Introduction
Clear cell renal cell carcinoma (ccRCC) is a cancer
in which malignant cells form in the tubules of the kidney
(1,2). ccRCC constitutes >85% of kidney
cancers, with an estimated 403,262 new cases (1,3,4), and 175,098 deaths in 2018, and the
mortality rate is likely to continue to grow (5,6).
Although there are several types of standard treatments, nearly
half of the diagnosed patients succumb within five years (3,7).
Therefore, a better understanding of the molecular mechanisms
underlying this disease could help identify new therapeutic
targets. In particular, insight into the networks that control
signaling cascades associated with cell proliferation and migration
may lead to the discovery of novel target genes for ccRCC
treatment.
Transcription factor AP-2β (TFAP2B) is a
member of the TFAP2 protein family, which includes TFAP2-A, -B, -C,
-D and -E (8). Previous studies have
revealed that TFAP2 proteins serve as retinoic acid-inducible
transcriptional activators and serve important roles in cell growth
and differentiation (9–11). A knockout of TFAP2B enhanced
apoptotic renal epithelial cell death in mice (12). However, TFAP2B overexpression
has been demonstrated to promote tumor growth, thus, contributing
to a poor prognosis in human lung adenocarcinoma (13). In the present study, a systematic
investigation of the effects of TFAP2B on ccRCC in
vitro is described for the first time.
MicroRNAs (miRNAs) are a family of endogenous
non-coding single-stranded RNA molecules with a length of 19–22
nucleotides that may act either as tumor suppressors or oncogenes,
according to the function of their target genes (14). In the present study it was
demonstrated that miRNA (miR)-142-5p serves as an onco-miRNA in
ccRCC by targeting TFAP2B and downregulating its expression,
promoting cell proliferation and migration. Thus, the findings of
the present study revealed that the miR-142-5p/TFAP2B
pathway may provide potential therapeutic targets for treatment of
ccRCC.
Methods and materials
Cell lines and cell culture
293T cells, the human kidney cell line HK-2 and the
ccRCC cell lines 786-O and A-498 were obtained from ATCC. All cells
were maintained in DMEM containing 10% FBS, 100 U/ml penicillin,
100 U/ml streptomycin, and 2 mM L-glutamine at 37°C in a 5%
CO2 atmosphere and 21% oxygen.
Bioinformatic analysis
The expression of the TFAP2B was analyzed in
The Cancer Genome Atlas (TCGA) ccRCC database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
The regulation network of the TFAP2B and the interaction
partners of TFAP2B were identified using the STRING
(https://string-db.org) software tool. TargetScan
screen database (http://www.targetscan.org/vert_72/) was used online to
screen the targets and binding sites of miR-142-5p. The
miR-142-5p-associated survival was analyzed on the ONCOMIR website
(http://www.oncomir.org) (15). The total number of patients for
analysis was 517, which was divided into the low expression group
(n=259) and the high expression group (n=258). The statistical
difference in the survival rate was analyzed by the log-rank
test.
Plasmid construction and lentiviral
infection
The genomic DNA of 786-O cells was isolated using a
FastPure Cell DNA Isolation Mini kit (cat. no. DC102; Vazyme
Biotech Co., Ltd.). The 3′UTR of human TFAP2B gene was
amplified from the genomic DNA by PCR using 2X Phanta Master Mix
(cat. no. P511-01; Vazyme Biotech Co., Ltd.). The primers used are
as follows: Forward, 5′-GAGCTCGCTAGCCTCGAGAAATTTTTAAAAAAAGAAGG-3′
and reverse, 5′-CATGCCTGCAGGTCGACTGAAGATGAAAACACAACAATC−3′. The
thermocycling conditions were as follows: 95°C for 5 min, followed
by 30 cycles of 95°C for 15 sec, 60°C for 15 sec and 72°C for 60
sec. The TFAP2B 3′UTR fragment was then cloned downstream to
the firefly luciferase reporter gene in the pmirGLO vector (Promega
Corporation), generating the pmirGLO-TFAP2B 3′UTR WT
plasmid. The mutant TFAP2B 3′UTR was generated using the
QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies,
Inc.) and was also cloned into the pmirGLO vector, generating the
pmirGLO-TFAP2B 3′UTR Mutant plasmid.
The lentivirus-based vector plv-EF1α-PGK-puro
(Xiamen Anti-hela Biological Technology Tarde Co., Ltd.) was used
to overexpress miR-142-5p. The precursor of miR-142-5p was
amplified from the genomic DNA of A-498 cells by PCR using 2X
Phanta Master Mix (cat. no. P511-01; Vazyme Biotech Co., Ltd.) and
cloned into the vector. The primers used were as follows: Forward,
5′-TCACGCGTGCGGCCGCAGCCTGAAGAGTACACGCCG-3′ and reverse,
5′-CTAGGGATCCGGGCCCGGGCGGGCGGCAGCAGTGGCGTG−3′. The thermocycling
conditions were as follows: 95°C for 5 min, followed by 30 cycles
of 95°C for 15 sec, 60°C for 15 sec and 72°C for 30 sec. To prepare
lentiviral particles, 9 µg of plv-EF1α-miR-142-5p-PGK-puro or empty
vector plasmid with packaging plasmids (3 µg pMD2G; 6 µg pspax2;
Xiamen Anti-hela Biological Technology Tarde Co., Ltd.) were
co-transfected into 293T cells in 10-cm dishes using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Lentivirus-containing medium was collected 48 h
after transfection and used to infect A-498 and 786-O cells
(1×106 cells/well, in six-well plates) at a multiplicity
of infection (MOI) of 30. Two days after lentiviral infection,
A-498 and 786-O cells were maintained in the presence of 1.0 µg/ml
puromycin (Sigma-Aldrich; Merck KGaA) for 5 days to generate stable
miR-142-5p-overexpressing cells and empty vector negative control
cells.
