow-up period after
CR (Fig. 2B-J). The value at the
observation point was >50% lower compared with the value
measured 3 months prior (50%; Fig.
2G-J). The turning points in the trend appeared 4–8 weeks
before abnormal plasma cells were obtained via MFC (Fig. 3A-I). Thus, when MFC results were
still negative, significant changes in circulating miR-451a
expression had occurred. Subsequently, clinical relapse occurred in
these nine patients.
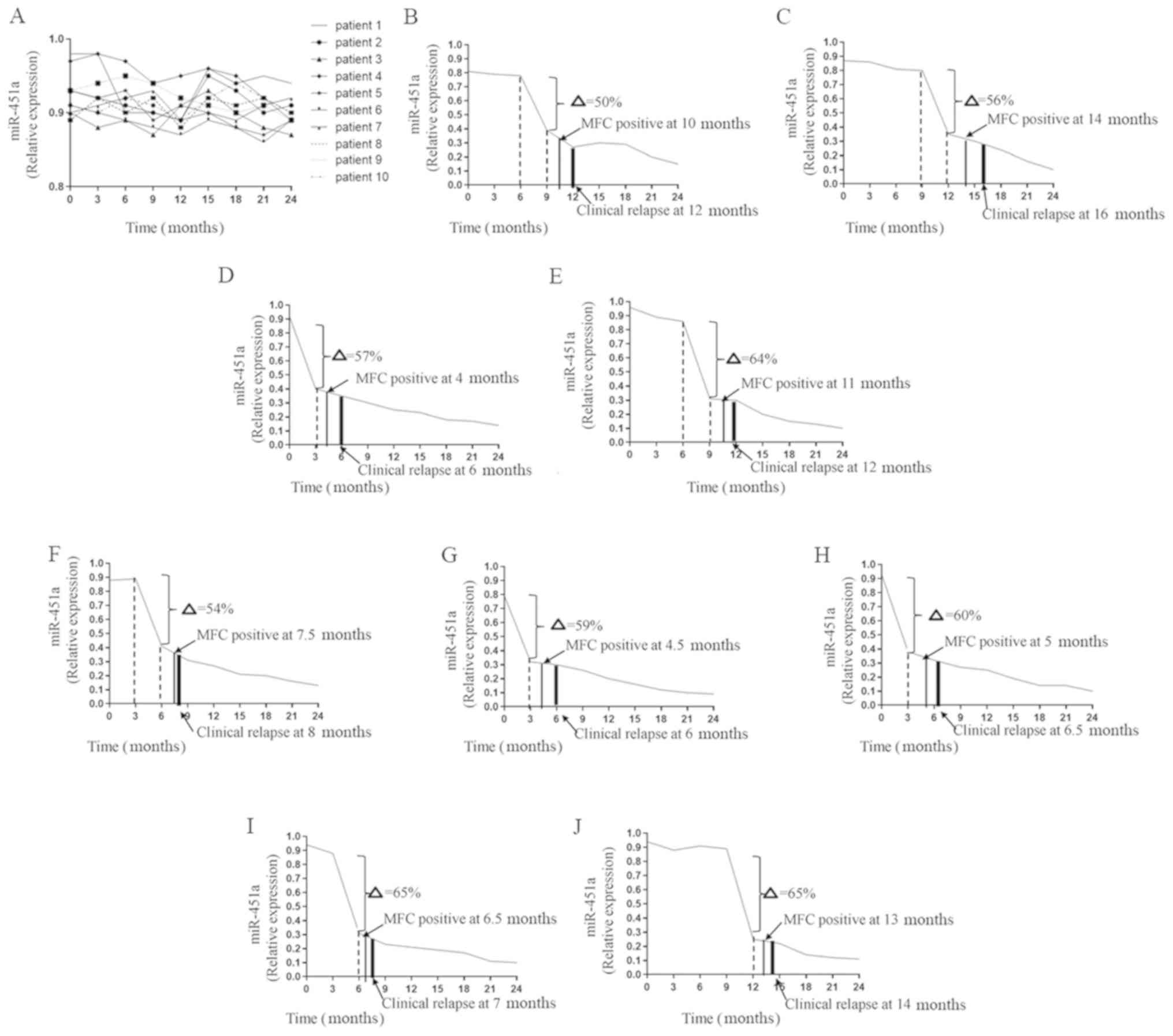 | Figure 2.Follow-up of patients who experienced
CR. (A) In total, 10 patients experienced long-term remission.