The plasmid pCDH-EF1α-TFAP2B-T2A-BSD for
TFAP2B overexpression was constructed using the
lentivirus-based vector pCDH-EF1α-MCS-T2A-BSD (Xiamen Anti-hela
Biological Technology Tarde Co., Ltd.). Lentiviral particles
carrying TFAP2B were produced as aforementioned. The cells
(1×106 cells/well, in six-well plates) stably
overexpressing miR-142-5p were infected with the TFAP2B
lentivirus at a MOI of 30. Two days after lentiviral infection, the
cells were maintained in the presence of 4.0 µg/ml blasticidin S
(Sigma-Aldrich; Merck KGaA) for 10 days to generate A-498 and 786-O
cells stably overexpressing both miR-142-5p and TFAP2B
(miR-142-5p + TFAP2B).
Transfection
786-O and A-498 cells were plated into a 6-well
plate (5×105 cells/well) at 37°C. The next day, the
pCDH-EF1α-MCS-T2A-BSD plasmid (4 µg/well) and the
pCDH-EF1α-TFAP2B-T2A-BSD plasmid (4 µg/well) were
transfected into the cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C. After a 24-h
period of transfection, the cells were harvested for further
experiments.
Reverse transcription-quantitative PCR
(RT-qPCR) assay
RNA isolated from cells were subjected to RT using
Superscript III Reverse Transcriptase (Invitrogen, Thermo Fisher
Scientific, Inc.) at 50°C for 30 min. TFAP2B and 18S were
transcribed using random RT primers; the miR-142-5p RT primer
sequence was
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTAGT-3′ and the U6
RT primer sequence was 5′-CGCTTCACGAATTTGCGTGTCAT-3′. qPCR was
conducted using a Bio-Rad CFX96 system with a ChamQ
SYBR® qPCR Master Mix kit (Vazyme Biotech Co., Ltd.) to
determine the mRNA or miRNA expression levels of the genes of
interest. The method of quantification was as described previously
(16). The thermocycling conditions
were as follows: 94°C for 3 min, followed by 40 cycles of 94°C for
15 sec, 60°C for 20 sec and 72°C for 20 sec. Each detection was
performed in triplicate. Expression levels were normalized to those
of U6 or 18S ribosomal RNA.
The following primers were used for qPCR: miR-142-5p
forward, 5′-GCGCGAACATAAAGTAGAAAGC-3′ and reverse,
5′-AGTGCAGGGTCCGAGGTATT-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; TFAP2B forward,
5′-GTTGAAGATGCCAATAACAGCGG-3′ and reverse,
5′-GGACGGAGCAAAACACCTCGC−3′; and 18S forward,
5′-CGACGACCCATTCGAACGTCT−3′ and reverse,
5′-CTCTCCGGAATCGAACCCTGA−3′.
Western blotting
Cells were lysed in ice-cold RIPA buffer (Sangon
Biotech Co., Ltd.) and the protein was quantified using a BCA
Protein Assay kit (Abcam). Lysates (20 µg/sample) were loaded on
8–12% denaturing SDS-PAGE gels and transferred to a PVDF membrane
(Roche Diagnostics GmbH). Tris-HCl buffer containing 5% bovine
serum albumin (BSA; Beijing Solarbio Science & Technology Co.,
Ltd.) was used to block the membranes at 28°C for 2 h. The membrane
was probed with the primary antibodies prepared in Tris-HCl buffer
containing 5% BSA at 4°C overnight. Subsequently, the membranes
were washed three times with Tris-HCl buffer containing 0.1%
Tween-20, followed by incubation with the appropriate secondary
antibodies prepared in Tris-HCl buffer at 28°C for 1 h. Finally,
the membranes were washed three times, detected and visualized by
an enhanced chemiluminescence detection system (Thermo Fisher
Scientific, Inc.). The antibodies used in this study included
anti-TFAP2B (1:500; cat. no. 13183-1-AP; ProteinTech Group,
Inc.), anti-GAPDH (1:5,000; cat. no. YM3029; ImmunoWay
Biotechnology Company), HRP-conjugated goat anti-rabbit IgG
(1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.) and
HRP-conjugated rabbit anti-mouse IgG (1:1,000; cat. no. 7076; Cell
Signaling Technology, Inc.).
Dual luciferase reporter assay
miR-142-5p mimics (5′-CAUAAAGUAGAAAGCACUACU−3′) and
negative control mimic (mimics ctrl; 5′-UUGUACUACACAAAAGUACUG−3′)
were obtained from Guangzhou RiboBio Co., Ltd. 293T cells were
seeded into a 6-well plate (106 cells/well) and
transfected with mimics ctrl or miR-142-5p mimics (200 pmol/well),
along with the pmirGLO-WT TFAP2B 3′UTR (4 µg/well) or
pmirGLO-Mutant TFAP2B 3′UTR plasmid (4 µg/well) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), and kept at 37 °C. The cells were lysed at 48 h
post-transfection and luciferase activity was measured using the
Dual-Glo Luciferase Assay System (Promega Corporation) according to
the manufacturer's instructions. The firefly luciferase activity
was calibrated to Renilla luciferase activity. Each
treatment was carried out in triplicate.