(B-J) In total, nine patients relapsed during the follow-up period.
(B) During the follow-up period, the expression of miR-451a at the
9th month after CR decreased by 50% compared with the value at the
6th month (δ=50%). MRD was reported positive using MFC at the 10th
month. The patient experienced a clinical relapse at the 12th
month. (C) MFC positive at the 14th month, clinical relapse at the
16th month, δ=56%. (D) δ=57%, MFC positive at the 4th month, and
clinical relapse at the 6th month. (E) δ=64%, MFC positive at the
11th month, and clinical relapse at the 12th month. (F) δ=54%, MFC
positive at the 7.5th month, and clinical relapse at the 8th month.
(G) δ=59%, MFC positive at the 4.5th month, and clinical relapse at
the 6th month. (H) δ=60%, MFC positive at the 5th month, and
clinical relapse at the 6.5th month. (I) δ=65%, MFC positive at the
6.5th month, and clinical relapse at the 7th month. (J) δ=65%, MFC
positive at the 13th month, and clinical relapse at the14th month.
CR, complete remission; MFC, multiparameter flow cytometry; miR,
microRNA. |
IL-6R is a direct target of
miR-451a
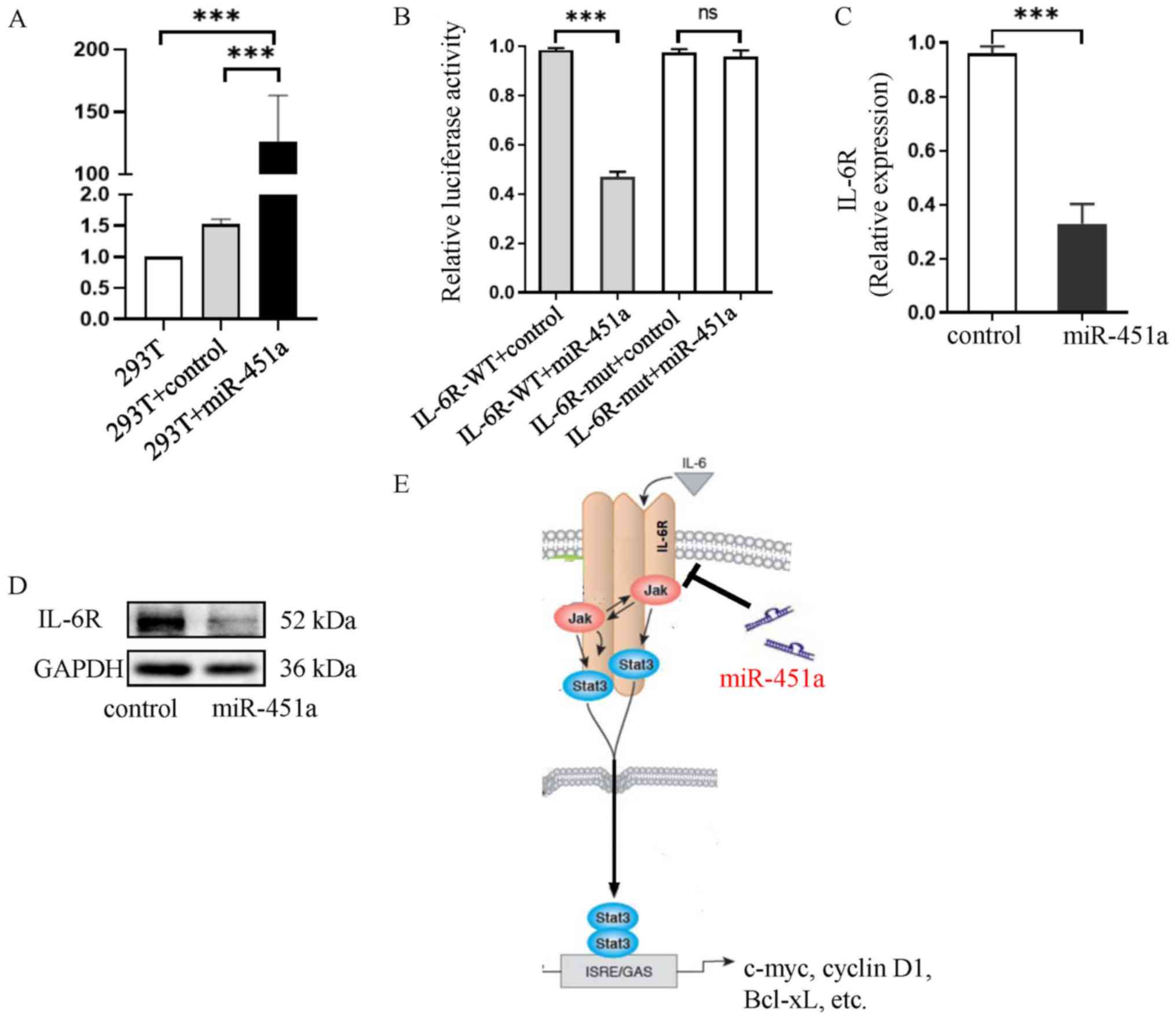
Significantly higher miR-451a expression was
observed in the mimic group compared with the mimic control cells
and in non-transfected cells (both P<0.001; Fig. 4A). The dual-luciferase reporter assay
results revealed that co-transfection of the miR-451a mimic and
IL-6R-wild-type (WT) resulted in decreased the luciferase activity
compared with co-transfection of the mimic control and IL-6R-WT
(P<0.001; Fig. 4B). However,
there was no effect of co-transfection of the miR-451a mimic and
IL-6R-mut had no effect on the luciferase activity (P>0.05;
Fig. 4B). Furthermore, the
overexpression of miR-451a significantly downregulated IL-6R
expression both at the mRNA (P<0.001; Fig. 4C) and protein (Fig. 4D) levels. Collectively, these results
suggested that IL-6R was a target gene of miR-451a and was
negatively regulated by miR-451a (Fig.
4E).
Discussion
With the wide application of novel drugs, the depth
and duration of remission in patients with MM are continuously
improving, but patients still eventually relapse or progress
(3). The heterogeneity of patients
who relapse requires an individualized assessment of these patients
to determine the timing and medication used for treatment. Due to
the biological complexity and dynamic nature of therapy, sampling a
single site of the disease is unable to fully assess the clonal
complexity of multisite metastatic disease in patients (7). Therefore, cell-free nucleic acids in
circulation are useful for detecting the genomic alterations
present in patients with genetically heterogeneous diseases. In the
current study, a circulating miRNA, miR-451a, was indicated to be
an efficient approach to monitor dynamic tumor load and predict
relapse.
MM has a unique characteristic, namely, myeloma
cells are not evenly distributed in BM, which are called foci; this
characteristic often causes false negative results for MRD
(7). Furthermore, MM is
characterized by a high level of heterogeneity, as subclones cause
clonal evolution. Thus, one type of mutation does not effectively
characterize most patients with MM (22). Although the R-ISS stratification
system is a powerful prognostic tool to predict outcomes in
patients with myeloma, it does not reflect the real time tumor load
in real time (3). Therefore, a new
marker that is universal, rapid and can be monitored in real time
is urgently required. To conduct follow-up tests more conveniently
and avoid false negatives, the present study attempted to develop a
non-invasive approach.