Cell proliferation assay
A total of 3×103 cells/well were seeded
onto 96-well plates. Cell proliferation was measured using a MTT
cell proliferation assay kit (cat. no. 11465007001; Shanghai Qcbio
Science & Technologies Co., Ltd.) at each time point (0, 24, 48
and 72 h) for 1 h at 37°C. The absorbance was measured at 450 nm.
All experiments were performed in triplicate.
Cell cycle assay
A total of 1×106 cells/well were seeded
onto 6-well plates. Next day, 786-O and A-498 cells were harvested
and fixed in 70% ethanol at 4°C overnight. The fixed cells were
then incubated with PBS containing 10 µg/ml RNase A (Sangon Biotech
Co., Ltd.) and 0.2% Triton X-100 for 30 min at 37°C, and then
stained with 20 µg/ml propidium iodide for 30 min in the dark at
room temperature. The stained cells were analyzed using the
NovoCyte flow cytometer with NovoCyte 1.4.1 software (ACEA
Biosciences, Inc.). All experiments were performed in
triplicate.
Wound healing assay
786-O and A-498 cells with stable overexpression of
TFAP2B were seeded into 6-well plates to reach 100%
confluence. Wound was generated using a 200 µl pipette tip with a
straight scratch. Cells were maintained in serum free medium for 24
h and were observed at ×40 magnification under a bright field
microscope (Motic Incoporation, Ltd.). The percentage of the wound
healing was quantified by ImageJ 1.8.0 software (National
Institutes of Health).
Transwell assay
Migration assays were performed using Transwell
plates with 8-µm pore size membranes. A total of 2.5×105
786-O and A498 cells with stable overexpression of TFAP2B
were plated in the upper chambers of the Transwell plates. After a
24-h period of incubation at 37°C, the migrated cells were stained
with 0.5% toluidine blue and photographed at ×100 magnification
under a bright field microscope (Motic Incoporation, Ltd.). The
migrated cells were counted using ImageJ in three random
fields.
Statistical analysis
All statistical analyses were conducted using SPSS
version 19.0 (IBM Corp.) and GraphPad Prism version 8.0 (GraphPad
Software, Inc.). Data are presented as the mean ± SD. Differences
between two groups were analyzed with Student's t-test, whereas
ANOVA followed by Tukey's post-hoc test was used for multiple
comparisons of three or more experimental groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Database analysis of the expression
profile of TFAP2B and its regulation
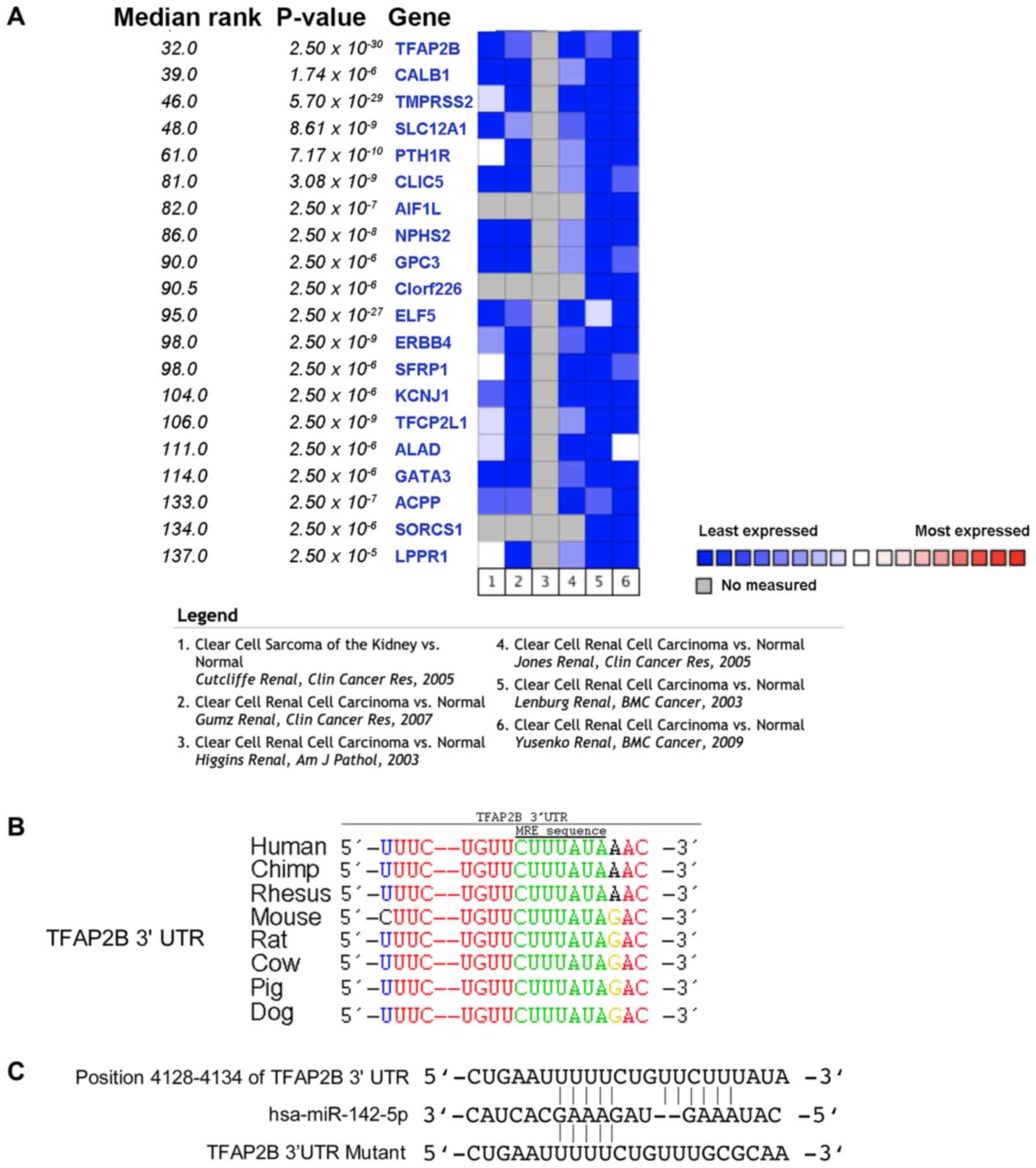
To identify genes that were lowly expressed in RCC,
a screen was conducted using Oncomine software. The TFAP2B
expression level in ccRCC was decreased compared with that in the
normal tissue group; thus, TFAP2B was selected for further
experiments based on the P-value and median expression rank
(Fig. 1A). It is possible that
TFAP2B protein levels may be regulated by miRNAs, as it
contains a conserved miRNA target site in the 3′ UTR region
(Fig. 1B). To confirm this
hypothesis and to determine the possible miRNAs that may target
TFAP2B, a screen using the TargetScan software was
performed, which lead to the identification of miR-142-5p as a
putative miRNA that can bind to TFAP2B 3′UTR regulating its
protein expression (Fig. 1C).
Furthermore, TCGA ccRCC database analysis demonstrated that high
levels of miR-142-5p is negatively associated with lower
TFAP2B mRNA expression in patient samples (Fig. S1A and B). Moreover, the miR-142-5p
expression was significantly associated (log-rank test) with the
hazard ratio of death status in various cancer types in the TCGA
public database using Kaplan-Meier analysis by long-rank tests
between the miR-142-5p high expression group and the miR-142-5p low
expression group (Fig. S1C). And
the expression level of miR-142-5p was significantly associated
with the survival rate in the kidney cancer (Fig. S1C). Furthermore, the STRING software
tool was used to analyze the TFAP2B interaction network,
which demonstrated that the Cbp/p300-interacting transactivator
with ED-rich tail family, the TF2P family, the small ubiquitin-like
modifier family and TP53 might be important regulators in the
expression and functions of TFAP2B due to their direct and
stronger interactions (Fig.
S1D).
Comparison of TFAP2B expression levels
in normal kidney cells and renal tumor cells
To identify suitable ccRCC cell lines for further
study, RT-qPCR and western blot assays were used to analyze mRNA
and protein expression levels of TFAP2B in ccRCC cell lines