A previous study reported that extracellular miRNA
levels are stable in body fluids, and are retrievable and
measurable from fresh or archived serum and plasma samples
(23). In patients with cancer, the
use of a less invasive method than tissue biopsy is needed, and it
may lead to a major difference in clinical outcomes. Hence, a large
number of studies have assessed the potential use of circulating
miRNAs as biomarkers of various types of cancer, including breast
cancer, gastric cancer, hepatocellular cancer and non-small cell
lung cancer (24–26). Moreover, miRNA signatures in blood
are similar in men and women, as well as in individuals of
different ages (27). Circulating
miRNAs originate from microvesicles (released by exocytosis),
exosomes (formed by the invagination of the early endosome and
released upon the fusion of the late endosome with the plasma
membrane) and apoptotic vesicles and/or senescent bodies (27–29). In
the current study, circulating miR-451a levels were measured using
RT-qPCR, which is more sensitive compared with MFC, and more
economical compared with next-generation sequencing. A stationary
state of circulating miR-451a expression suggested remission, but
an abrupt decrease in expression predicted relapse in the present
study. This change often occurred prior to MFC and clinical
relapse. However, the definition of an ‘abrupt decrease’ remains
unclear. Based on the current results, 50% may be the rate of
change in miR-451a expression. However, the number of samples must
be increased to identify the exact threshold.
The abnormal expression of miR-451a is observed in
multiple solid malignant tumor types, and its expression is
negatively correlated with the degree of malignancy (30–32). In
the current research, the downregulation of miR-451a expression in
the serum of patients with MM distinguished patients with MM from
healthy individuals. Moreover, a decline in miR-451a expression is
significantly correlated with poor clinical prognosis (33). This finding is consistent with the
present study. However, the mechanism has not yet been elucidated.
In humans and rats, miR-451a displays a tissue-specific expression
pattern, and it is highly expressed in BM (34). As BM is part of the hematological
system, miR-451a release into the blood may be more likely to
occur. Long non-coding RNAs (lncRNAs) have been revealed to sponge
miRNAs to decrease their expression (35). In terms of miR-451a, LINC00657 and
AC084082.3 are shown to interact with miR-451a in BM (36). In MM, IL-6, a vital growth factor for
myeloma cells, can induce the upregulation of several lncRNAs
(37), and these lncRNAs may cause
miR-451a dysregulation. However, which lncRNA can sponge miR-451a
in MM remains unknown. In the present study, IL-6R was identified
to be a target of miR-451a. Thus, it was indicated that miR-451a
was associated with the IL-6R/JAK2/STAT3 pathway. However, whether
the lncRNA that may sponge miR-451a is also associated with this
pathway is yet to be elucidated, and will be examined in future
studies.
The downstream mechanism of downregulated miR-451a
that influences MM is not fully understood. In osteosarcoma,
miR-451a inhibits the proliferation, migration and angiogenesis of
osteosarcoma cells by silencing IL-6R (21). In the present study, miR-451a was
negatively associated with the R-ISS stage, and negatively
correlated with IL-6 and IL-6R. The target verification experiment
identified the relationship between miR-451a and IL-6R. Therefore,
miR-451a may function via the IL-6R/JAK2/STAT3 pathway. Previous
studies have reported that miR-451a could change the levels of JAK2
and STAT3 phosphorylation to induce apoptosis in myeloma cells
(38,39). Therefore, this mechanism could apply
to clinical transformation, and miR-451a has the potential to be a
biomarker to predict the therapy outcome of a patient. In addition
to the JAK2/STAT3 pathway, IL-6 could also promote the
proliferation of myeloma cells via the Ras/MAPK pathway (40) and regulate the PI3K/AKT pathway to
reduce apoptosis in MM cells (41).
IL-6 interacts with VEGF to promote angiogenesis, migration and
invasion (42). Moreover, numerous
pathways activated by IL-6 could activate NF-Кb (43). Therefore, miR-451a may serve a role
in other mechanisms, but these have yet to be confirmed.
In conclusion, miR-451a was associated with the
R-ISS stage and tracked dynamic changes in the tumor. Circulating
miR-451 could reflect the tumor load in real time and may be used
to monitor MRD in patients with MM. An early warning sign,
especially a non-invasive method, will allow for the anticipation
of the disease. For instance, patients may start treatment if there
is a dramatic and rapid decrease in miR-451a. However, if the rate
of decline is slow, observation is recommended. Although the number
of cases in the current study are limited, the available data
demonstrate the value of miR-451a in early recurrence after
chemotherapy in patients with MM. In the future, the sample size
will continue to further enlarged to clarify and determine the
prognostic value of miR-451a.