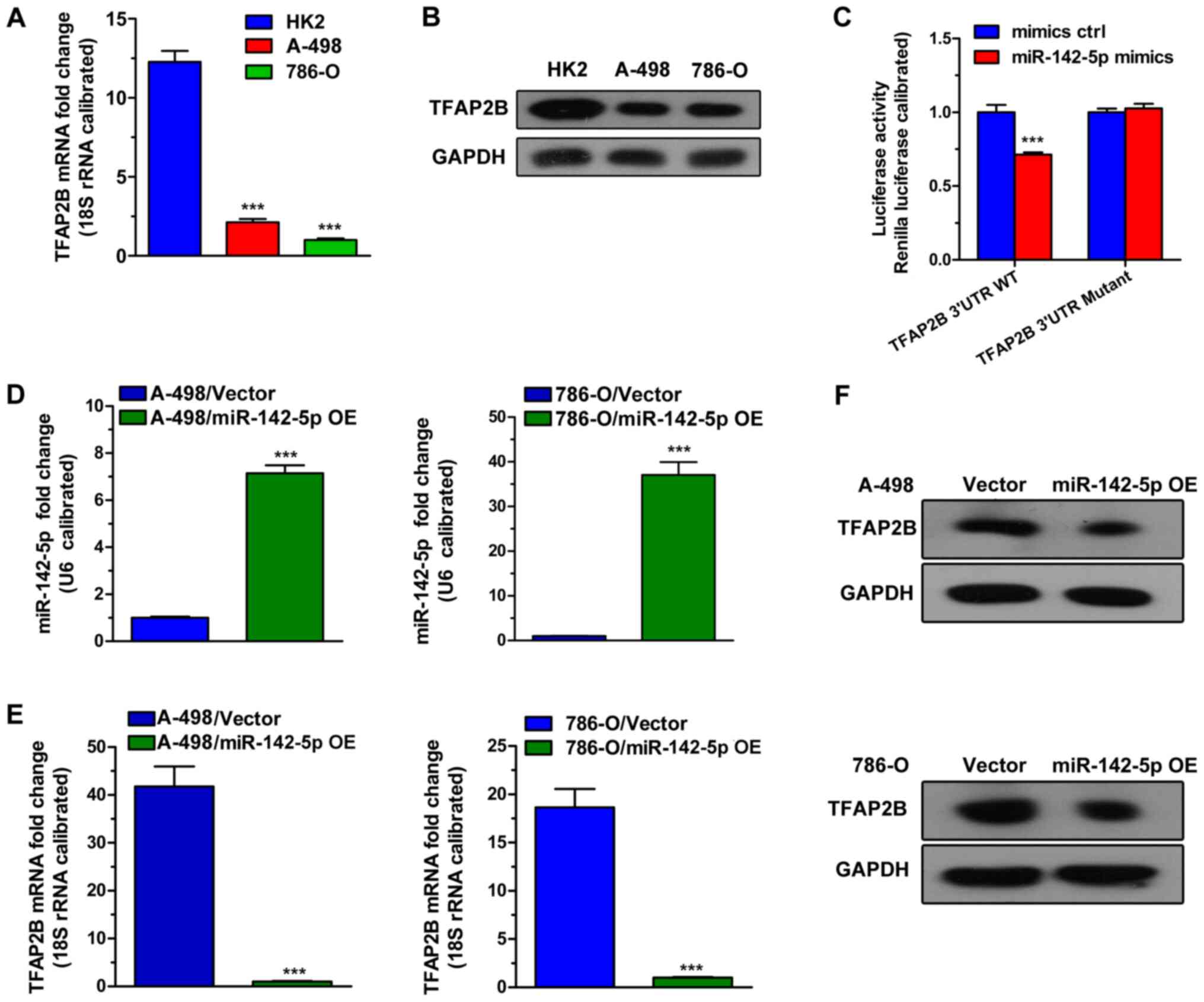
compared with the normal kidney cell line HK-2. As shown in
Fig. 2A, the results indicated that
the mRNA expression levels of TFAP2B was lower in both renal
tumor cells 786-O and A498 compare with expression HK-2. In
addition, TFAP2B protein expression levels were lower in the
ccRCC cell lines compared with HK-2 (Fig. 2B). These results suggested that 786-O
and A498 cells can recapitulate the effects observed with Oncomine
software screen, and therefore were selected as a suitable model
for subsequent studies.
miR142-5p targets TFAP2B
Based on the TargetScan screen database analysis,
the interaction between miR-142-5p and its putative target
TFAP2B was examined (Fig.
1B). The results demonstrated that the miR-142-5p
mediated-decrease of TFAP2B protein level was rescued by the
TFAP2B overexpression vector, compared with miR-142-5p group
(Fig. S2A and B). In addition, as
shown in Fig. S2C and D,
TFAP2B overexpression vector could induce the overexpression
of TFAP2B alone, compared with the empty vector negative
control cells (Fig. S2A and B).
Furthermore, a dual luciferase reporter assay revealed a
significant decrease in the luciferase activity in 293T cells
co-transfected with miR-142-5p and the TFAP2B−3′UTR WT
luciferase reporter vector compared with the luciferase activity in
cells co-transfected with the negative control (mimics ctrl) and
the reporter vector. Besides, the TFAP2B−3′UTR Mutant
abolished the interaction between the miR-142-5p and the
TFAP2B−3′UTR region as no significant differences were
observed in the luciferase activity between miR-142-5p and mimics
control (Fig. 2C). Moreover, the
effects of miR-142-5p overexpression on TFAP2B in 786-O and
A-498 cell lines were examined by RT-qPCR and western blot assays.
As shown in Fig. 2D, the level of
miR-142-5p was higher in miR-142-5p-overexpression (OE) 786-O and
A-498 cells compared with expression levels in the negative control
group. TFAP2B mRNA and protein levels were markedly
decreased in miR-142-5p-OE cells (Fig.
2E and F, respectively). Based on the above results, it was
concluded that miR142-5p targeted TFAP2B and suppressed its
expression in these cell lines.
miR142-5p promotes proliferation of
ccRCC cells
To study the effects of the miR142-5p on ccRCC cell
proliferation, the proliferation abilities of 786-O and A-498 cell
lines overexpressing miR-142-5p or co-overexpressing miR-142-5p +
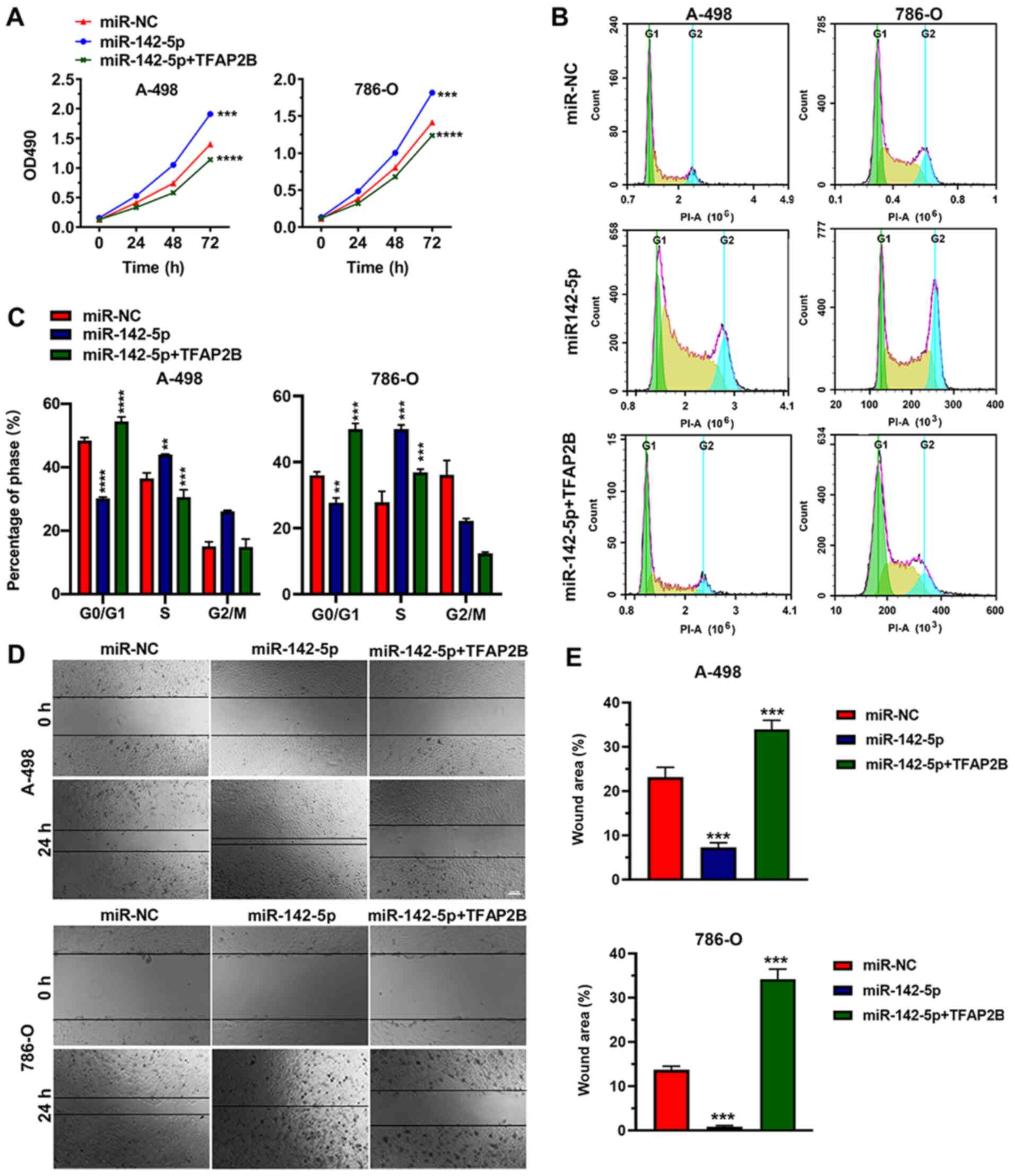
TFAP2B were evaluated with an MTT assay (Fig. 3A). An increased proliferation rate
was observed in miR-142-5p-OE cells compared with the empty vector
negative control group in each cell line. In addition, the
overexpression of TFAP2B could eliminate the
miR142-5p-induced increase in cell proliferation (Fig. 3A).
Because miR142-5p promoted 786-O and A-498 cell
proliferation, we evaluated the effects of miR142-5p on 786-O and
A-498 cell cycle progression by flow cytometry. The results
demonstrated that the percentage of cells in phase
G0/G1 was significantly lower in
miR-142-5p-OE 786-O and A-498 cells, whereas the proportion of S
phase cells was significantly higher in miR-142-5p-OE 786-O and
A-498 cells compared with the negative control group (Fig. 3B and C). This effect was eliminated
by TFAP2B overexpression, which lead to an increased
percentage of A-988 cells in phase G0/G1 and
reduced the percentage of 786-O cells in S phase. These results
indicated that miR142-5p could prevent G1 phase arrest
in 786-O and A-498 cells and significantly promote ccRCC cell
proliferation.