Acknowledgements
The manuscript was based on an abstract appearing in
the 2019 Abstract Proceedings book of the ISLH (International
Society of Laboratory Hematology; Volume 41, Issue S2; abstract no.
208). The authors appreciate the advice provided by Mr. Yi Shi
(Sichuan Academy of Medical Sciences and Sichuan Provincial
People's Hospital, School of Medicine, University of Electronic
Science and Technology of China), who assisted with analyzing
circulating miR-451a expression.
Funding
This work was supported by the Health Commission of
Sichuan Province (grant no. 150193), the Commission of the Cardre
Health Care in Sichuan Province (grant no. 2017-228), the Science
and Technology Department of Sichuan Province (grant no. 19YJ0593),
the Sichuan Provincial People's Hospital (grant no. 2018LY03), the
Chengdu Science and Technology Bureau (grant no.
2015-HM01-00470-SF) and the Science and Technology Department of
Sichuan Province (grant no. 2020YFS0433).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ, HJ, ZX, MZ, DC, YH, JZ, TJ and JC participated
in the design, interpretation of the studies, analysis of the data
and review of the manuscript. LZ, XJ and ZX conducted the
experiments. MZ conducted statistical analysis. DC and YH followed
up the patients. LZ, JZ, TJ and JC wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was performed with the approval of the
Institutional Review Board of Sichuan Provincial People's Hospital
of China. Informed written consent was collected from each eligible
patient, and the whole study was performed in accordance with the
Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Palumbo A and Anderson K: Multiple
myeloma. N Enq J Med. 364:1046–1060. 2011. View Article : Google Scholar
|
|
2
|
Rajkumar SV: Multiple myeloma: Every year
a new standard? Hematol Oncol. 37 (Suppl 1):S62–S65. 2019.
View Article : Google Scholar
|
|
3
|
Kumar SK, Rajkumar V, Kyle RA, van Duin M,
Sonneveld P, Mateos MV, Gay F and Anderson KC: Multiple myeloma.
Nat Rev Dis Primers. 3:170462017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tandon N, Rajkumar SV, LaPlant B,
Pettinger A, Lacy MQ, Dispenzieri A, Buadi FK, Gertz MA, Hayman SR,
Leung N, et al: Clinical utility of the revised international
staging system in unselected patients with newly diagnosed and
relapsed multiple myeloma. Blood Cancer J. 7:e5282017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martin T and Huff CA: Multiple myeloma:
Current advances and future directions. Clin Lymphoma Myeloma Leuk.
19:255–263. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kumar S, Paiva B, Anderson KC, Durie B,
Landgren O, Moreau P, Munshi N, Lonial S, Bladé J, Mateos MV, et
al: International myeloma working group consensus criteria for
response and minimal residual disease assessment in multiple
myeloma. Lancet Oncol. 17:e328–e346. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paiva B, García-Sanz R and San Miguel JF:
Multiple myeloma minimal residual disease. Cancer Treat Res.
169:103–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Furukawa M, Ohkawara H, Ogawa K, Ikeda K,
Ueda K, Shichishima-Nakamura A, Ito E, Imai JI, Yanagisawa Y, Honma
R, et al: Autocrine and paracrine interactions between multiple
myeloma cells and bone marrow stromal cells by growth
arrest-specific gene 6 cross-talk with interleukin-6. J Biol Chem.
292:4280–4292. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chong PSY, Zhou J, Lim JSL, Hee YT, Chooi
JY, Chung TH, Tan ZT, Zeng Q, Waller DD, Sebag M and Chng WJ: IL6
promotes a STAT3-PRL3 feedforward loop via SHP2 repression in
multiple myeloma. Cancer Res. 79:4679–4688. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jurczyszyn A, Czepiel J, Biesiada G,
Gdula-Argasińska J, Cibor D, Owczarek D, Perucki W and Skotnicki
AB: HGF, sIL-6R and TGF-β1 play a significant role in the
progression of multiple myeloma. J Cancer. 5:518–524. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Altuvia Y, Landgraf P, Lithwick G, Elefant
N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T and Margalit H:
Clustering and conservation patterns of human microRNAs. Nucleic
Acids Res. 33:2697–2706. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang R, Wang ZX, Yang JS, Pan X, De W and
Chen LB: MicroRNA-451 functions as a tumor suppressor in human
non-small cell lung cancer by targeting ras-related protein 14
(RAB14). Oncogene. 30:2644–2658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang JY, Zhang K, Chen DQ, Chen J, Feng
B, Song H, Chen Y, Zhu Z, Lu L, De W, et al: MicroRNA-451:
Epithelial-mesenchymal transition inhibitor and prognostic
biomarker of hepatocelluar carcinoma. Oncotarget. 6:18613–18630.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim Y, Powathil G, Kang H, Trucu D, Kim H,
Lawler S and Chaplain M: Strategies of eradicating glioma cells: A
multi-scale mathematical model with MiR-451-AMPK-mTOR control. PLoS
One. 10:e1143702015.
|
|
15
|
Yuan J, Lang J, Liu C, Zhou K, Chen L and
Liu Y: The expression and function of miRNA-451 in osteosarcoma.
Med Oncol. 32:3242015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gits CM, van Kuijk PF, Jonkers MB, Boersma
AW, Smid M, van Ijcken WF, Coindre JM, Chibon F, Verhoef C,
Mathijssen RH, et al: MicroRNA expression profiles distinguish
liposarcoma subtypes and implicate miR-145 and miR-451 as tumor
suppressors. Int J Cancer. 135:348–361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu X, Zhang A, Xiang J, Lv Y and Zhang X:
miR-451 acts as a suppressor of angiogenesis in hepatocellular
carcinoma by targeting the IL-6R-STAT3 pathway. Oncol Rep.
36:1385–1392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dimopoulos M, Terpos E, Comenzo RL, Tosi
P, Beksac M, Sezer O, Siegel D, Lokhorst H, Kumar S, Rajkumar SV,
et al: IMWG. International myeloma working group consensus
statement and guidelines regarding the current role of imaging
techniques in the diagnosis and monitoring of multiple myeloma.
Leukemia. 23:1545–1556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Flores-Montero J, Sanoja-Flores L, Paiva
B, Puig N, García-Sánchez O, Böttcher S, van der Velden VHJ,
Pérez-Morán JJ, Vidriales MB, García-Sanz R, et al: Next generation
flow for highly sensitive and standardized detection of minimal
residual disease in multiple myeloma. Leukemia. 31:2094–2103. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu SY, Deng SY, He YB and Ni GX: MiR-451
inhibits cell growth, migration and angiogenesis in human
osteosarcoma via down-regulating IL 6R. Biochem Biophys Res Commun.
482:987–993. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Corre J, Cleynen A, Robiou du Pont S,
Buisson L, Bolli N, Attal M, Munshi N and Avet-Loiseau H: Multiple
myeloma clonal evolution in homogeneously treated patients.
Leukemia. 32:2636–2647. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schwarzenbach H, Nishida N, Calin GA and
Pantel K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Filipów S and Łaczmański Ł: Blood
circulating miRNAs as cancer biomarkers for diagnosis and surgical
treatment response. Front Genet. 10:1692019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oura K, Fujita K, Morishita A, Iwama H,
Nakahara M, Tadokoro T, Sakamoto T, Nomura T, Yoneyama H, Mimura S,
et al: Serum microRNA-125a-5p as a potential biomarker of
HCV-associated hepatocellular carcinoma. Oncol Lett. 18:882–890.
2019.PubMed/NCBI
|
|
26
|
Hagrass HA, Sharaf S, Pasha HF, Tantawy
EA, Mohamed RH and Kassem R: Circulating microRNAs-a new horizon in
molecular diagnosis of breast cancer. Genes Cancer. 6:281–287.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui
R and Zhang X: Circulating MicroRNAs in cancer: Potential and
challenge. Front Genet. 10:6262019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mollashahi B, Aghamaleki FS and Movafagh
A: The roles of miRNAs in medulloblastoma: A systematic review. J
Cancer Prev. 24:79–90. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schwarzenbach H and Gahan PB: MicroRNA
shuttle from cell-to-cell by exosomes and its impact in cancer.