miR142-5p enhances migration of ccRCC
cells
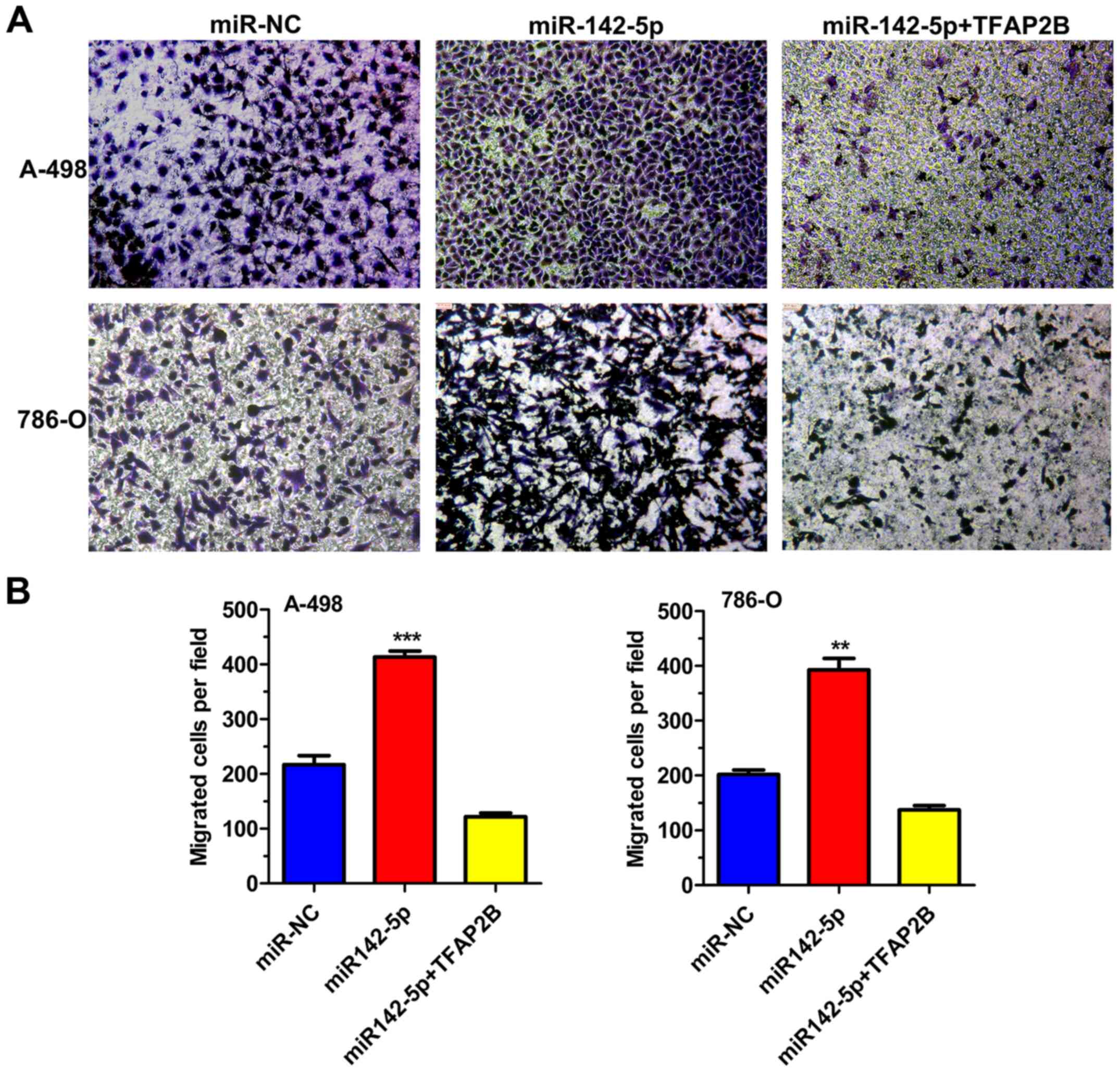
Results from wound healing and Transwell migration
assays demonstrated that miR-142-5p-OE 786-O and A-498 cells
exhibited significantly increased cell migration compared with the
negative control cells (Figs. 3D and
E and 4). As expected,
overexpression of TFAP2B significantly removed the influence
of increased miR142-5p expression on cell migration (Fig. 3E). These data suggested that the
migratory ability of ccRCC cells was enhanced by miR-142-5p but was
weakened by TFAP2B.
Discussion
In the present study, TFAP2B expression was
found to be markedly low in renal cancer, indicating that
TFAP2B might function as a tumor suppressor in ccRCC.
miR-142-5p was found to directly target TFAP2B and
downregulate its expression, indicating that miR-142-5p may act as
an oncogenic microRNA. Finally, the effect of miR-142-5p and
TFAP2B on ccRCCs was systematically investigated through a
series of cell function experiments. The results demonstrated that
miR-142-5p increased cell proliferation and migration and that
TFAP2B could reverse these effects. Cell cycle assays showed
that overexpression of miR-142-5p reduced the population of
G0/G1 phase cells, which indicated that
miR-142-5p may promote cell growth by forcing them to enter the S
phase.
A miRNA can target numerous genes and act as either
a tumor suppressor or oncogenic miRNA, depending on the function of
targeted genes. Liu et al (17), reported that miR-142-5p promotes
development of colorectal cancer by targeting succinate
dehydrogenase complex iron sulfur subunit B. However, a study by
Wang et al (18) showed that
miR-142-5p targets PI3K-α to suppress tumorigenesis in non-small
cell lung cancer. In human osteosarcoma, miR-142-5p also suppresses
proliferation and promotes apoptosis by targeting phospholipase A
and acyltransferase 3 (19). In the
present study, miR-142-5p was shown to promote proliferation and
migration of ccRCC cells by targeting TFAP2B, which is
consistent with study results reported by Liu et al
(20) that revealed that miR-142-5p
promotes cell growth and migration of ccRCCs by targeting BTG
anti-proliferation factor 3. These results revealed the importance
of the miR-142-5p in ccRCC development.
TFAP2B expression has been previously
detected in embryonic renal tissues and was found to be important
for kidney development in mice (12). In 2004, immunohistochemical evidence
first suggested that transcription factor AP-2 may play a role in
carcinogenesis (21). Furthermore,
using comprehensive bioinformatic analyses, RT-qPCR and
immunohistochemistry, downregulation of TFAP2B has been
found to be important not only for normal renal development and
epithelial differentiation but also for mesenchymal/adipogenic
transdifferentiation and pluripotent mesenchymal stem cell-like
differentiation (22). To explore
the regulation network of the TFAP2B in renal cancer, the
current study also analyzed the interaction partners of
TFAP2B. The results with STRING software indicated that p53,
the well-known tumor suppressor gene, might be important for the
downstream and upstream regulation of TFAP2B due to their
interaction, which required further experiments to be confirmed.