Noncoding RNA. 5:282019.
|
|
30
|
Zeng T, Peng L, Chao C, Fu B, Wang G, Wang
Y and Zhu X: miR-451 inhibits invasion and proliferation of bladder
cancer by regulating EMT. Int J Clin Exp Pathol. 7:7653–7662.
2014.PubMed/NCBI
|
|
31
|
Liu F, Bu Z, Zhao F and Xiao D: Increased
T-helper 17 cell differentiation mediated by exosome-mediated
microRNA-451 redistribution in gastric cancer infiltrated T cells.
Cancer Sci. 109:65–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang W, Zhang L, Wang Y, Ding Y, Chen T,
Wang Y, Wang H, Li Y, Duan K, Chen S, et al: Involvement of miR-451
in resistance to paclitaxel by regulating YWHAZ in breast cancer.
Cell Death Dis. 8:e30712017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rastgoo N, Abdi J, Hou J and Chang H: Role
of epigenetics-microRNA axis in drug resistance of multiple
myeloma. J Hematol Oncol. 10:1212017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ludwig N, Leidinger P, Becker K, Backes C,
Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E
and Keller A: Distribution of miRNA expression across human
tissues. Nucleic Acids Res. 44:3865–3877. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Panoutsopoulou K, Avgeris M and Scorilas
A: miRNA and long non-coding RNA: Molecular function and clinical
value in breast and ovarian cancers. Expert Rev Mol Diagn.
18:963–979. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Balakrishnan I, Yang X, Brown J,
Ramakrishnan A, Torok-Storb B, Kabos P, Hesselberth JR and Pillai
MM: Genome-wide analysis of miRNA-mRNA interactions in marrow
stromal cells. Stem Cells. 32:662–673. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Binder S, Zipfel I, Friedrich M, Riedel D,
Ende S, Kämpf C, Wiedemann K, Buschmann T, Puppel SH, Reiche K, et
al: Master and servant: LINC00152-a STAT3-induced long noncoding
RNA regulates STAT3 in a positive feedback in human multiple
myeloma. BMC Med Genomics. 13:222020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
De Oliveira MB, Fook-Alves VL, Eugenio
AIP, Fernando RC, Sanson LFG, de Carvalho MF, Braga WMT, Davies FE
and Colleoni GWB: Anti-myeloma effects of ruxolitinib combined with
bortezomib and lenalidomide: A rationale for JAK/STAT pathway
inhibition in myeloma patients. Cancer Lett. 403:206–215. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Brown R, Yang S, Weatherburn C, Gibson J,
Ho PJ, Suen H, Hart D and Joshua D: Phospho-flow detection of
constitutive and cytokine-induced pSTAT3/5, pAKT and pERK
expression highlights novel prognostic biomarkers for patients with
multiple myeloma. Leukemia. 29:483–490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gocke CB, McMillan R, Wang Q, Begum A,
Penchev VR, Ali SA, Borrello I, Huff CA and Matsui W: IQGAP1
Scaffold-MAP kinase interactions enhance multiple myeloma
clonogenic growth and self-renewal. Mol Cancer Ther. 15:2733–2739.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mimura N, Hideshima T, Shimomura T, Suzuki
R, Ohguchi H, Rizq O, Kikuchi S, Yoshida Y, Cottini F, Jakubikova
J, et al: Selective and potent Akt inhibition triggers anti-myeloma
activities and enhances fatal endoplasmic reticulum stress induced
by proteasome inhibition. Cancer Res. 74:4458–4469. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Berenstein R, Nogai A, Waechter M, Blau O,
Kuehnel A, Schmidt-Hieber M, Kunitz A, Pezzutto A, Dörken B and
Blau IW: Multiple myeloma cells modify VEGF/IL-6 levels and
osteogenic potential of bone marrow stromal cells via
Notch/miR-223. Mol Carcinog. 55:1927–1939. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Matthews GM, de Matos Simoes R, Dhimolea
E, Sheffer M, Gandolfi S, Dashevsky O, Sorrell JD and Mitsiades CS:
NF-κB dysregulation in multiple myeloma. Semin Cancer Biol.
39:68–76. 2016. View Article : Google Scholar : PubMed/NCBI
|