However, more experiments should be conducted to confirm this
hypothesis in the future.
There are some limitations in the present study,
such as lack of rescue experiments, interference experiments of
TFAP2B in cell lines, the limited number of ccRCC cell lines
studied for functional assessment of the effects observed and the
lack of reproductivity in the western blotting assays in other
ccRCC cell lines. Although the detailed regulatory mechanistic
functions of TFAP2B in driving cancer remain to be further
explored, to the best of our knowledge, this study is the first to
demonstrate TFAP2B regulation by miR-142-5p in ccRCC. In
conclusion, miR-142-5p/TFAP2B pathway in ccRCC is described
for the first time, which may provide novel targets for ccRCC
therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This research was supported by The Provincial
Natural Science Foundation of Fujian (grant no. 2018D0022) and The
Xiamen Science and Technology Guiding Program of China (grant no.
3502Z20189043).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MZ, LZ, JF and YL designed and performed the
experiments. BS, KY, FL, LY and ML performed the experiments. All
authors critically revised the manuscript. JF and YL supervised the
project. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Disease Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cho E, Adami HO and Lindblad P:
Epidemiology of renal cell cancer. Hematol Oncol Clin North Am.
25:651–665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Makhov P, Joshi S, Ghatalia P, Kutikov A,
Uzzo RG and Kolenko VM: Resistance to systemic therapies in clear
cell renal cell carcinoma: Mechanisms and management strategies.
Mol Cancer Ther. 17:1355–1364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tippu Z, Au L and Turajlic S: Evolution of
renal cell carcinoma. Eur Urol Focus S2405-S4569. 30383–30389.
2020.
|
|
7
|
Mennitto A, Verzoni E, Grassi P, Ratta R,
Fucà G and Procopio G: Multimodal treatment of advanced renal
cancer in 2017. Expert Rev Clin Pharmacol. 10:1395–1402. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eckert D, Buhl S, Weber S, Jäger R and
Schorle H: The AP-2 family of transcription factors. Genome Biol.
6:2462005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lüscher B, Mitchell PJ, Williams T and
Tjian R: Regulation of transcription factor AP-2 by the morphogen
retinoic acid and by second messengers. Genes Dev. 3:1507–1517.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeng YX, Somasundaram K and el-Deiry WS:
AP2 inhibits cancer cell growth and activates p21WAF1/CIP1
expression. Nat Genet. 15:78–82. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Auman HJ, Nottoli T, Lakiza O, Winger Q,
Donaldson S and Williams T: Transcription factor AP-2gamma is
essential in the extra-embryonic lineages for early
postimplantation development. Development. 129:2733–2747.
2002.PubMed/NCBI
|
|
12
|
Moser M, Pscherer A, Roth C, Becker J,
Mücher G, Zerres K, Dixkens C, Weis J, Guay-Woodford L, Buettner R
and Fässler R: Enhanced apoptotic cell death of renal epithelial
cells in mice lacking transcription factor AP-2beta. Genes Dev.
11:1938–1948. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu L, Shi K, Wang J, Chen W, Shi D, Tian
Y, Guo W, Yu W, Xiao X, Kang T, et al: TFAP2B overexpression
contributes to tumor growth and a poor prognosis of human lung
adenocarcinoma through modulation of ERK and VEGF/PEDF signaling.
Mol Cancer. 13:892014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Konta T, Ichikawa K, Suzuki K, Kudo K,
Satoh H, Kamei K, Nishidate E and Kubota I: A microarray analysis
of urinary microRNAs in renal diseases. Clin Exp Nephrol.
18:711–717. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wong NW, Chen Y, Chen S and Wang X:
OncomiR: An online resource for exploring pan-cancer microRNA
dysregulation. Bioinformatics. 34:713–715. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu S, Xiao Z, Ai F, Liu F, Chen X, Cao K,
Ren W, Zhang X, Shu P and Zhang D: miR-142-5p promotes development
of colorectal cancer through targeting SDHB and facilitating
generation of aerobic glycolysis. Biomed Pharmacother.
92:1119–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Liu Z, Fang X and Yang H:
MiR-142-5p suppresses tumorigenesis by targeting PIK3CA in
non-small cell lung cancer. Cell Physiol Biochem. 43:2505–2515.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheng D, Li J, Zhang L and Hu L:
miR-142-5p suppresses proliferation and promotes apoptosis of human
osteosarcoma cell line, HOS, by targeting PLA2G16 through the
ERK1/2 signaling pathway. Oncol Lett. 17:1363–1371. 2019.PubMed/NCBI
|
|
20
|
Liu L, Liu S, Duan Q, Chen L, Wu T, Qian
H, Yang S, Xin D, He Z and Guo Y: MicroRNA-142-5p promotes cell
growth and migration in renal cell carcinoma by targeting BTG3. Am
J Transl Res. 9:2394–2402. 2017.PubMed/NCBI
|
|
21
|
Oya M, Mikami S, Mizuno R, Miyajima A,
Horiguchi Y, Nakashima J, Marumo K, Mukai M and Murai M:
Differential expression of activator protein-2 isoforms in renal
cell carcinoma. Urology. 64:162–167. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tun HW, Marlow LA, von Roemeling CA,
Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ and
Copland JA: Pathway signature and cellular differentiation in clear
cell renal cell carcinoma. PLoS One. 5:e106962010. View Article : Google Scholar : PubMed/NCBI
|